Introduction
Prostate cancer (PCa) represents the most common
malignancy in males in the United States other than skin cancer and
the second leading cause (after lung cancer) of cancer-related
deaths (1). The majority of
patients with PCa behave in an indolent manner, but a subset is
highly aggressive (2). Locally
defined disease is often successfully treated with surgery and/or
radiotherapy; however, disease recurs in an estimated 15–30% of
patients (3). Treatment decisions
are currently based on clinical and histological features only.
Although radical prostatectomy (RP) and radiation therapy are the
most appropriate treatment for patients with localized PCa, the
risk of biochemical disease recurrence in RP patients is ~25%
(4). Since PCa often progresses
rapidly and most death is caused by metastases that are resistant
to conventional therapies, the optimal therapy for patients with
this cancer has yet to be defined. As the molecular etiology of PCa
becomes better understood, it is clear that PCa progression is
influenced by a multistep process, involving both genetic insults
to epithelial cells and changes in epithelial-stromal interactions
(5). Therefore, it is extremely
necessary to better understand the biology of the disease which may
lead to the identification of novel prognostic biomarkers and
efficient treatment options.
Cell cycle-specific transcription factor E2F1 was
originally identified as a member of the E2F family of
transcription factors, which consists of eight proteins commonly
classified as activators (E2F1-E2F3a) or repressors (E2F3b-E2F8),
based on their ability to either activate or repress target gene
transcription (6). The activator
E2F family proteins are required for cell proliferation, whereas
the repressor E2Fs play a role in cell cycle exit and
differentiation (7). A puzzling
feature of the activator E2Fs, especially E2F1, is the ability to
regulate the seemingly opposing processes of proliferation and
apoptosis (8). Genome-wide
analysis of targets of E2F1 using microarrays has also identified a
large number of genes that are regulated by E2F1 in diverse
cellular processes, such as replication, apoptosis, checkpoint
control and DNA repair (9). In
tumor biology, aberrant expression of E2F1 has been detected in
various cancer cell lines and clinical tissues, including
amplification of the E2F1 gene in erythroleukemia cell lines,
elevated expression of E2F1 protein in breast cancer cell lines and
head and neck carcinoma cell lines, and overexpression of E2F1 in
invasive ductal breast cancer and non-small cell lung cancer, where
high E2F1 expression associates with advanced disease and poor
prognosis (10–14). Particularly, Ren et al
(15) observed that E2F1
expression was elevated in advanced PCa and found that E2F1
knockdown could inhibit prostate tumor growth in vitro and
in vivo through sensitizing tumor cells to ICAM-1 mediated
anti-immunity by NF-κB modulation, highlighting the potential of
E2F1 as a therapeutic target; Davis et al (16) indicated that elevated E2F1
expression, through its ability to repress androgen receptor (AR)
transcription, might contribute to the progression of
hormone-independent PCa; Zheng et al (17) found that E2F1 could induce cancer
cell survival via NF-κB-dependent induction of EGR1 transcription
in PCa cells; Libertini et al (18) described E2F1-dependent
modifications of androgen dependence, differentiation, and
sensitivity to apoptotic stimuli in PCa cell lines. It is, however,
unclear whether E2F1 promotes metastasis of PCa cells and which of
the E2F1 targets are key metastatic mediators in this cancer.
To address this problem, we determined the
expression patterns of E2F1, at both mRNA and protein levels, in
human PCa tissues, and evaluated the associations of E2F1
expression with clinicopathological characteristics and patient
prognosis. Then, the roles of E2F1 on malignant phenotypes of PCa
cells were investigated. Furthermore, the combined bioinformatic
binding site prediction with chromatin immunoprecipitation-PCR
(ChIP-PCR) and western blot analysis was performed to identify the
E2F1 targets in PCa cells.
Materials and methods
Patients and tissue samples
The present study was approved by the Research
Ethics Committee of the Guangzhou First People's Hospital,
Guangzhou Medical University, China. Informed consent was obtained
from all the patients. The specimens were handled and made
anonymous according to the ethical and legal standards.
Cell culture
Human PCa cell lines, LNCaP and PC3 were purchased
from Guangzhou Land Biosciences Co., Ltd., (Guangzhou, China) and
were cultured in RPMI-1640 medium (Cellgro, Manassas, VA, USA)
supplemented with 10% fetal bovine serum (FBS; Cellgro), 2 mM
L-glutamine and antibiotics. All cell lines were maintained at 37°C
in a humidified chamber supplemented with 5% CO2.
Animals
Animal experiments in the present study were
performed in compliance with the guidelines of the Institute for
Laboratory Animal Research at Guangzhou Medical University
(Guangzhou, China). A total of 8 BALB/c nude mice (6–8-week-old
males) were purchased from Guangdong Medical Laboratory Animal
Center and were housed 4 per cage in wire-top cages with sawdust
bedding in an isolated, clean, air-conditioned room at a
temperature of 25–26°C and a relative humidity of ~50%, lit 12
h/day.
Cell transfection
The siRNAs of E2F1 and control (cat nos.
siB11715100401 and siN05815122147; Guangzhou RiboBio Co., Ltd.,
Guangzhou, China) were respectively transfected in LNCaP cells with
Lipofectamine 2000. The expression of E2F1 was detected by qPCR and
western blot analyis. The results showed that the siRNA with
sequence (5′-CCUGAUGAAUAUCUGUACUdTdT-3′) inhibited the E2F1
expression significantly and selected to construct the sh-E2F1
plasmid as an insert sequence. The pGreenPuro Scramble Hairpin
Control as sh-RNA negative control was purchased from SBI (cat. no.
MZIP000-PA-1; System Biosciences, Mountain View, CA, USA) and the
shRNA-E2F1 was constructed using the pGreenPuro Scramble Hairpin
Control as the bone plasmid. To make the virus, 293TN cells were
transfected with shRNA-E2F1 or sh-RNA negative control with plasmid
and Lipofectamine 2000, and then after 3 days, the virus particles
were collected according to the packaging protocol of SBI with the
Lenti-Concentin virus precipitation solution (cat. no. LV810A-1;
System Biosciences). LNCaP and PC3 cells were infected by virus
solution with TransDux virus transduction reagent (cat. no.
LV850A-1; System Biosciences). The infected cells were detected by
fluorescence microscope and isolated with a flow cytometer to
construct the stable sh-E2F1 and control cell lines.
Western blot analysis
Proteins were extracted 48 h post-transfection for
western blot analyses. Proteins (40 μg) were fractioned on SDS-PAGE
and transferred onto Hybond nitrocellulose membranes (GE
Healthcare). The membranes were blocked with 5% skim milk in
PBS-Tween 20 and probed with anti-E2F1 (ab-64161; Abcam Co., Ltd.,
Hong Kong), anti-CD147 (ab-108308; Abcam) and anti-GAPDH (P30008;
Abmart, Co., Ltd., Shanghai, China). The results were visualized
with the SuperSignal West Pico chemiluminescent detection system
(Pierce Biotechnology, Rockford, IL, USA). GAPDH was used as an
internal loading control.
