Introduction
Lung cancer is the leading cause of cancer deaths.
Majority (80%) of lung cancer are non-small cell lung cancer, 60%
of which are resistant to chemotherapy. Small molecule targeted
therapies have been developed for lung cancers carrying epidermal
growth factor mutation, but the efficacy has been limited by drug
resistance. A breakthrough in lung cancer therapy field is the
immunotherapy targeting molecules which suppress the immune
surveillance against cancer, as exemplified by blocking antibodies
to PD1 molecule, a molecule expressed by tumor-killing lymphocytes
which suppresses the lymphocyte activation (1). Anti-PD1 antibody has been approved in
the United States for treatment of melanoma. It has also shown
clear efficacy in non-small cell lung cancer. Another blocking
antibody, anti-PDL1, specific for PD1 ligand 1, which is a molecule
expressed by tumor and suppresses immune activation through binding
to PD1, has also shown efficacy in treating non-small cell lung
cancer (2).
Drug targets like PD1 and PDL1 are highly sought
after because they are not limited by drug-resistance observed in
small molecule targeted therapy. Mucins are cancerous proteins
which promote tumor growth through binding to signaling molecules
in apoptosis pathway (3,4), such as the binding of MUC1 protein to
BH3 domain of BAX protein. Mucins also bind to galectins expressed
on surface of tumor-killing lymphocytes and trigger their apoptosis
to subvert immune surveillance (5–8). The
expression of mucins in lung cancer cell lines and tissue sections
have been studied by staining with a few monoclonal antibodies
(9–12), however, the big picture of mucins
in lung cancer patients is still lacking.
Mucins are highly expressed by healthy epithelial
cells, and MUC1 peptide vaccines based on mucin protein backbones
have not led to significant objective responses in cancer treatment
(13–16). Abnormally glycosylated mucins in
malignant cells are current research focus because of the unique
post-translational modification of mucin backbones by
carbohydrates. In this study, by analyzing lung cancer microarray
data available in public domain and computer predicting of
glycopeptide TR sequence, we identified TR glycopeptides bearing Tn
and sialyl-Tn. Using MUC1 as a model, we report three disciplines
used by monoclonal IgG antibodies to recognize glycopeptide
antigens at molecular level.
Materials and methods
mRNA array data source for different
types of lung carcinomas
mRNA array data for all lung cancer types were
acquired from www.genome.wi.mit.edu/MPR/lung (17). All data were evaluated by an R
package, Simpleaffy, using relative log expression (RLE) boxplot
and normalized unscaled standard error (NUSE) boxplot as previously
described (18). There were 216
cases in total, but four of them were discarded due to their
abnormal performance in the quality control process. The 212
remaining array data include the following patients: 150
adenocarcinoma, 20 bronchial carcinoid, 5 small cell lung cancer,
21 squamous lung cancer, and 16 healthy individuals.
mRNA data array source for different
stages of lung adenocarcinomas
mRNA array data were acquired from caArray
(https://array.nci.nih.gov/caarray/project/details.action?project.experiment.publicIdentifier=jacob-00182).
The array experiment was performed by multiple laboratories in
North America and yielded 442 array data on lung adenocarcinomas
(19). Based on RLE and NUSE plot
evaluation by Bhattacharjee et al (19), array data from the Dana-Farber
Cancer Institute (CAN/CF) is systematically different from the
other sites. Thus, we discarded the data from CAN/CF and a few more
data with incomplete information regarding cancer stages. In the
end, we collected 358 array dataset (132 for T1 stage, 188 for T2
stage, 26 for T3 stage, and 12 for T4 stage in stage category; 241
for N0 stage, 64 for N1 stage, 53 for N2 stage in metastasis stage
category).
Analysis of the mRNA quantitation
Array data were processed by Robust Multiarray
Average normalization (20).
Data collection of membrane proteins with
repeating sequences
XML R package (21, http://www.omegahat.org/RSXML/) was used to collect
sequence and annotation information of human membrane protein with
repeating sequence from Uniprot Database (http://www.uniprot.org/).
Prediction of glycopeptidome sequences by
computational analysis
All calculations were by programs using R
(http://www.R-project.org/). The programs
were designed to read the peptide sequence of each mucin as input,
with the output as the numbers of all possible GalNAc (Tn) and
NeuAcα2,6GalNAc (sialyl Tn) glycosylation patterns.
Staining of human lung adenocarcinoma
cell lines by mAbs 14A, 16A, and B72.3
Lung adenocarcinoma cell lines, NCI-H1395, HCC4019,
H838, H1573, H1703, H2030, and H3255 were from Dr Sam Hanash, the
University of Texas MD Anderson Cancer Center. Cell lines were
cultured in RMPI1640 medium supplemented with 10% FCS. Cell surface
expression of Tn antigen and MUC1 was assessed by flow cytometry
staining. Monoclonal antibodies 14A (22), which binds to MUC1 peptide part
RPAPGSTAPPAHG; 16A, which binds to MUC1 glycopeptide
RPAPGS(GalNAc)TAPPAHG; and B72.3, which binds to clustered Tn
antigen (23, 24) were used as primary antibodies. Goat
anti-mouse IgG (Allophycocyanin-conjugated), and mouse IgG isotype
control were from Southern Biotech (Birmingham, AL, USA).
Synthetic peptides and glycopeptides
Biotinylated peptides and glycopeptides were
synthesized by Peptide International (Louisville, KY, USA) as
previously described (22), and
Wuxi Apptech Shanghai (China). Bovine Serum Albumin-GalNAc (each
BSA carries 23 GalNAc residue) were from Vector Labs (UK). The
MUC1-106 amino acid long peptide containing 5 tandem repeat (TR)
sequences was described in previous studies (13).
