Introduction
Lung cancer is one of the common malignant cancers
in China with the highest mortality, constituting 18% of all
cancer-related deaths (1).
According to the American Cancer Society (ACS), it is estimated
that non-small cell lung cancer (NSCLC) accounts for 27% of all
male cancer deaths and 26% of all female cancer deaths in 2016
(2). NSCLC includes lung
adenocarcinoma (LAD), lung squamous carcinoma (LSCLC) and large
cell lung cancer (LCLC) which accounts for ~85% of lung cancer
(3). The development of NSCLC in
cancer surgeries, radiotherapy, and anticancer drugs has become
more rapid recently. However, the 5-year survival rate of the NSCLC
patients still remains low, at 16.6%. It may be caused by the
cancer cell invasion and the presence of metastatic disease that is
not apparent at the time of surgery (4,5).
Therefore, it is urgent to exploit the underlying mechanisms to
identify the altered gene expression during the tumor development
and establishing new biological biomarker with high sensitivity and
specificity to improve the early diagnosis and treatment of
NSCLC.
According to the ENCODE Consortium, 70% of the human
genome has been transcribed for non-coding RNA molecules (6). Non-coding RNAs were previously
considered as transcriptional noise and now they have been proved
to have functional roles in structure, catalysis and regulation.
Based on the transcript size, non-coding RNAs are classified into
two major groups: small non-coding RNAs (sncRNA, <200 bp) and
long non-coding RNAs (lncRNA, >200 bp) (7). MicroRNAs (miRNAs), representatives of
small non-coding RNAs, are a group of short, single-stranded RNA
molecules that have been confirmed to have great significance in
numerous human cancers (8–11). Long non-coding RNAs (lncRNAs) are
evolutionarily conserved non-coding RNAs with low expression level
and no protein-coding capacity (12,13).
Accumulating studies have demonstrated that cellular functions of
lncRNAs include the regulation of cell proliferation and
differentiation or cancer progression and invasion, through which
they may play important roles in multiple cancers (14,15).
The potential regulation pattern of lncRNAs to modulate the
expression of associated genes may be at transcriptional,
post-transcriptional and epigenetic levels (16). In recent years, accumulating
studies have concluded that lncRNA dysregulation played a vital
role in the development and progression of NSCLC (17,18).
Moreover, researchers have found that lncRNAs can be stably
detected in plasma or even urine for earlier diagnosis (19–22).
Due to the limited sensitivity and specificity of traditional tumor
biomarkers for early stage NSCLC such as carcinoembryonie antigen
(CEA), squamous cell carcinoma (SCC), neuron specific enolase (NSE)
and cytokeratin 19 fragments (CA211) (23,24),
it becomes essential to screen for other alternative molecules
functioning as diagnostic, prognostic and predictive biomarkers for
NSCLC.
The lncRNA growth arrest-specific transcript 5
(GAS5) has been reported to be associated with proliferation and
apoptosis of NSCLC (25), implying
that it may serve as a new biomarker for NSCLC patient diagnosis.
Our study here mainly focused on the circulating lncRNA expression
in NSCLC patient tissues and plasma to further verify its
diagnostic and prognostic value in NSCLC. The results showed that
the expression levels of GAS5 were significantly downregulated in
NSCLC tissues and the lower expression level has correlated with
tumor size of NSCLC patients. We further ascertained its
contribution in plasma and found that GAS5 was differentially
expressed in early NSCLC plasma and the levels changed with
surgery. Moreover, its expression level was negatively linked to
CEA and CA199. These data might imply that circulating GAS5 can not
only serve as a diagnosis biomarker for earlier NSCLC, but is also
valuable in assessment for the postoperative condition of
NSCLC.
Materials and methods
Tissue samples
Eighty NSCLC tissues and their morphologically
normal tissues (located >3 cm away from the tumor) utilized in
this study were obtained from patients with NSCLC (59 men and 21
women; mean age, 61±15) who underwent surgical resection without
any therapy in Zhongnan Hospital of Wuhan University between
October 2014 and January 2016. All fresh tissue specimens were
snap-frozen in liquid nitrogen and stored at −80°C until total RNA
extraction.
Plasma samples
Whole blood samples of 111 patients (86 men and 25
women; mean age 52±10) with NSCLC were recruited before surgery
during the same period, including 55 patients whose tissue samples
had been collected. Moreover, 57 paired postoperative blood samples
were obtained ~1 month after surgery. Additionally, 78 healthy
controls (52 men and 26 women; mean age, 55±10) were randomly
collected from the Medical Examination Center at the same stage
without any cancers and other health problems. All plasma samples
were isolated from peripheral blood specimens in ethylene diamine
tetraacetic acid (EDTA) anti-coagulation tubes followed with a
two-step centrifugation protocol to fully spin down the blood cells
(2000 g for 5 min at 4°C, 12,000 g for 5 min at 4°C). Then, they
were stored at −80°C until use.