Immunohistochemistry
Expression pattern and subcellular localization of
E2F1 protein in clinical PCa tissues were detected by
immunohistochemistry and the immunoreactivity scores (IRS) were
calculated according to the protocol of our previous studies
(19,20). IRS were scored by two independent
experienced pathologists, who were blinded to the
clinicopathological data and clinical outcomes of the patients. The
number of positive-staining cells in ten representative microscopic
fields was counted and the percentage of positive cells was
calculated. Given the heterogeneity of the staining of the target
genes or proteins, tumor specimens were scored in a
semi-quantitative manner. The percentage was grouped as follows: 0
(0%), 1 (1–10%), 2 (11–50%) and 3 (>50%). The staining intensity
was categoried as follows: 0 (negative), 1 (weak), 2 (moderate) and
3 (strong). A final score was obtained for each case by the sum of
the percentages and the intensity scores. The antibodies used in
this study were: E2F1 antibody (ab-64161; Abcam); CD147 antibody
(ab-108308; Abcam); vimentin antibody (za-0511; ZSGB-BIO Co., Ltd.,
Beijing, China); E-cadherin antibody (za-0565; ZSGB-BIO).
Cell cycle analysis
The effects of E2F1 on cell cycle progression were
determined by fluorescence-activated cell sorter (FACS) analysis
using the Cell Cycle and Apoptosis Analysis kit (Beyotime Institute
of Biotechnology, Shanghai, China) according to the manufacturer's
instructions. Ten thousand cells were counted per sample. All
experiments were done in triplicate. Mean normalized gene
expression ± SE was calculated from independent experiments.
Cell invasion assays
The Transwell inserts (8-μm pores) were filled with
50 μl of a mixture of serum-free RPMI-1640 medium (Cellgro) and
Matrigel (1:10; BD Biosciences, San Jose, CA, USA). The inserts
were then placed in 24-well tissue culture plates (Transwell;
Corning Incorp., Corning, NY, USA) containing 10% FBS-medium. After
solidification by incubation in 37°C for 4 h, 5×104
cells in 200 μl medium were placed in upper chambers. Following 48
h of incubation at 37° with 5% CO2 and in culture medium
with mitomycin to stop the mitosis, the membranes were fixed with
10% formalin and stained with 0.05% crystal violet. The number of
cells that migrated through the pores was assessed and the data
were expressed as mean ± SD of three independent experiments.
Migration assays
For the scratch wound-healing motility assay, a
scratch was made with a pipette tip when the cells reach
confluence. After being cultured under standard conditions with
mitomycin for 24 h, plates were washed twice with fresh medium to
remove non-adherent cells and then photographed. The cell migrated
from the wound edge were counted and the data were expressed as the
mean ± SD of three independent experiments.
Generation of the in vivo xenograft
model
For the in vivo tumor formation assays, PC3
or LNCaP cells transfected with shRNA-E2F1 were trypsinized and
suspended in phosphate-buffered saline (PBS). Then, the cells were
subcutaneously injected into the flanks of each nude mouse (4 per
group). PC3 cells were subcutaneously injected with 0.1 ml volume
of 1×106 cells. LNCaP cells were subcutaneously injected
at a 0.1 ml mixture of 1×106 cells and an 0.05 ml
Matrigel (cat. no. 356234; BD Biosciences), reaching a total
concentration of 10 mg/ml. The tumor sizes were measured at 4-day
intervals, as soon as the tumors were measurable and the tumor
volumes were calculated: V(mm3) =
width2(mm2) × length(mm)/2. On day 44 for
LNCaP and day 36 for PC3 groups, the mice were sacrificed. The mice
were manipulated and housed according to the protocols approved by
the Institute for Laboratory Animal Research at Guangzhou Medical
University.
Bioinformatics prediction of E2F1 binding
sites in sets of genes
Three online bioinformatics software including
oPOSSUM (http://www.cisreg.ca/oPOSSUM/, Last updated: Jan 31st,
2007) (21), ConSite (http://consite.genereg.net/) and PROMO (version 3.0.2)
(22) were used to predict E2F1
binding sites in sets of genes.
CHIP-PCR
ChIP was conducted with Chromatin
immunoprecipitation assay kit (cat. no. 26156; Thermo Fisher
Scientific, Waltham, MA, USA) according to the manufacturer's
instructions. In brief, cells were fixed in 1% formaldehyde at room
temperature for 10 min; the nuclei were isolated with nuclear lysis
buffer (Thermo Fisher Scientific) supplemented with protease
inhibitor cocktail (Thermo Fisher Scientific). Chromatin DNA was
sonicated and sheared to a length between 200 and 500 bp. The
sheared chromatin was immunoprecipitated at 4°C overnight with
anti-E2F1 (Cell Signaling Technology). Normal rabbit IgG was used
as a negative control and genome DNA was used as an input control.
Protein/DNA complex was reverse cross-linked and DNA was purified
using spin columns. Purified DNA was detected with quantitative
PCR. The sequences used in the present study were: E2F1/CD147 F1,
5′-ATGTGCGTCTCGCCATGTT-3′; E2F1/CD147 R1,
5′-TGTGGCTCACGTCTGTACTC-3′.
Statistical analysis
The version 13.0 SPSS for Windows (SPSS, Inc., IL,
USA) and SAS 9.1 (SAS Institute, Cary, NC, USA) software was used
for statistical analysis. Continuous variables were expressed as
mean ± SD. Statistical analyses of PCR and western blot analysis
were conducted using Wilcoxon signed-rank test. Statistical
analysis was performed independently by two biostatisticians with
Fisher's exact test for any 2×2 tables and Pearson χ2
test for non-2×2 tables. Kaplan-Meier method was used for the
survival analyses. The Pearson's correlation was calculated between
the expression levels of E2F1 and CD147 in PCa tissues. Differences
were considered statistically significant when the P-value was
<0.05.