ELISA measurement of Ab binding to
glycopeptides
The biotinylated (glyco)-peptide,
RPAPGS(GalNAc)TAPPAHG-dPEG™11-Biotin, (1 μg/ml) was bound to
streptavidin-coated plates (2 μg/ml) and incubated with 16A
monoclonal Ab (mAb) for 2 h. Binding of 16A was visualized by a
secondary Ab (goat anti-mouse IgG) followed by colorimetric
detection. One percent BSA was used as blank for determining the
cutoff value. To measure the inhibitory effects of competing
ligands, ligands were mixed with the 16A mAb at 0–500 μM for 1 h,
before incubation with plate-bound glycopeptide
RPAPGS(GalNAc)TAPPAHG-dPEG.
Surface plasmon resonance (SPR)
measurement of Ab binding affinity
SPR measurement of Ab affinity toward consecutive TR
peptides were as previously described (22). Interactions of peptides with
immobilized 14A and 16A mAbs were determined by using a Biacore
T-200 (GE Healthcare, Pittsburgh, PA, USA). The 14A and 16A were
immobilized on a research-grade, CM5 sensor chip (GE Healthcare)
until 5000 RU was reached. Immobilizations were carried out at
protein concentrations of 50 μg/ml in 10 mM acetate, pH 5.0 and 10
mM acetate, pH 5.5 for 14A and 16A, respectively, using an amine
coupling kit supplied by the manufacturer. In all cases, analyses
were carried out at 25°C in 10 mM Hepes, pH 7.4 containing 150 mM
NaCl and 0.005% surfactant P20 at a flow rate of 40 μl/min. The
surface was regenerated with 4M MgCl2 then washed with
the running buffer. Data were analyzed with BIA evaluation software
(GE Healthcare).
Flow cytometry analysis of
MUC1-transfected cells
Ag104 cells, which express only Tn and sialyl-Tn
O-linked glycans (25), were
transfected by a MUC1 gene as described using a
pcDNA3.1-hMUC1-IRES-eGFP plasmid (22). MUC1-expressing cells were selected
by sorting of GFP-positive cells. Stably established
MUC1-expressing cells and Ag104 cells transfected by mock
pcDNA3.1-IRES-eGFP plasmid were stained by B72.3 mAb (ATCC,
Manassas, VA, USA) and Sambucus Nigra Lectin (Vector Labs, UK)
specific to α-2,6 linked sialyl acid (26). Flow cytometry data were acquired by
FACS Canto instrument (San Jose, CA, USA) and analyzed by FlowJo
software (Ashland, OR, USA).
Results
Expression of mucin mRNA in four subtypes
of lung cancers
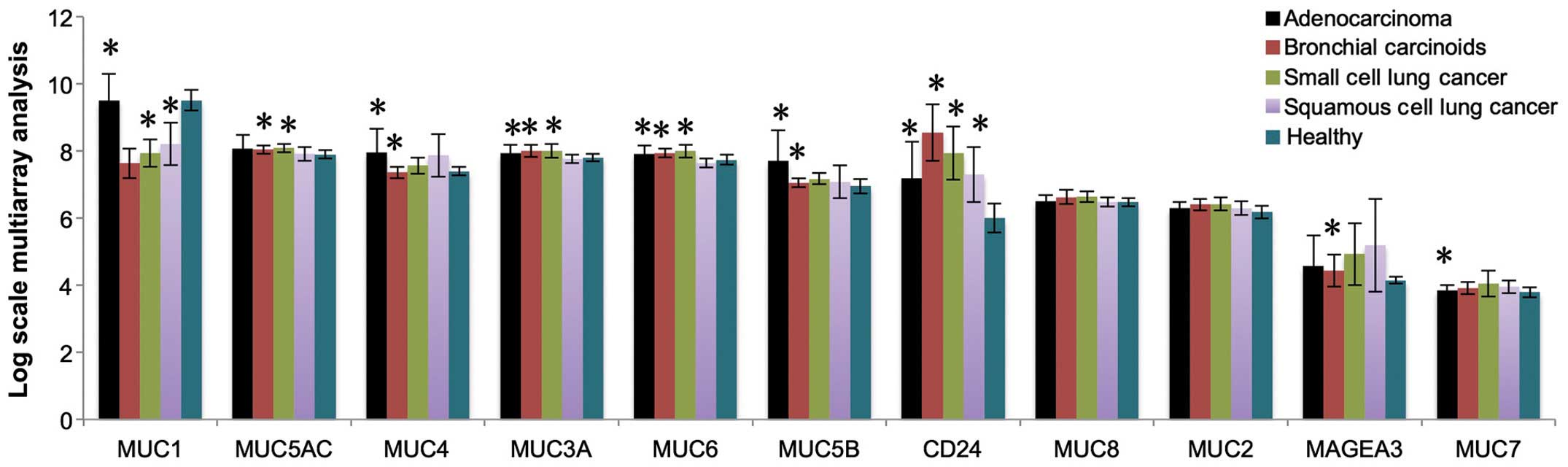
By analyzing healthy control and cancer patients, we
found nine mucins (MUC1, MUC2, MUC3A, MUC4, MUC5AC, MUC5B, MUC6,
MUC7, and MUC8) expressed in lung cancer patients (Fig. 1). CD24 and MAGEA3, two well-known
lung cancer biomarkers, were used as controls for evaluating the
mRNA expression. Fig. 1 shows the
expression of cancerous mucins in healthy control, lung
adenocarcinoma, bronchial carcinoid cancer, small cell lung cancer,
and squamous cell lung cancer. Notably, all mucins found in lung
cancer are also found in healthy control.