Data collection
Clinical characteristics of these patients including
sex, age, smoking history, tumor traditional biomarker levels (CEA,
NSE, SCC, CA211, CA199, CA125), tumor size, lymph node metastasis
and histological grade were collected (Tables I and II). The basic information such as
gender, age, CEA, CA199 and CA125 of healthy controls was gathered
from LIS of hospital clinical laboratory for further data
analysis.
 | Table ICorrelation clinicopathological
factors and GAS5 level (2−ΔCT) in NSCLC patients. |
Table I
Correlation clinicopathological
factors and GAS5 level (2−ΔCT) in NSCLC patients.
| Variable | Nos. | GAS5-lowa | GAS-high | P-valueb |
|---|
| Gender | | | | 0.204 |
| Male | 59 | 32 | 27 | |
| Female | 21 | 8 | 13 | |
| Age (years) | | | | 0.740 |
| ≤60 | 41 | 19 | 22 | |
| >60 | 39 | 21 | 18 | |
| Smoking
(years) | | | | 0.075 |
| Never | 24 | 9 | 15 | |
| Ever | 56 | 31 | 25 | |
| Tumor size
(cm) | | | |
0.026c |
| ≤3 | 21 | 7 | 14 | |
| >3 | 59 | 33 | 26 | |
| Histological | | | | 0.109 |
| I | 38 | 16 | 22 | |
| II | 13 | 5 | 8 | |
| III | 29 | 19 | 10 | |
| T-stage | | | | 0.319 |
| T1 | 20 | 7 | 13 | |
| T2 | 39 | 20 | 19 | |
| T3 | 11 | 6 | 5 | |
| T4 | 10 | 7 | 3 | |
| Regional lymph
nodes | | | | 0.592 |
| N0 | 50 | 23 | 27 | |
| N1 | 8 | 4 | 4 | |
| N2 | 22 | 13 | 9 | |
 | Table IIThe plasma GAS5 level alteration
among stages of NSCLC. |
Table II
The plasma GAS5 level alteration
among stages of NSCLC.
| Group | −ΔΔCt | Percentage
(%)a |
|---|
| Stage I vs. stage
II | 0.06697 | 4.536 |
| Stage I vs. stage
III | 0.48683 | 28.64 |
| Stage II vs. stage
III | 0.41986 | 25.25 |
| Stage I+II vs.
stage III | 0.46557 | 27.58 |
| Stage I vs. stage
II +III | 0.32546 | 19.64 |
Cell lines and culture
Four NSCLC cell lines (including NSCLC cell lines
H1299, 95-D and ADC cell lines A549, SPC-A-1) and human embryonic
lung fibroblast (HELF) were kindly provided by Public Health of
Wuhan University. All cell lines were cultured in DMEM medium
(Hyclone, China) added with 10% (v/v) fetal bovine serum, 100 IU/ml
penicillin and streptomycin (100 U/ml), then they were incubated at
37°C in a humidified chamber with 5% (v/v) CO2. The
cellular RNA was extracted when all the cells were at the best
state and repeated three times.
Ethics approval
All the patients signed the informed consent form.
The use of the tissues and plasma samples for the experiment was
approved by the Ethics Committee of Zhongnan Hospital of Wuhan
University (Wuhan, China).
Total RNA isolation, RT and QRT-PCR
For tissues and cell lines, the total RNA was
extracted using TRIzol reagent (Invitrogen, CA, USA) according to
the manufacturer's instructions. For plasma samples, it was
extracted from 300 µl cell-free plasma using blood/liquid
sample Total RNA Rapid Extraction kit (spin-column) (Bioteke,
Beijing, China) and eluted with 30 µl RNase-free water
eventually. All the RNA was then transcribed to cDNA at once.
The QRT-PCR assay was conducted to detect the level
of RNA transcripts. For reverse transcription (RT) reaction, the
reaction mixture (20 µl) for tissues and cell lines
containing 1 µg total RNA was reversely transcribed to cDNA
by using PrimeScript RT reagent kit with gDNA Eraser (Takara,
Dalian, China), while 7 µl plasma RNA was used in the RT
reaction (20 µl), followed the protocol of 42°C for 2 min,
then 37°C for 15 min and 85°C for 5 sec. Then all cDNA samples were
detected immediately by QRT-PCR using the SYBR®Premix Ex
Taq™ II real-time PCR kit (Takara) in a 20-µl reaction
volume, which contained 10 µl of SYBR-Green master PCR mix,
0.8 µl each of forward and reverse primers, 2 µl of
diluted cDNA template, and appropriate amounts of sterile distilled
water. The cycling conditions were initial denaturation at 95°C for
5 min; 40 cycles of denaturation at 95°C for 30 sec, annealing at
62.5°C for 30 sec, and elongation at 72°C for 30 sec. Given that
the level of glyceraldehyde-phosphate dehydrogenase (GAPDH) mRNA
was found to be relatively stable in plasma (26,27),
we chose GAPDH as the endogenous control for data normalization to
determine the relative expression of the target genes. The sequence
of PCR primers are as follows: GAS5, forward,
5′-GTGTGGCTCTGGATAGCAC-3′; reverse, 5′-ACCCAAGCAAGTCATCCATG-3′.