Results
Overexpression of E2F1 in human PCa
tissues
Expression pattern and sub-cellular localization of
E2F1 protein in 97 PCa and 81 adjacent non-cancerous prostate
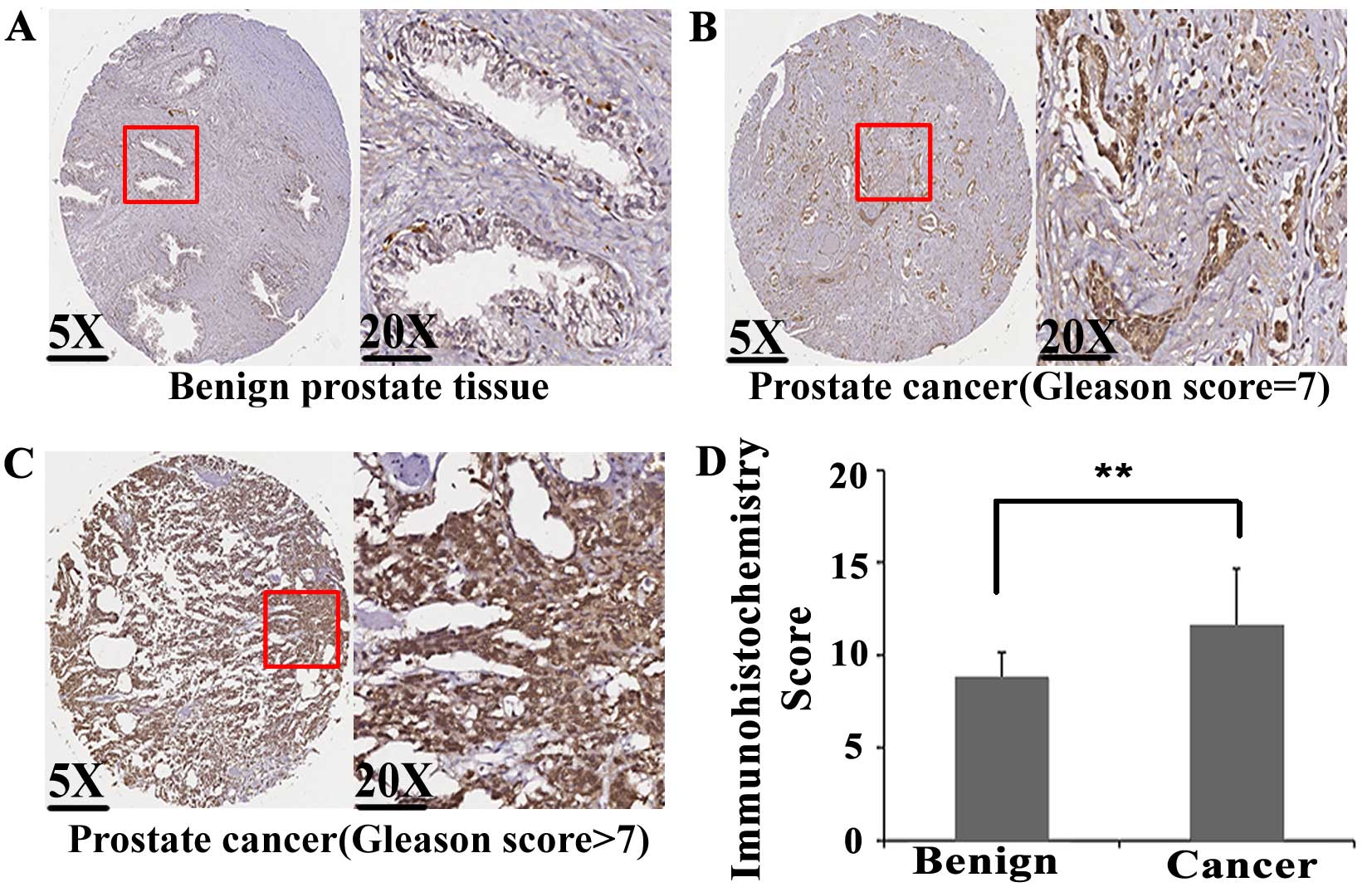
tissues were detected by immunohistochemistry. As shown in Fig. 1A–C, E2F1 protein immunostaining
occurred mainly in the cytoplasm and nuclei of PCa cells, but
weakly or negatively in adjacent non-cancerous prostate tissues.
Statistical analysis found that the expression levels of E2F1
protein in PCa tissues were significantly higher than those in
adjacent non-cancerous prostate tissues (IRS, PCa, 6.60±0.76 vs.
Benign, 5.67±1.18, P<0.001; Fig.
1D).
Overexpression of E2F1 associates with
the aggressive progression of human PCa
Table I summarizes
the associations between E2F1 protein expression and various
clinicopathological features of PCa patients. We found that E2F1
protein overexpression in PCa tissues was significantly associated
with high Gleason score (P=0.005) and advanced pathological stage
(P=0.005), which was consistent with the statistical results of
E2F1 mRNA expression based on Taylor dataset (Gleason score ≥8 vs.
Gleason score <8, P=0.013; pathological stage >T3A vs.
pathological stage <T3A, P=0.002). According to the Taylor
dataset, we also found that PCa patients with positive metastasis
and short overall survival time had higher expression levels of
E2F1 mRNA expression than those with negative metastasis
(P<0.001) and long overall survival time (P=0.003).
 | Table IAssociations of E2F1 protein and mRNA
expression with various clinicopathological characteristics of
human prostate cancer (PCa). |
Table I
Associations of E2F1 protein and mRNA
expression with various clinicopathological characteristics of
human prostate cancer (PCa).
| E2F1 in our tissue
microarray | E2F1 expression in
Taylor dataset |
|---|
|
|
|
|---|
| Clinical