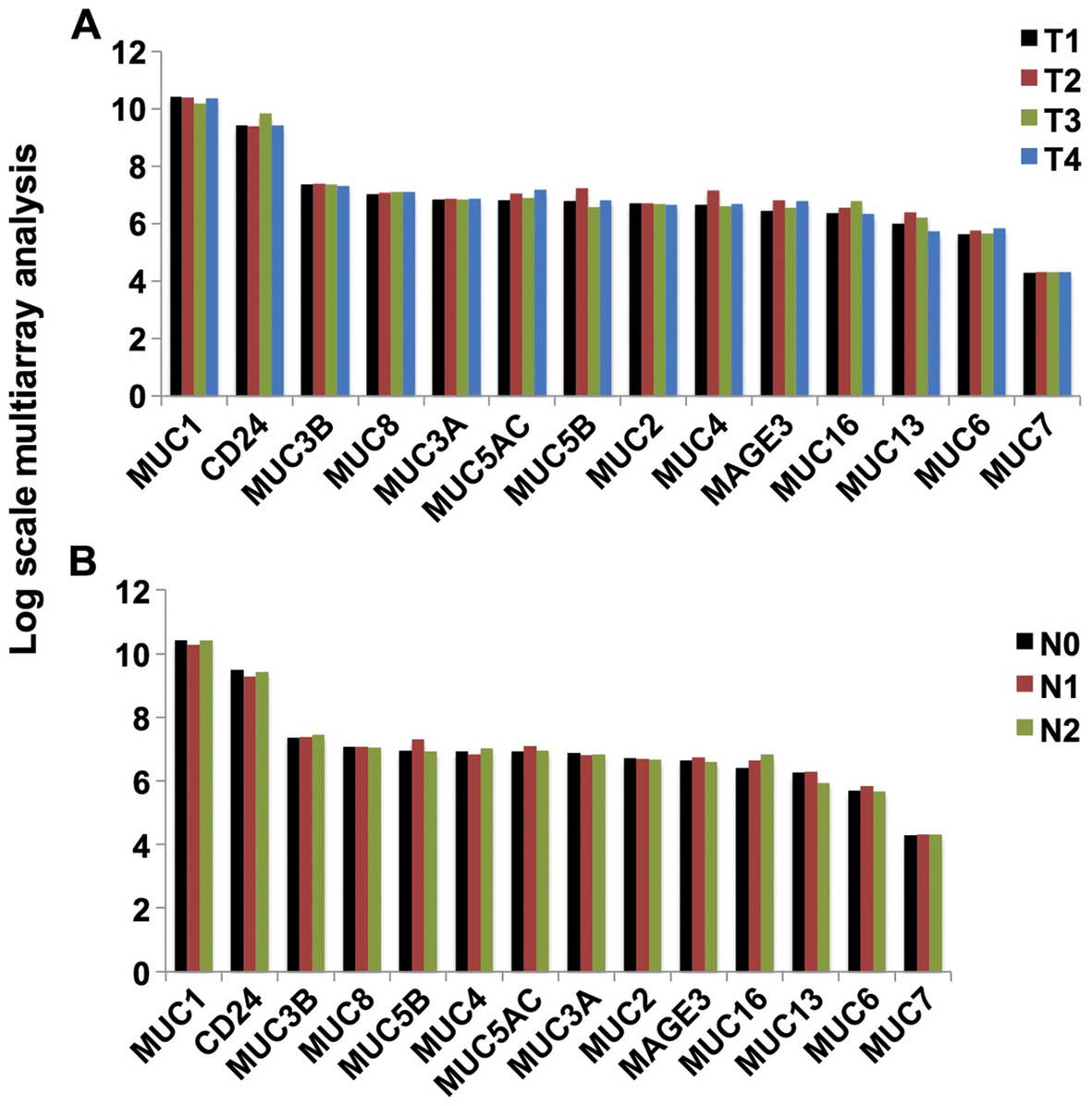
Expression of mucin mRNA in all stages of
lung adenocarcinoma
Fig. 2 shows the
expression of the cancer-associated mucins in different stages of
lung adenocarcinoma. No significant differences in expression were
identified among the various stages.
Mucin glycopeptide TR sequences predicted
by R program
We used an R program to predict the possible
glycopeptide TR sequences of mucins that may bear Tn and sialyl-Tn
antigens; a large number of extremely diverse glycopeptide
sequences were predicted for each mucin TR alone. The glycopeptide
sequences predicted for MUC1 TR was published previously (22). TRs of MUC2, MUC3A, MUC3B, MUC5B,
MUC6, MUC16, and MUC17 showed more than 10,000 sequence results
when bearing one sugar (Tn) alone (data not shown). When the
disaccharide sequence (sialyl-Tn) was included in this analysis,
more than 100,000 structures were found for such mucin TRs (data
not shown).
A glycopeptide epitope on lung cancer
cell surface is preferably recognized by mAb 16A
To examine whether MUC1 protein is expressed at
protein level, we stained 7 patient-derived lung adenocarcinoma
cells with 14A and 16A monoclonal antibodies (22). The 14A antibody only binds to a
peptide backbone of MUC1, while the 16A antibody preferentially
binds to a glycopeptide of MUC1 (22). The results (Fig. 3) showed that 6 out of 7 human lung
adenocarcinoma cell lines could be stained by 14A and 16A antibody,
and 16A showed stronger binding in every cell line studied. This
suggests that a glycosylated MUC1 epitope, RPAPGS(GalNAc)TAPPAHG,
is better recognized than its non-glycosylated backbone
RPAPGSTAPPAHG.