GAPDH, forward, 5′-GGTCTCCTCTGACTTCAACA-3′; reverse,
5′-GTGAGGGTCTCTCTCTTCCT-3′. The reaction conditions were set
according to the kit instructions. After completion of the
reaction, the amplification curve and melting curve were analyzed
to measure the specificity of the amplified products.
Statistical analysis
The relative expression of GAS5 was calculated using
the comparative cycle threshold (Ct) method (2−ΔCt).
Samples with a Ct >40 were considered as negative. All
statistical analyses were performed using the Statistical Product
and Service Solutions (SPSS) 17.0 software (SPSS, Chicago, IL,
USA). Data analysis was performed for pairs of tissues and plasma
samples by using paired-sample t-test. The analysis between NSCLC
plasma and controls was applied by using independent two-tailed
t-test. Two-sided Chi-square test and one-way ANOVA were used to
assess the association between the expression level and
clinicopathological factors. A receiver operating characteristic
(ROC) curve was applied to evaluate the diagnostic value. P<0.05
was considered statistically significant.
Results
Cloning and sequencing of qRT-PCR
products
The qRT-PCR products of GAS5 and GAPDH were purified
using the AxyPrep DNA Gel Extraction kit (Axygen, USA) and then
were cloned into the PMD 19-T vector (Takara). The melting curve
analysis (Fig. 1) and their PCR
products sequence results were available as expected, which were
completely consistent with the entries from the NCBI database
(Fig. 2).
GAS5 expression level in NSCLC tissues
and cell lines
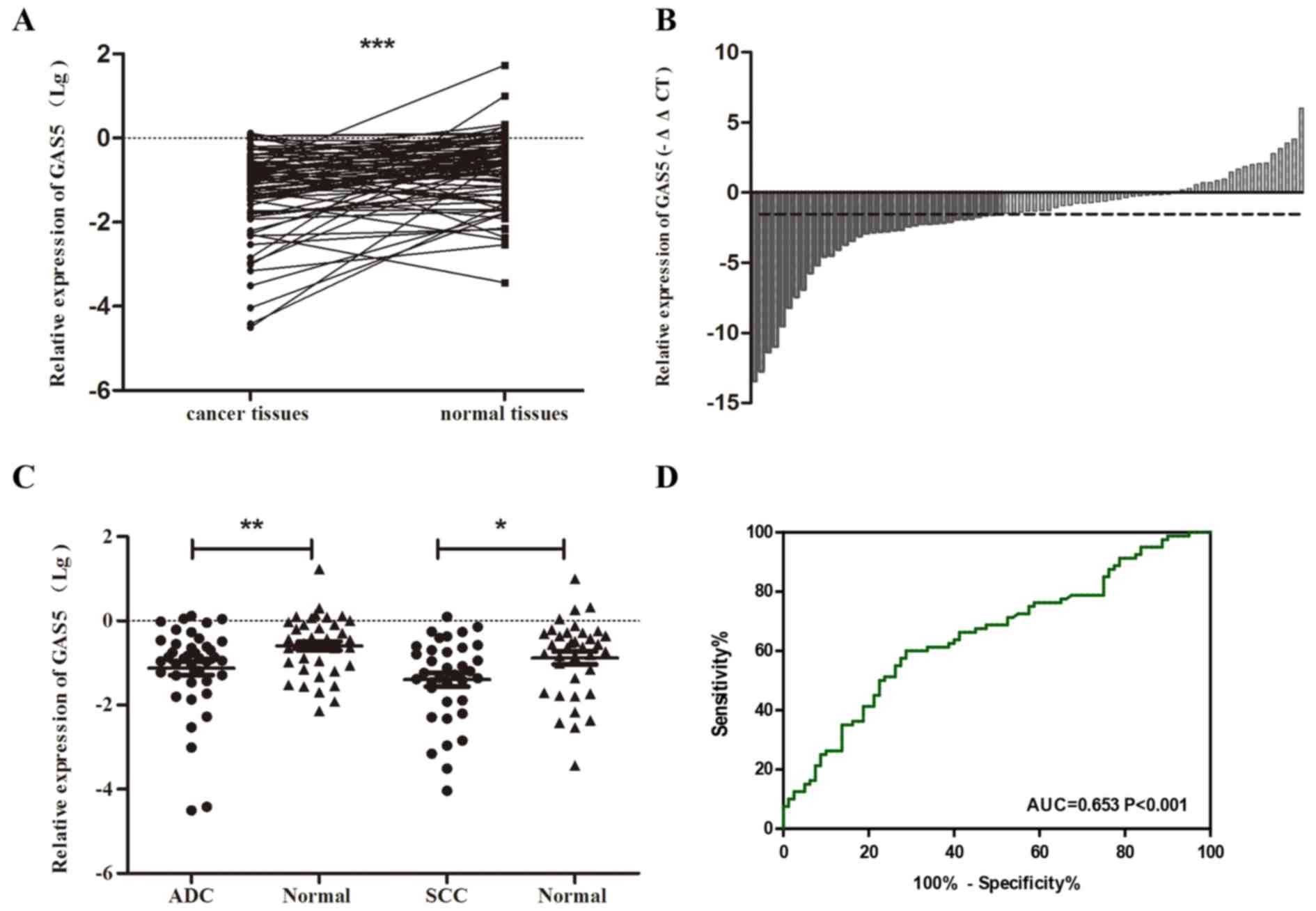
To further validate the importance of GAS5 in NSCLC,
its expression level was detected in 80 pairs of NSCLC tissue