features | No. of cases | Mean ± SD | P-value | No. of cases | Mean ± SD | P-value |
|---|
| Age (years) |
| <70 | 43 | 6.47±0.94 | 0.162 | 144 | 7.59±0.20 | 0.770 |
| ≥70 | 54 | 6.70±0.57 | | 6 | 7.61±0.13 | |
| Serum PSA
(ng/ml) |
| <4 | - | - | - | 24 | 7.59±0.21 | 0.212 |
| ≥4 | - | - | | 123 | 7.58±0.19 | |
| Gleason score |
| <8 | 70 | 6.50±0.85 | 0.005 | 117 | 7.56±0.25 | 0.013 |
| ≥8 | 27 | 8.85±0.36 | | 22 | 7.68±0.25 | |
| Pathological
stage |
| <T3A | 70 | 6.50±0.85 | 0.005 | 86 | 7.54±0.18 | 0.002 |
| ≥T3A | 27 | 8.85±0.36 | | 55 | 7.64±0.20 | |
| Metastasis |
| No | 97 | 6.60±0.76 | - | 122 | 7.56±0.18 | <0.001 |
| Yes | 0 | - | | 28 | 7.74±0.22 | |
| Overall
survival |
| Alive | - | - | - | 131 | 7.57±0.18 | 0.003 |
| Die | - | - | | 19 | 7.71±0.28 | |
| PSA failure |
| Negative | - | - | - | 104 | 7.56±0.18 | 0.064 |
| Positive | - | - | | 36 | 7.63±1.18 | |
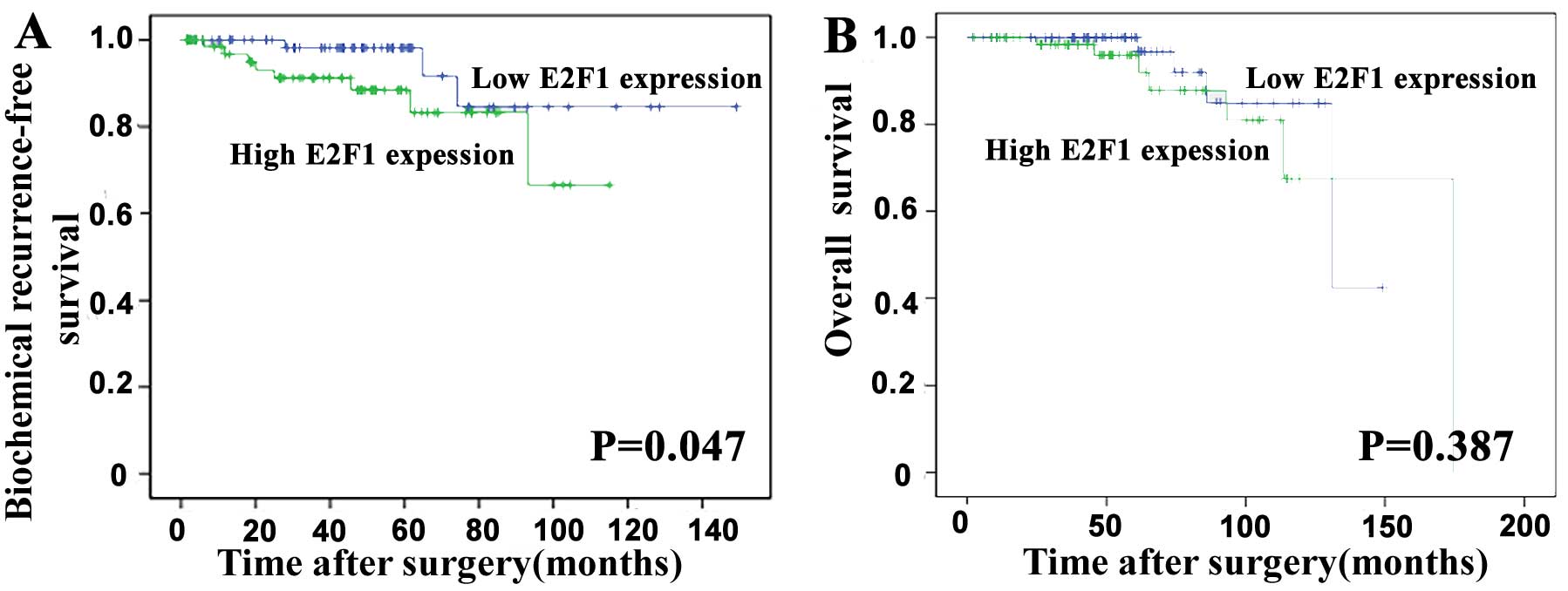
Overexpression of E2F1 predicts poor
prognosis of human PCa
To evaluate the prognostic value of E2F1 expression
in PCa, the Kaplan-Meier method was performed to analyze the
correlation of E2F1 expression with biochemical recurrence
(BCR)-free survival and overall survival of PCa patients based on
Taylor dataset. The median value of E2F1 mRNA expression in all PCa
tissues was used as the cut-off point to divide all cases into high
(n=75) and low (n=74) E2F1 expression groups. Pairwise comparisons
showed significant differences in the BCR-free survival (P=0.047;
Fig. 2A) between patients with
high and low E2F1 expression groups, except the overall survival
(P=0.387; Fig. 2B).
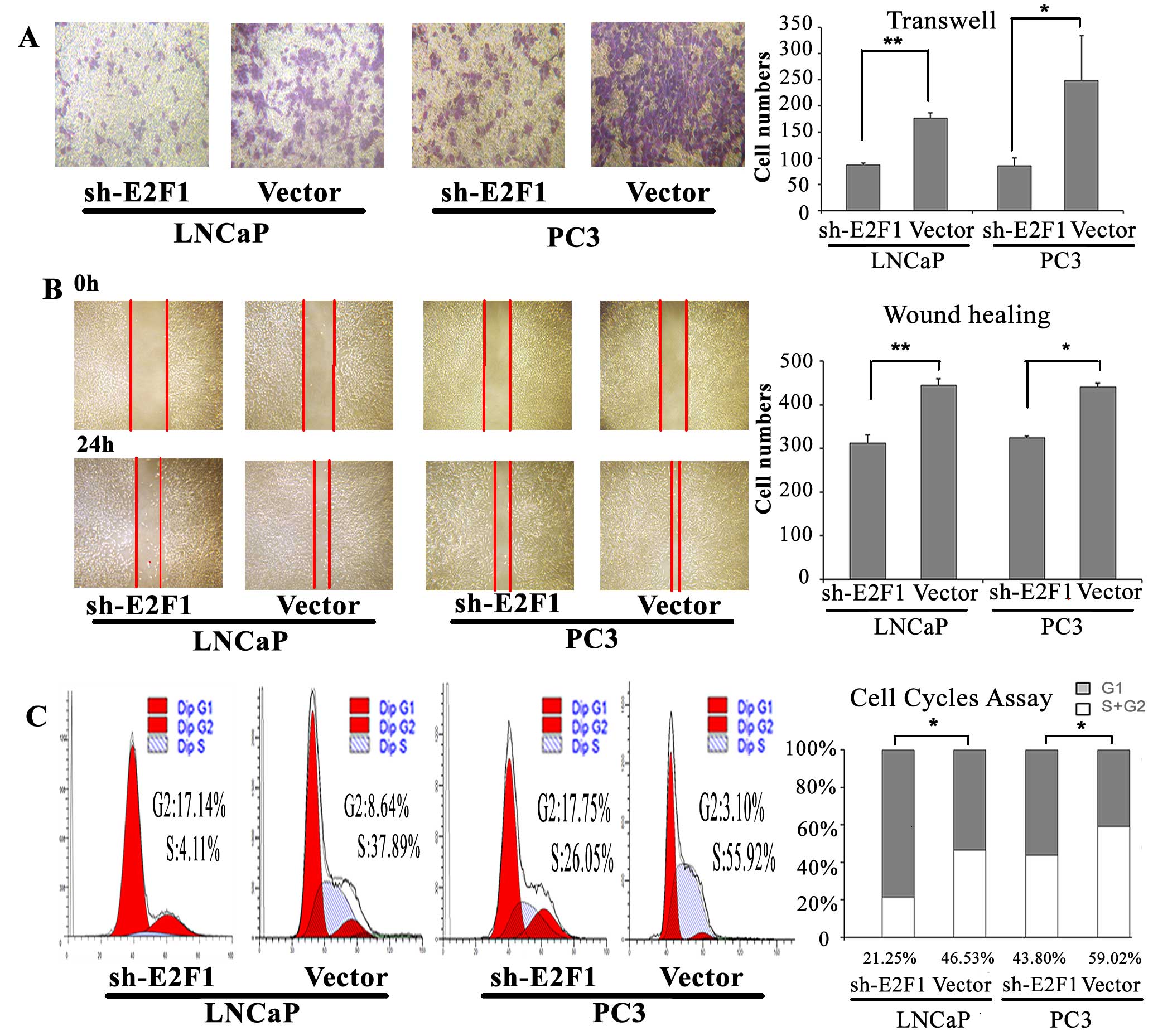
Knock-down of E2F1 inhibits cell cycle
progression, invasion and migration of PCa cell lines in vitro
To determine the oncogenic roles of E2F1 in PCa, we
detected the cell cycle progression, invasion and migration
abilities of PC3 and LNCaP cells with the knock-down of E2F1.