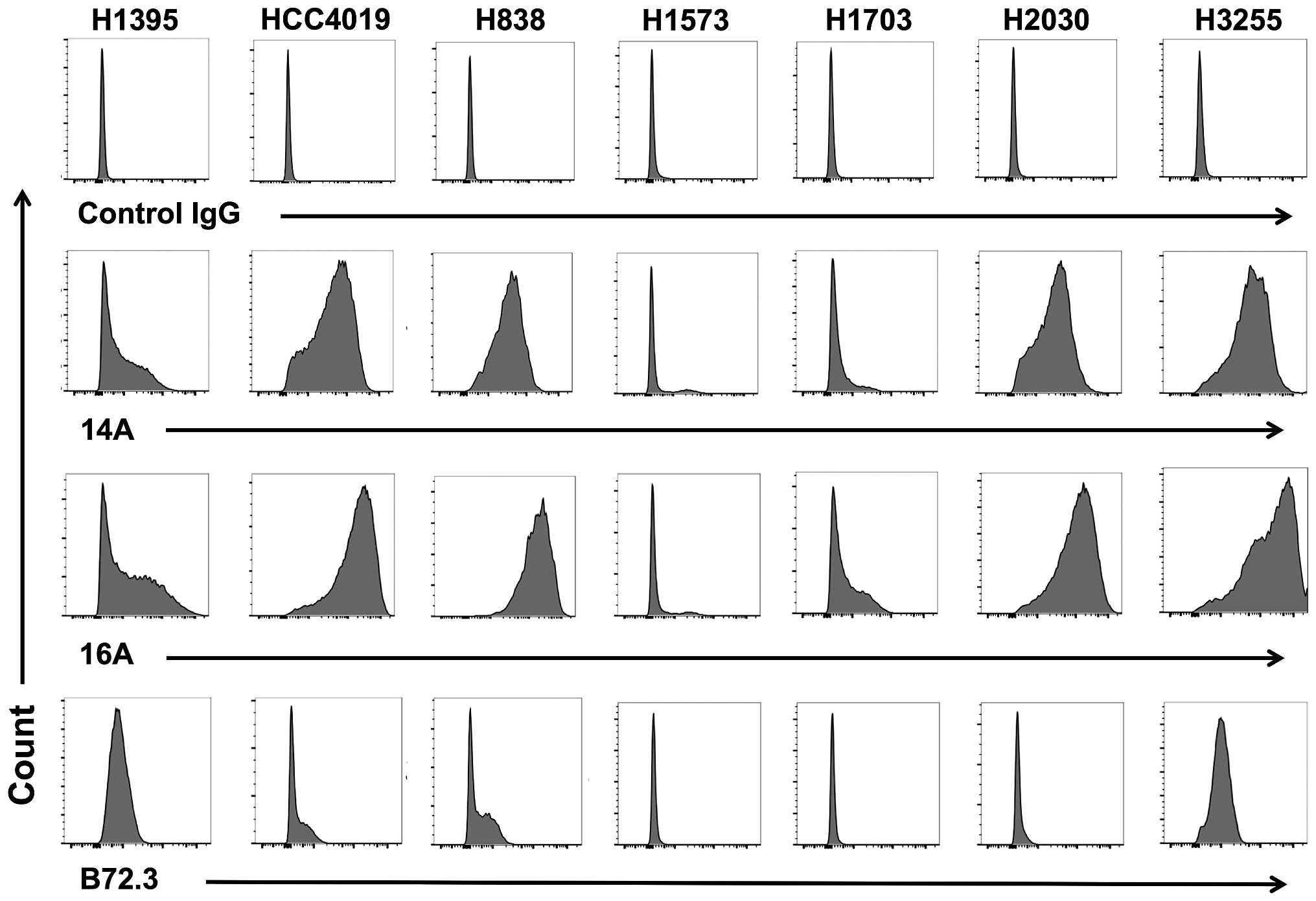 | Figure 3A glycopeptide epitope on lung cancer
cell surface is preferably recognized by mAb 16A. Lung
adenocarcinoma cell lines, NCI-H1395, HCC4019, H838, H1573, H1703,
H2030, and H3255 were studied by flow cytometry staining.
Monoclonal antibodies 14A, which binds to MUC1 peptide part only
(RPAPGSTAPPAHG); 16A, which binds to MUC1 glycopeptide
RPAPGS(GalNAc)TAPPAHG; and B72.3, which binds to sugars only
(clustered Tn antigen), were used as primary antibodies. Goat
anti-mouse IgG (Allophycocyanin-conjugated), and mouse IgG isotype
control were from Southern Biotech (Birmingham, AL, USA). |
We also examined the binding of mAb B72.3, which is
specific for clustered Tn and sialyl-Tn antigens independent of the
peptide backbone sequence (23,24).
The binding of human lung adenocarcinoma cells to B72.3 antibody
could be observed in 4 of 7 cell lines, suggesting the expression
of clustered Tn antigens.
16A mAb binds to both peptide and sugar
parts of a glycopeptide
We previously generated a mAb, 16A, that
preferentially binds to a MUC1 peptide modified by a Tn residue
(22). We also generated a 14A mAb
which only binds to the MUC1 peptide backbone. However, the exact
role of peptide or sugar part in contributing to the binding of 16A
antibody is not clear. To further understand how the 16A mAb binds
to the MUC1 glycopeptide, we used peptide, glycopeptide, and sugar
structures to inhibit its binding to RPAPGS(GalNAc)TAPPAHG. The
peptide RPAPGSTAPPAHG inhibits the 16A Ab binding to
RPAPGS(GalNAc)TAPPAHG at a half maximal inhibitory concentration
(IC50) of 5.79 μM, while the RPAPGS(GalNAc) TAPPAHG inhibits at an
IC50 of 2.89 μM (Fig.
4).
In clear contrast, the GalNAc inhibits the binding
of 16A Ab to RPAPGS(GalNAc)TAPPAHG at a much higher IC50
of 11.13 mM. We also tested polymeric GalNAc attached to a BSA
molecule (each BSA carries 23 GalNAc residue); it inhibits the
binding to RPAPGS(GalNAc)TAPPAHG at an IC50 of 69.25
μM.
Not surprisingly, neither GalNAc nor polymeric
GalNAc inhibited the binding of 14A mAb to glycopeptide
RPAPGS(GalNAc)TAPPAHG, indicating that only peptide part is
recognized by 14A mAb (data not shown).