According to the western blot analysis, the expression of E2F1
protein was successfully suppressed by sh-E2F1 vector in both PC3
and LNCaP cells (Fig. 6B). Then,
we found that the knockdown of E2F1 could lead to substantial
accumulation of the cell population at the G1 stage of the cell
cycle in PC3 and LNCaP cells (P=0.015 and P=0.010, respectively;
Fig. 3C). Moreover, the invasive
and migration activities of both PC3 and LNCaP cells were inhibited
by the downregulating of E2F1 expression (all P<0.05; Fig. 3A and B).
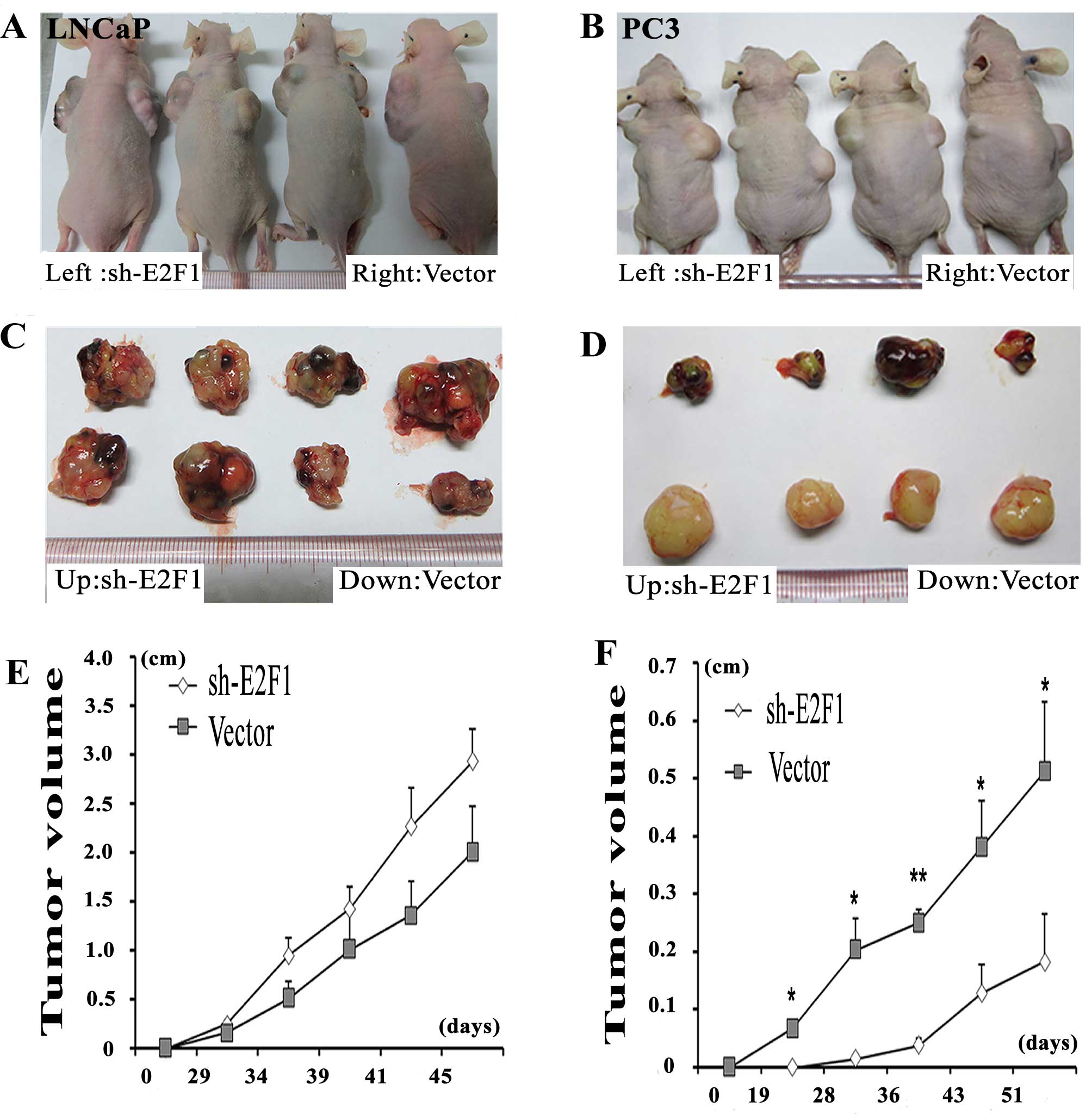
Knockdown of E2F1 suppresses tumor growth
and invasion in vivo
To better evaluate the biological functions of E2F1
in vivo, LNCaP and PC3 cell lines transfected with sh-E2F1
were used and subcutaneously injected into the flank of each male
nude mouse, simultaneously, the vector control PCa cells were
subcutaneously injected into the other flank of the same mouse. The
PC3 cells with the knockdown of E2F1 formed significantly smaller
tumor nodules and remarkably slowed tumor xenograft growth compared
with the controls (Fig. 4B, D and
F). However, there is no significantly difference between the tumor
size of E2F1 knockdown group and the control group in LNCaP cells
(Fig. 4A, C and E).
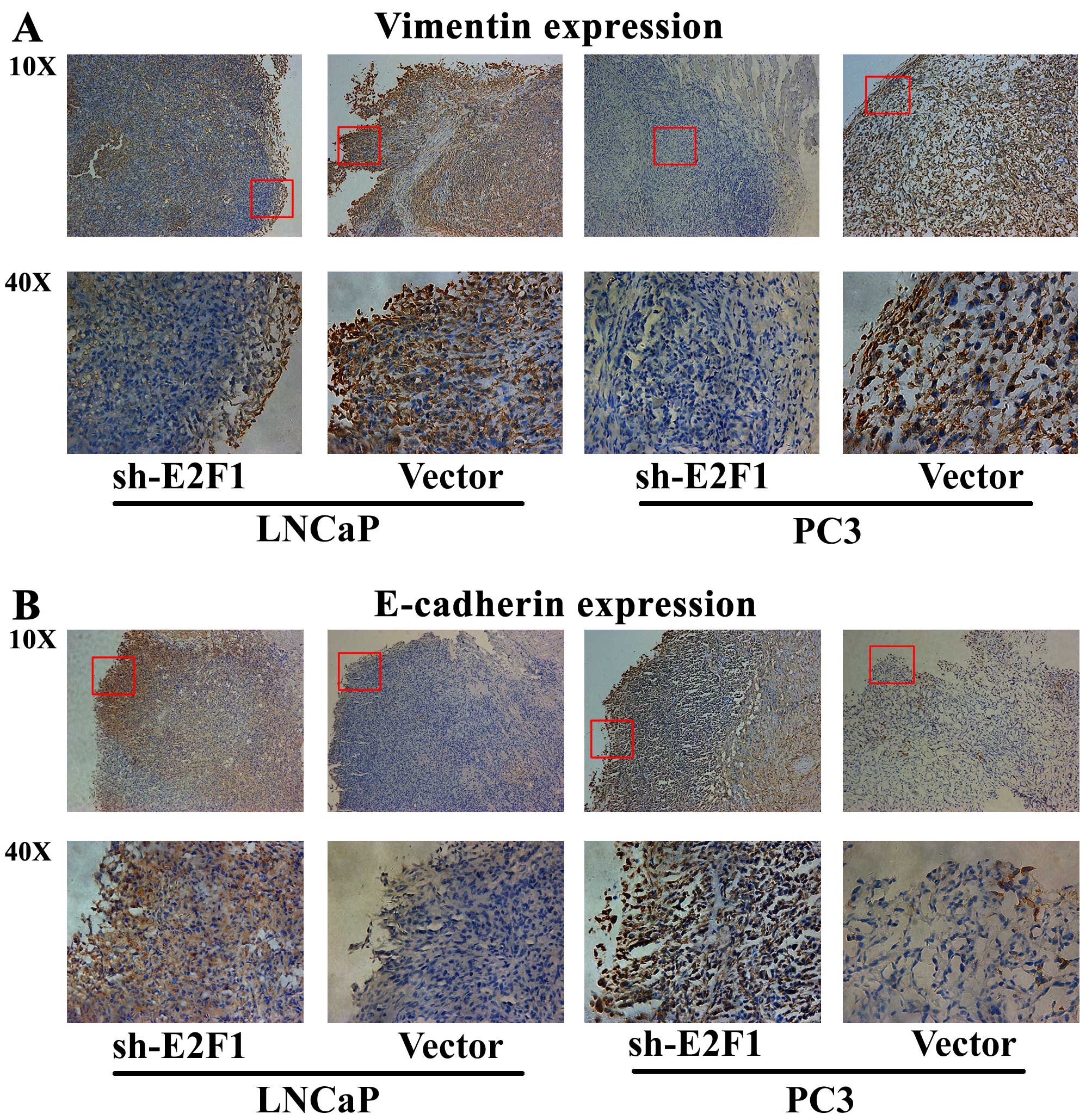
Next, immunohistochemical analysis was performed to
detect the expression patterns of epithelial marker E-cadherin and
mesenchymal marker vimentin in the tumor xenografts, in order to
evaluate the tumor invasive tendency in different groups. The
results indicated that the expression level of vimentin protein in
the tumor xenografts established by LNCaP or PC3 cells transfected
with sh-E2F1 was remarkably lower than that in the xenografts
established by cells transfected with control vectors (Fig. 5A). In contrast, the tumor
xenografts established by LNCaP or PC3 cells with low E2F1
expression presented significantly more E-cadherin protein than the
control xenografts (Fig. 5B).
These results strongly demonstrated that the knockdown of E2F1
could significantly inhibit tumor growth and invasion in
vivo.
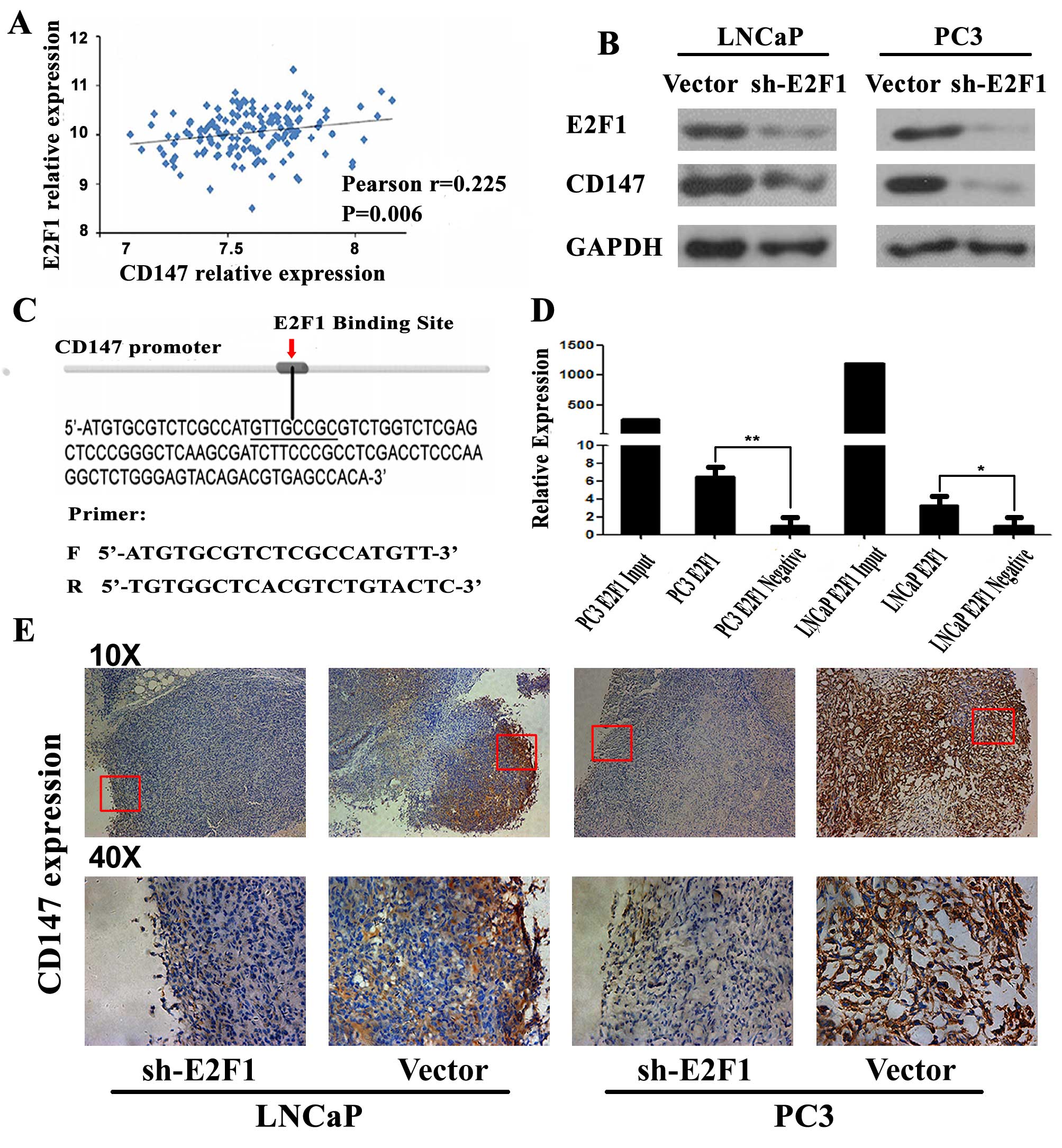
Positive correlation between expression
of E2F1 and CD147
In a previous study, we found that CD147 expression
in human PCa tissues was significantly higher than those in
adjacent non-cancerous prostate tissues and might be an important
factor in predicting the prognosis of the PCa patients (23). Based on the Taylor dataset, the
Pearson correlation analysis clearly showed a positive correlation
between CD147 and E2F1 mRNA expression in PCa tissues (r=0.225,
P=0.006; Fig. 6A). To identify
E2F1 target genes in the human genome, 3 online bioinformatics
software including oPOSSUM, ConSite and PROMO were used and
commonly predicted CD147 as a potential target of E2F1. As shown in
Fig. 6C, E2F1 and CD147 could
directly bind to the putative E2F1-CD147 binding region (sequence:
GTTGCGG). To validate this prediction, we detected the expression
of CD147 at protein level in 2 PCa cell lines (PC3 and LNCAP)
transfected with sh-E2F1. As shown in Fig. 6B, knocking down E2F1 resulted in
significant down-regulation of CD147 protein expression. Thus, we
speculated that E2F1 might upregulate CD147 expression in PC3 and
LNCAP cells. To confirm our speculation, we analyzed the binding
site of CD147 promoter by ChIP-PCR. Compared to the negative
control group, E2F1 could directly bind to a putative site in the
promoter of CD147 (Fig. 6D). Also,
the tumor xenografts established by LNCaP or PC3 cells with low
E2F1 expression significantly suppressed the expression of CD147
compared to the control xenografts (Fig. 6E).