14A and 16A mAbs bind to MUC1-106aa
polyvalent vaccine with 10-fold higher affinity than consecutive TR
sequence
Because IgG molecules bind to bivalent antigen
epitopes with higher affinity, we designed consecutive TR
sequences, 2014C, 2015C, and 2016C (Table I) and measured the dissociation
constant by SPR analysis. Both 2014C and 2016C showed much higher
affinity (20- and 40-fold, respectively) binding to 16A and 14A, as
compared with the RPAPGSTAPPAHG single TR sequence alone (Table II). Of note, the 2015C did not
show stronger binding to 16A or 14A, suggesting the underlined PA
sequence RPAPGSTAPPAHG must be recognized by each arm of IgG
molecule.
 | Table ISequences of peptides and
glycopeptides. |
Table I
Sequences of peptides and
glycopeptides.
| Name | Sequence |
|---|
| Pep1 |
RPAPGS(GalNAc)TAPPAHG |
| Pep2 | RPAPGSTAPPAHG |
| 2014C |
PDTRPAPGSTAPPAHGVTSA
PDTRPAPGSTAPPAHGVTSA |
| 2015C |
AHGVTSAPDTRPAPGSTAPP
AHGVTSAPDTRPAPGSTAPP |
| 2016C |
RPAPGSTAPPAHGVTSAPDTR
PAPGSTAPPAHGVTSAPDT |
| MUC106aa |
GVTSAPDTRPAPGSTAPPAHGVTSAPDTRPAPGSTAPPAHGVTSAPDTRPAPGSTAPPAHGVTSA
PDTRPAPGSTAPPAHGVTSAPDTRPAPGSTAPPAHGVTSA |
 | Table IISPR measurement of Kd for the binding
of 14A and 16A mAbs to glycopeptides. |
Table II
SPR measurement of Kd for the binding
of 14A and 16A mAbs to glycopeptides.
| (Glyco)peptide | Ka (1/Ms) | Kd (1/s) | KD (nM) | Chi2
(RU2) |
|---|
| 16A |
|
RPAPGS(GalNAc)TAPPAHG | 3.353E+4 | 0.03120 | 930.4 | 23.1 |
| RPAPGSTAPPAHG | 7208 | 0.003898 | 540.8 | 4.95 |
| 2014C | 4.908E+5 | 0.01524 | 31.05 | 3 |
| 2015C | 1.500E+4 | 0.005081 | 338.8 | 12 |
| 2016C | 1.158E+6 | 0.01660 | 14.34 | 5 |
| MUC106 | 5.235E+5 | 8.062E-4 | 1.540 | 6 |
| 14A |
|
RPAPGS(GalNAc)TAPPAHG | 1.172E+5 | 0.05141 | 438.6 | 19.4 |
| RPAPGSTAPPAHG | 1.218E+4 | 0.002851 | 234.0 | 3.35 |
| 2014C | 1.411E+6 | 0.02046 | 14.50 | 5 |
| 2015C | 2.563E+4 | 0.004151 | 161.9 | 15 |
| 2016C | 3.622E+6 | 0.02433 | 6.718 | 9 |
| MUC106 | 1.022E+6 | 6.674E-4 | 0.653 | 8 |
Trivalent (14) and
pentavalent (13) TR sequences for
MUC1 were previously designed as cancer vaccines, and IgG responses
have been reported in vaccinated patients. The MUC1-106aa showed
extremely high affinity to the 14A and 16A mAbs, with a Kd of 0.653
and 1.54 nM, respectively. This strongly suggests that two
non-consecutive bivalent TR epitopes are preferably recognized than
the two consecutive bivalent TR sequence (Table II).
Transfection of COSMC-deficient cells
with MUC1 gene caused 100-fold higher binding to B72.3, a
sialyl-Tn-specific mAb
To investigate whether clustered TR sequences also
play a role in Abs that bind to Tn and sialy-Tn antigens, we
studied B72.3, a mAb that binds to clustered Tn epitopes (23,24).
We overexpressed the human MUC1 gene in Ag104 cells, a mouse
fibrosarcoma cell line with a known mutation in the COSMC gene
(25). Transfection of the human
MUC1 gene caused 100-fold (two log) stronger binding to B72.3 mAb,
but the binding to Sambucus Nigra Lectin specific to α-2,6 linked
sialyl acid (26) remained
unchanged, indicating that the increased binding to B72.3 mAb is
not caused by the increased synthesis of total sialyl Tn epitopes
(Fig. 5). Therefore, MUC1 serves
as the preferred backbone to display the sugar epitopes for Ab
recognition, 100-fold more efficiently as compared to other
membrane proteins.
Discussion
By analyzing mucin expression in lung cancer at
transcriptome level, we found expression of nine mucins in both
lung cancer and healthy controls (Fig.
1). There is no rationale to use non-glycosylated mucin
peptides as cancer vaccines to induce CD8 T cell responses, as no
difference exists for the processing of MUC1 proteins in MHC class
I pathway by tumor cells versus healthy cells. The value of mucins
as diagnostic markers for lung cancer can only be based on their
posttranslational modification, such as the well-known abnormal
glycosylation exemplified by Tn, sialyl-Tn, and CA19-9. Future
proteomics studies must be focused on the abnormal glycosylation of
mucins. A recent breakthrough in this field is the glycopeptidome
analysis on a CHO cell line mutant deficient of Core-1 O-glycan
elongation, by an electron transfer dissociation (ETD) ionization
method (27–29). We attempted to apply this
technology to a Jurkat cell line transfected with the human MUC1
gene; however, this approach failed because mucins are resistant to
trypsin digestion, which is critical to generate glycopeptide
fragments to be read by mass spectrometry. The use of other
proteases may allow us to cleave mucins to short glycopeptides
amenable to mass spectrometry analysis.