Discussion
The transcription factor E2F1 expression has been
detected in several human cancers, and oncogenic and
tumor-suppressive roles have been proposed for E2F1 depending on
the cancer type and, in some cases, the subtype. For example, Li
et al (24) demonstrated
the increased expression of E2F1 in small cell lung cancer (SCLC),
and confirmed that E2F1 could directly enhance matrix
metalloproteinase (MMP) transcription by binding to the E2F1
binding sequences in the promoter, or indirectly activate MMPs
through enhanced Sp1 and NF-κB as a consequence of E2F1 activation
in SCLC; Ma et al (25)
indicated that the overexpression of E2F1 might promote tumor
malignancy and correlates with TNM stages in clear cell renal cell
carcinoma; Engelmann et al (26) also revealed that E2F1 might
function as an enhancer of angiogenesis via regulation of
VEGF-C/VEGFR-3 signaling in tumors to cooperatively activate PDGF-B
expression, and targeting this pathway might be reasonable to
complement standard anti-angiogenic treatment of cancers with
deregulated E2F1; on the contrary, E2F1 has been revealed to be a
mediator of apoptosis, and in clinical trials where its high
expression leads to improved survival of patients with adjuvant
chemo-radiation therapy (27).
These findings suggest that E2F1 exhibits dual properties, acting
as a tumor suppressor and oncogene.
Previous studies have reported the oncogenic role of
E2F1 in the control of PCa cell cycle, and the regulation of
proliferation and apoptosis (15–18).
However, its clinical significance in PCa remains unclear. In the
present study, our clinical evidence revealed that the upregulation
of E2F1 was remarkably associated with the tumor aggressive
clinicopathological features and might act as an important
prognostic factor for BCR-free survivals in patients with PCa,
which prompted us to determine the roles of E2F1 in malignant
phenotypes of PCa in vitro and in vivo models. Our
data showed that the downregulation of E2F1 expression by
shRNA-E2F1 could suppress PCa cell cycle progression, invasion and
migration in vitro, and caused a dramatic decrease in PC3
tumor growth and suppressed tumor invasion in vivo. These
findings delineate an as yet unrecognized function of E2F1 as
enhancer of tumor invasion and migration of PCa.
Furthermore, we determined that E2F1 could directly
target CD147 and be implicated in carcinogenesis of PCa through the
regulation of CD147, which is a transmembrane glycoprotein, and
member of the immunoglobulin superfamily molecules expressed on the
cell surface of most tumor cells (28). Functionally, CD147 may exert an
oncogenic role by stimulating the synthesis of several MMPs,
activating tumor angiogenesis and cell survival signaling (29). Our previous studies have reported
the involvement of CD147 in human PCa. In 2008, we found that
CD147, MMP-1, MMP-2 and MMP-9 expression levels were all
significantly higher in PCa tissues than in benign tissues, and
there was a significant positive correlation between CD147 and
MMP-1, MMP-2 and MMP-9 expression in PCa tissues (30). Then, we identified CD147 as an
independent prognostic factor for patients with PCa (31). Moreover, in 2012, we examined
whether the expression of CD147 can be used as a prognostic marker
for predicting PCa progression based on a large cohort of clinical
cases. As a result, our data revealed that the increased expression
of CD147 was correlated with higher Gleason scores, positive
surgical margin, prostate-specific antigen failure, metastasis and
reduced overall survival. Importantly, higher expression of CD147
can serve as an independent prognostic predictor for PSA
failure-free survival in PCa patients when they are stratified by
Gleason scores (31). In the
present study, we confirmed that CD147 might function as a direct
target for E2F1 in PCa cells through bioinformatics binding site
prediction, combined with ChIP-PCR, and western blot analysis. The
expression of CD147 mRNA in PCa tissues was positively correlated
with that of E2F1 mRNA. Then, knocking down the expression of E2F1
was able to suppress the expression of CD147 in PCa cells in
vitro and in vivo. Given that CD147 has been reported to
be significantly upregulated in advanced PCa and could promote cell
migration and invasion, the oncogenic role of E2F1 in PCa might be
exerted by regulating CD147 expression.
In conclusion, E2F1 may promote tumor cell invasion
and migration through the regulation of CD147 in PCa cells.
Importantly, E2F1 may function as a biomarker that can
differentiate patients with biochemical recurrent and
non-biochemical recurrent disease following RP, highlighting its
potential as a therapeutic target.
Acknowledgements
The present study was supported by grants from the
National Natural Science Foundation of China (81170699 and
81272813), the Natural Science Foundation of Guangdong Province
(2014A030310501) and the Science and Technology Project of Bureau
of Health in Guangzhou Municipality (20141A011006).
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nandana S and Chung LW: Prostate cancer
progression and metastasis: Potential regulatory pathways for
therapeutic targeting. Am J Clin Exp Urol. 2:92–101.
2014.PubMed/NCBI
|
|
3
|
Han M, Partin AW, Zahurak M, Piantadosi S,
Epstein JI and Walsh PC: Biochemical (prostate specific antigen)
recurrence probability following radical prostatectomy for
clinically localized prostate cancer. J Urol. 169:517–523. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lapointe J, Li C, Higgins JP, van de Rijn
M, Bair E, Montgomery K, Ferrari M, Egevad L, Rayford W, Bergerheim
U, et al: Gene expression profiling identifies clinically relevant
subtypes of prostate cancer. Proc Natl Acad Sci USA. 101:811–816.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
D'Amico AV, Moul J, Carroll PR, Sun L,
Lubeck D and Chen MH: Cancer-specific mortality after surgery or
radiation for patients with clinically localized prostate cancer
managed during the prostate-specific antigen era. J Clin Oncol.
21:2163–2172. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ogawa H, Ishiguro K, Gaubatz S, Livingston
DM and Nakatani Y: A complex with chromatin modifiers that occupies
E2F- and Myc-responsive genes in G0 cells. Science. 296:1132–1136.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ishida S, Huang E, Zuzan H, Spang R, Leone
G, West M and Nevins JR: Role for E2F in control of both DNA
replication and mitotic functions as revealed from DNA microarray
analysis. Mol Cell Biol. 21:4684–4699. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ginsberg D: E2F1 pathways to apoptosis.