Analysis of mRNA expression in published databases
clearly suggests that MUC1 should be a prioritized mucin for cancer
immunological research because it is expressed at high levels in
all stages of lung cancer. In light of the extremely diverse
structures of the MUC1 glycopeptidome, we designed experiments to
answer three major questions of antibody recognition: i) Is the
sugar or peptide part of a glycopeptide preferentially recognized
by an antibody? When designing MUC1 glycopeptide vaccines for
cancer therapy, a main observation in mouse models is that the
majority (90%) of Abs induced by glycopeptide vaccination can be
absorbed by non-glycosylated peptide backbones (30). Clearly the peptide part of a
glycopeptide is more immunogenic than the sugar part. The majority
of Abs may be induced toward the non-glycosylated peptide backbone.
Alternatively, the majority of the Abs bind to both the peptide
part and sugar part, and the peptide binding contributes to most of
the affinity.
The inhibition of 16A Ab binding to the glycopeptide
by free sugar (GalNAc) clearly shows that the sugar part of the
glycopeptide directly binds to the 16A mAb (Fig. 4). This is different from previously
reported mAbs SM3 and C595 (31–33).
SM3 binds to an amino acid repeat region (sequence PDTR) of MUC1,
which contains a glycosylated threonine residue. Although X-ray
crystallography and NMR studies reveal that glycosylation is not
required for binding, the GalNAc O-glycosylation induces
conformational changes in the peptide that enhances its
interactions with the Ab (31–33).
Similarly, mAb C595 raised against another peptide epitope in MUC1
(sequence RPAP) has enhanced affinity because of conformational
changes induced by Tn glycosylation; this affinity is attributed to
the stabilizing of a left-handed polyproline II helix by di- or
tri-glycosylation of the peptide (33).
Other mAbs that are specific for the sugar part only
but not the peptide backbone have been reported. For example, B72.3
mAb[23–24] binds to the disaccharide sialyl-Tn antigen but is not
dependent on the peptide backbone. Similarly, MSL128 mAb binds to
clustered Tn antigens independent of the sequence of peptide
backbones (34). Unfortunately,
very low titers of sialyl-Tn-specific Abs were induced in patients
vaccinated with Theratope vaccine, clustered sialyl-Tn sugars
conjugated to KLH (35). The
median titer against sialyl-Tn was only 1:300. Sugars are known to
be poorly immunogenic. Furthermore, the Tn and sialyl-Tn antigens
are self-epitopes that cause deletion of the B cells specific to
them at high affinity during B cell development in the bone
marrow.
Thus, our findings suggest that B cells
preferentially recognize the peptide backbone of a glycopeptide. In
our preliminary analysis, we found that 16A mAb binds to the
peptide backbone through mutations in complementarity determining
region (CDRs) of B cell receptor, while the sugar-binding
specificity is further acquired through mutations in frame work of
the heavy chain (unpublished data).
ii) Is the consecutive or non-consecutive TR
sequence in TR clusters preferentially recognized by B cells? IgGs
are bivalent, and IgMs are decavalent. The greater an
immunoglobulin's valency (number of antigen-binding sites), the
greater the amount of antigen it can bind. Similarly, antigens can
demonstrate multivalency because they can bind to more than one Ab.
Multimeric interactions between an Ab and an antigen help their
stabilization. Mucins that contain clustered TR sequences are ideal
backbones that trigger Ab responses. The heavy glycosylation may be
a mechanism for mucins to evade self Ab induction, which may cause
severe autoimmune diseases. Under cancer conditions, abnormally
glycosylated or non-glycosylated TR sequences are exposed, which
may trigger strong Ab responses. However, Ab titers toward mucin
peptides are not detected in cancer patients by ultra-dense peptide
arrays (36). Using a 100 amino
acid MUC1 peptide containing five consecutive TR sequences to
monitor Ab responses, circulating anti-MUC1 IgG Abs were reported
as a favorable prognostic factor for pancreatic cancer (13), and MUC1 vaccine containing 3
consecutive TR sequences have been tested in breast cancer patients
(14). Our data (Table II) clearly show that the clustered
TR sequences of MUC1 are 300 times more efficient in binding to
mAbs 14A and 16A as compared with a single TR repeat. Clustered TR
sequences of MUC1 are 10 times more efficient in binding as
compared with consecutive TR sequences (2014C and 2016C).
Thus, our data suggest that B cells preferentially
recognize consecutive TR sequences, but favor clustered TR
sequences containing more than three TR sequences even more. Future
attempts at immune-epitope discovery should be focused on designing
clustered TR epitopes that contain more than 3 TR sequences.
iii) Is a mucin backbone required for optimal
binding of Tn and sialyl-Tn antigens? Previous studies of
vaccinating patients with the sialyl-Tn Theratope vaccine were
based on the assumption that clustered sialyl-Tn epitopes mimic the
silyl-Tn epitopes expressed on the surface of a cancer cell. In our
study, through comparison of Ag104 cells and Ag104 cells
transfected with a human MUC1 gene, we found that the expression of
sialyl-Tn antigen alone is not sufficient for the optimal binding
to B72.3 Ab, a mAb originally considered as independent of the
peptide backbone. Thus, the conformation of the MUC1 backbone, but
not other membrane proteins, is clearly essential for optimal B
cell recognition of Tn and Sialyl Tn sugars. Future
immuno-monitoring must be based on rational design of sugar
epitopes displayed on proper protein backbones such as MUC1.
Herein, we report three disciplines used by
monoclonal antibodies to bind glycopeptide antigens: i) a
glycopeptide epitope on the surface of lung cancer adenocarcinoma
cell lines is preferably recognized, as compared to its
peptide-alone or sugar-alone counterpart; ii) the clustered
epitopes with more than 3 TR sequences are preferentially
recognized than consecutive TR sequences; iii) sugar epitopes
displayed on MUC1 is preferentially recognized as compared to those
on other O-glycosylated protein carriers. Our findings may indicate
common rules for monoclonal antibody binding to mucin glycopeptides
on cancer cell surface, and provide critical clues for designing
cancer vaccines and biosimilars targeting mucins.
Acknowledgements
D.Z. is supported by the National Natural Science
Foundation of China grant 81570007, US NIH R01 grant AI079232,
Shanghai Pulmonary Hospital, Tongji University and PRC's 1000 Plan
Recruitment Program for Young Talents.
References
|
1
|
Hamid O, Robert C, Daud A, Hodi FS, Hwu
WJ, Kefford R, Wolchok JD, Hersey P, Joseph RW, Weber JS, et al:
Safety and tumor responses with lambrolizumab (anti-PD-1) in
melanoma. N Engl J Med. 369:134–144. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Brahmer JR, Tykodi SS, Chow LQ, Hwu WJ,
Topalian SL, Hwu P, Drake CG, Camacho LH, Kauh J, Odunsi K, et al:
Safety and activity of anti-PD-L1 antibody in patients with
advanced cancer. N Engl J Med. 366:2455–2465. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ahmad R, Alam M, Rajabi H and Kufe D: The
MUC1-C oncoprotein binds to the BH3 domain of the pro-apoptotic BAX
protein and blocks BAX function. J Biol Chem. 287:20866–20875.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Xu X, Wells A, Padilla MT, Kato K, Kim KC
and Lin Y: A signaling pathway consisting of miR-551b, catalase and
MUC1 contributes to acquired apoptosis resistance and
chemoresistance. Carcinogenesis. 35:2457–2466. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang K, Sikut R and Hansson GC: A MUC1
mucin secreted from a colon carcinoma cell line inhibits target
cell lysis by natural killer cells. Cell Immunol. 176:158–165.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ogata S, Maimonis PJ and Itzkowitz SH:
Mucins bearing the cancer-associated sialosyl-Tn antigen mediate
inhibition of natural killer cell cytotoxicity. Cancer Res.
52:4741–4746. 1992.PubMed/NCBI
|
|
7
|
Moreno M, Bontkes HJ, Scheper RJ, Kenemans
P, Verheijen RH and von Mensdorff-Pouilly S: High level of MUC1 in
serum of ovarian and breast cancer patients inhibits huHMFG-1
dependent cell-mediated cytotoxicity (ADCC). Cancer Lett.
257:47–55. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Belisle JA, Horibata S, Jennifer GA,
Petrie S, Kapur A, André S, Gabius HJ, Rancourt C, Connor J,
Paulson JC, et al: Identification of Siglec-9 as the receptor for
MUC16 on human NK cells, B cells, and monocytes. Mol Cancer.
9:1182010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kuemmel A, Single K, Bittinger F, Faldum
A, Schmidt LH, Sebastian M, Micke P, Taube C, Buhl R and Wiewrodt
R: TA-MUC1 epitope in non-small cell lung cancer. Lung Cancer.
63:98–105. 2009. View Article : Google Scholar
|
|
10
|
Devine PL, Birrell GW, Quin RJ and Shield
PW: Monoclonal antibodies recognising sialyl-Tn: Production and
application to immunochemistry. Dis Markers. 12:175–186. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Longenecker BM, Willans DJ, MacLean GD,
Selvaraj S, Suresh MR and Noujaim AA: Monoclonal antibodies and
synthetic tumor-associated glycoconjugates in the study of the
expression of Thomsen-Friedenreich-like and Tn-like antigens on
human cancers. J Natl Cancer Inst. 78:489–496. 1987.PubMed/NCBI
|
|
12
|
Nguyen PL, Niehans GA, Cherwitz DL, Kim YS
and Ho SB: Membrane-bound (MUC1) and secretory (MUC2, MUC3, and
MUC4) mucin gene expression in human lung cancer. Tumour Biol.
17:176–192. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Snijdewint FG, von Mensdorff-Pouilly S,
Karuntu-Wanamarta AH, Verstraeten AA, Livingston PO, Hilgers J and
Kenemans P: Antibody-dependent cell-mediated cytotoxicity can be
induced by MUC1 peptide vaccination of breast cancer patients. Int
J Cancer. 93:97–106. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Vassilaros S, Tsibanis A, Tsikkinis A,
Pietersz GA, McKenzie IF and Apostolopoulos V: Up to 15-year
clinical follow-up of a pilot Phase III immunotherapy study in
stage II breast cancer patients using oxidized mannan-MUC1.
Immunotherapy. 5:1177–1182. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kimura T, McKolanis JR, Dzubinski LA,
Islam K, Potter DM, Salazar AM, Schoen RE and Finn OJ: MUC1 vaccine
for individuals with advanced adenoma of the colon: A cancer
immunoprevention feasibility study. Cancer Prev Res (Phila).
6:18–26. 2013. View Article : Google Scholar
|
|
16
|
Ramanathan RK, Lee KM, McKolanis J,
Hitbold E, Schraut W, Moser AJ, Warnick E, Whiteside T, Osborne J,
Kim H, et al: Phase I study of a MUC1 vaccine composed of different
doses of MUC1 peptide with SB-AS2 adjuvant in resected and locally
advanced pancreatic cancer. Cancer Immunol Immunother. 54:254–264.