FEBS Lett. 529:122–125. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Johnson DG: The paradox of E2F1: Oncogene
and tumor suppressor gene. Mol Carcinog. 27:151–157. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Saito M, Helin K, Valentine MB, Griffith
BB, Willman CL, Harlow E and Look AT: Amplification of the E2F1
transcription factor gene in the HEL erythroleukemia cell line.
Genomics. 25:130–138. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Montenegro MF, Collado-González MM,
Fernández-Pérez MP, Hammouda MB, Tolordava L, Gamkrelidze M and
Rodríguez-López JN: Promoting E2F1-mediated apoptosis in oestrogen
receptor-α-negative breast cancer cells. BMC Cancer. 14:5392014.
View Article : Google Scholar
|
|
12
|
Lu M, Liu Z, Yu H, Wang LE, Li G, Sturgis
EM, Johnson DG and Wei Q: Combined effects of E2F1 and E2F2
polymorphisms on risk and early onset of squamous cell carcinoma of
the head and neck. Mol Carcinog. 51(Suppl 1): E132–E141. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rizwani W, Schaal C, Kunigal S, Coppola D
and Chellappan S: Mammalian lysine histone demethylase KDM2A
regulates E2F1-mediated gene transcription in breast cancer cells.
PLoS One. 9:e1008882014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hung JJ, Hsueh CT, Chen KH, Hsu WH and Wu
YC: Clinical significance of E2F1 protein expression in non-small
cell lung cancer. Exp Hematol Oncol. 1:182012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ren Z, Kang W, Wang L, Sun B, Ma J, Zheng
C, Sun J, Tian Z, Yang X and Xiao W: E2F1 renders prostate cancer
cell resistant to ICAM-1 mediated antitumor immunity by NF-κB
modulation. Mol Cancer. 13:842014. View Article : Google Scholar
|
|
16
|
Davis JN, Wojno KJ, Daignault S, Hofer MD,
Kuefer R, Rubin MA and Day ML: Elevated E2F1 inhibits transcription
of the androgen receptor in metastatic hormone-resistant prostate
cancer. Cancer Res. 66:11897–11906. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zheng C, Ren Z, Wang H, Zhang W,
Kalvakolanu DV, Tian Z and Xiao W: E2F1 Induces tumor cell survival
via nuclear factor-kappaB-dependent induction of EGR1 transcription
in prostate cancer cells. Cancer Res. 69:2324–2331. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Libertini SJ, Tepper CG, Guadalupe M, Lu
Y, Asmuth DM and Mudryj M: E2F1 expression in LNCaP prostate cancer
cells deregulates androgen dependent growth, suppresses
differentiation, and enhances apoptosis. Prostate. 66:70–81. 2006.
View Article : Google Scholar
|
|
19
|
Lin ZY, Huang YQ, Zhang YQ, Han ZD, He HC,
Ling XH, Fu X, Dai QS, Cai C, Chen JH, et al: MicroRNA-224 inhibits
progression of human prostate cancer by downregulating TRIB1. Int J
Cancer. 135:541–550. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen G, Liang YX, Zhu JG, Fu X, Chen YF,
Mo RJ, Zhou L, Fu H, Bi XC, He HC, et al: CC chemokine ligand 18
correlates with malignant progression of prostate cancer. Biomed
Res Int. 2014:2301832014.PubMed/NCBI
|
|
21
|
Kwon AT, Arenillas DJ, Worsley Hunt R and
Wasserman WW: oPOSSUM-3: Advanced analysis of regulatory motif
over-representation across genes or ChIP-Seq datasets. G3
(Bethesda). 2:987–1002. 2012. View Article : Google Scholar
|
|
22
|
Farré D, Roset R, Huerta M, Adsuara JE,
Roselló L, Albà MM and Messeguer X: Identification of patterns in
biological sequences at the ALGGEN server: PROMO and MALGEN.
Nucleic Acids Res. 31:3651–3653. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhong WD, Liang YX, Lin SX, Li L, He HC,
Bi XC, Han ZD, Dai QS, Ye YK, Chen QB, et al: Expression of CD147
is associated with prostate cancer progression. Int J Cancer.
130:300–308. 2012. View Article : Google Scholar
|
|
24
|
Li Z, Guo Y, Jiang H, Zhang T, Jin C,
Young CY and Yuan H: Differential regulation of MMPs by E2F1, Sp1
and NF-kappa B controls the small cell lung cancer invasive
phenotype. BMC Cancer. 14:2762014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ma X, Gao Y, Fan Y, Ni D, Zhang Y, Chen W,
Zhang P, Song E, Huang Q, Ai Q, et al: Overexpression of E2F1
promotes tumor malignancy and correlates with TNM stages in clear
cell renal cell carcinoma. PLoS One. 8:e734362013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Engelmann D, Mayoli-Nüssle D, Mayrhofer C,
Fürst K, Alla V, Stoll A, Spitschak A, Abshagen K, Vollmar B, Ran
S, et al: E2F1 promotes angiogenesis through the VEGF-C/VEGFR-3
axis in a feedback loop for cooperative induction of PDGF-B. J Mol
Cell Biol. 5:391–403. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lee J, Park CK, Park JO, Lim T, Park YS,
Lim HY, Lee I, Sohn TS, Noh JH, Heo JS, et al: Impact of E2F-1
expression on clinical outcome of gastric adenocarcinoma patients
with adjuvant chemoradiation therapy. Clin Cancer Res. 14:82–88.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Grupp K, Höhne TS, Prien K, Hube-Magg C,
Tsourlakis MC, Sirma H, Pham T, Heinzer H, Graefen M, Michl U, et
al: Reduced CD147 expression is linked to ERG fusion-positive
prostate cancers but lacks substantial impact on PSA recurrence in
patients treated by radical prostatectomy. Exp Mol Pathol.
95:227–234. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pértega-Gomes N, Vizcaíno JR,
Miranda-Gonçalves V, Pinheiro C, Silva J, Pereira H, Monteiro P,
Henrique RM, Reis RM, Lopes C, et al: Monocarboxylate transporter 4
(MCT4) and CD147 overexpression is associated with poor prognosis
in prostate cancer. BMC Cancer. 11:3122011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhong WD, Han ZD, He HC, Bi XC, Dai QS,
Zhu G, Ye YK, Liang YX, Qin WJ, Zhang Z, et al: CD147, MMP-1, MMP-2
and MMP-9 protein expression as significant prognostic factors in
human prostate cancer. Oncology. 75:230–236. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Han ZD, Bi XC, Qin WJ, He HC, Dai QS, Zou
J, Ye YK, Liang YX, Zeng GH, Chen ZN, et al: CD147 expression
indicates unfavourable prognosis in prostate cancer. Pathol Oncol
Res. 15:369–374. 2009. View Article : Google Scholar
|