2005. View Article : Google Scholar
|
|
17
|
Shedden K, Taylor JM, Enkemann SA, Tsao
MS, Yeatman TJ, Gerald WL, Eschrich S, Jurisica I, Giordano TJ,
Misek DE, et al; Director's Challenge Consortium for the Molecular
Classification of Lung Adenocarcinoma. Gene expression-based
survival prediction in lung adenocarcinoma: A multi-site, blinded
validation study. Nat Med. 14:822–827. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wilson CL and Miller CJ: Simpleaffy: A
BioConductor package for Affymetrix Quality Control and data
analysis. Bioinformatics. 21:3683–3685. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bhattacharjee A, Richards WG, Staunton J,
Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, et
al: Classification of human lung carcinomas by mRNA expression
profiling reveals distinct adenocarcinoma subclasses. Proc Natl
Acad Sci USA. 98:13790–13795. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Irizarry RA, Bolstad BM, Collin F, Cope
LM, Hobbs B and Speed TP: Summaries of Affymetrix GeneChip probe
level data. Nucleic Acids Res. 31:e152003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
R Development Core Team. R: A Language and
Environment for Statistical Computing: the R Foundation for
Statistical Computing. Vienna: 2011
|
|
22
|
Song W, Delyria ES, Chen J, Huang W, Lee
JS, Mittendorf EA, Ibrahim N, Radvanyi LG, Li Y, Lu H, et al: MUC1
glycopeptide epitopes predicted by computational glycomics. Int J
Oncol. 41:1977–1984. 2012.PubMed/NCBI
|
|
23
|
Reddish MA, Jackson L, Koganty RR, Qiu D,
Hong W and Longenecker BM: Specificities of anti-sialyl-Tn and
anti-Tn monoclonal antibodies generated using novel clustered
synthetic glycopeptide epitopes. Glycoconj J. 14:549–560. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Colcher D, Hand PH, Nuti M and Schlom J: A
spectrum of monoclonal antibodies reactive with human mammary tumor
cells. Proc Natl Acad Sci USA. 78:3199–3203. 1981. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Schietinger A, Philip M, Yoshida BA, Azadi
P, Liu H, Meredith SC and Schreiber H: A mutant chaperone converts
a wild-type protein into a tumor-specific antigen. Science.
314:304–308. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Vierbuchen MJ, Fruechtnicht W, Brackrock
S, Krause KT and Zienkiewicz TJ: Quantitative lectin-histochemical
and immunohistochemical studies on the occurrence of alpha(2,3)-
and alpha(2,6)-linked sialic acid residues in colorectal
carcinomas. Relation to clinicopathologic features. Cancer.
76:727–735. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Steentoft C, Vakhrushev SY, Joshi HJ, Kong
Y, Vester-Christensen MB, Schjoldager KT, Lavrsen K, Dabelsteen S,
Pedersen NB, Marcos-Silva L, et al: Precision mapping of the human
O-GalNAc glycoproteome through SimpleCell technology. EMBO J.
32:1478–1488. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yang Z, Halim A, Narimatsu Y, Jitendra
Joshi H, Steentoft C, Schjoldager KT, Alder Schulz M, Sealover NR,
Kayser KJ, Paul Bennett E, et al: The GalNAc-type O-Glycoproteome
of CHO cells characterized by the SimpleCell strategy. Mol Cell
Proteomics. 13:3224–3235. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Levery SB, Steentoft C, Halim A, Narimatsu
Y, Clausen H and Vakhrushev SY: Advances in mass spectrometry
driven O-glycoproteomics. Biochim Biophys Acta. 1850.33–42.
2015.
|
|
30
|
Lakshminarayanan V, Thompson P, Wolfert
MA, Buskas T, Bradley JM, Pathangey LB, Madsen CS, Cohen PA,
Gendler SJ and Boons GJ: Immune recognition of tumor-associated
mucin MUC1 is achieved by a fully synthetic aberrantly glycosylated
MUC1 tripartite vaccine. Proc Natl Acad Sci USA. 109:261–266. 2012.
View Article : Google Scholar :
|
|
31
|
Dokurno P, Bates PA, Band HA, Stewart LM,
Lally JM, Burchell JM, Taylor-Papadimitriou J, Snary D, Sternberg
MJ and Freemont PS: Crystal structure at 1.95 A resolution of the
breast tumour-specific antibody SM3 complexed with its peptide
epitope reveals novel hypervariable loop recognition. J Mol Biol.
284:713–728. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Möller H, Serttas N, Paulsen H, Burchell
JM, Taylor-Papadimitriou J and Meyer Bernd: NMR-based determination
of the binding epitope and conformational analysis of MUC-1
glycopeptides and peptides bound to the breast cancer-selective
monoclonal antibody SM3. Eur J Biochem. 269:1444–1455. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Spencer DI, Missailidis S, Denton G,
Murray A, Brady K, Matteis CI, Searle MS, Tendler SJ and Price MR:
Structure/activity studies of the anti-MUC1 monoclonal antibody
C595 and synthetic MUC1 mucin-core-related peptides and
glycopeptides. Biospectroscopy. 5:79–91. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Matsumoto-Takasaki A, Hanashima S, Aoki A,
Yuasa N, Ogawa H, Sato R, Kawakami H, Mizuno M, Nakada H, Yamaguchi
Y, et al: Surface plasmon resonance and NMR analyses of anti
Tn-antigen MLS128 monoclonal antibody binding to two or three
consecutive Tn-antigen clusters. J Biochem. 151:273–282. 2012.
View Article : Google Scholar
|
|
35
|
Ibrahim NK, Murray JL, Zhou D, Mittendorf
EA, Sample D, Tautchin M and Miles D: Survival Advantage in
Patients with Metastatic Breast Cancer Receiving Endocrine Therapy
plus Sialyl Tn-KLH Vaccine: Post Hoc Analysis of a Large Randomized
Trial. J Cancer. 4:577–584. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Forsström B, Axnäs BB, Stengele KP, Bühler
J, Albert TJ, Richmond TA, Hu FJ, Nilsson P, Hudson EP, Rockberg J,
et al: Proteome-wide epitope mapping of antibodies using
ultra-dense peptide arrays. Mol Cell Proteomics. 13:1585–1597.
2014. View Article : Google Scholar : PubMed/NCBI
|