samples (including 40 ADC, 36 SCC and 4 LSLC) and corresponding
normal tissues by using quantitative real-time PCR. The results
showed that GAS5 was distinctly decreased in NSCLC tissues
(P<0.001) (Fig. 3A). Waterfall
plot further demonstrated that GAS5 was reduced ≥3-fold in 46.3%
(37/80) of the NSCLC patients (Fig.
3B). Data analysis showed the GAS5 expression level presented a
similar trend and declined both in ADC and SCC (P<0.05)
(Fig. 3C). In addition, the ROC
curve illustrated strong separation between the tumor tissues and
control group, with an area under curve (AUC) of 0.653 (95% CI,
0.568–0.738; P<0.001) (Fig.
3D).
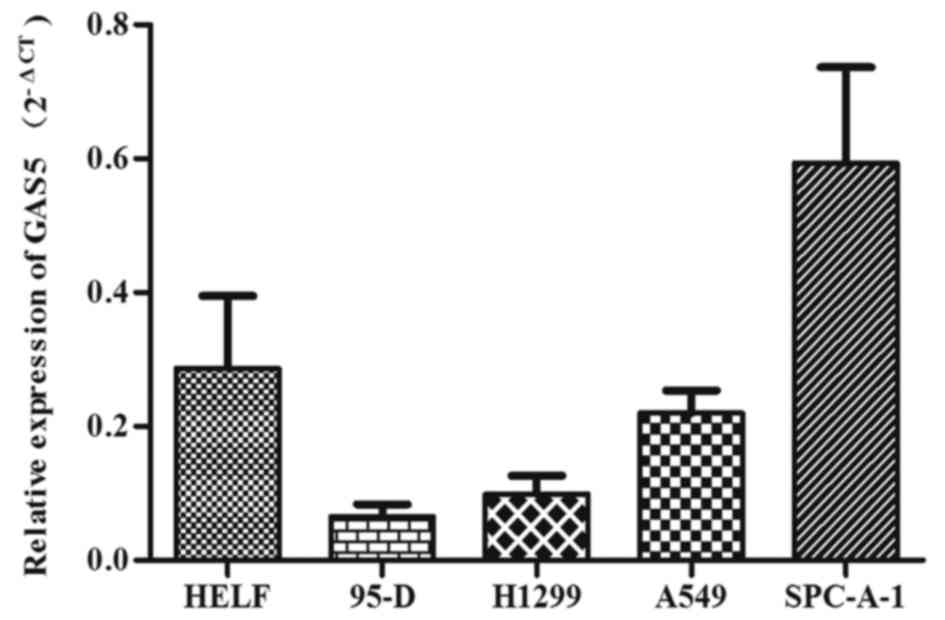
We further explored its relative expression in four
NSCLC cell lines (H1299, 95-D, A549, SPC-A-1) and human embryonic
lung fibroblast (HELF) by QRT-PCR, and results indicated a lower
expression of GAS5 in H1299, 95-D, A549 cell lines compared with
HELF cell line except for SPC-A-1 cell line (Fig. 4). Among the three NSCLC cell lines,
GAS5 decreased the most in 95-D and H1299 cell lines.
The correlation of GAS5 expression level in tissues
with clinicopathological features was assessed as shown in Table I. The low expression level of GAS5
was associated with tumor size of NSCLC patients (P<0.05).
However, no other significant differences were found between the
expression level and the clinical characteristics of the patients
including age, gender, smoking status, TNM stage, advanced T-stage
and regional lymph node metastasis (P>0.05). Presumably, these
results suggested that the downregulation of GAS5 might have
bearing on the disease status of human NSCLC.
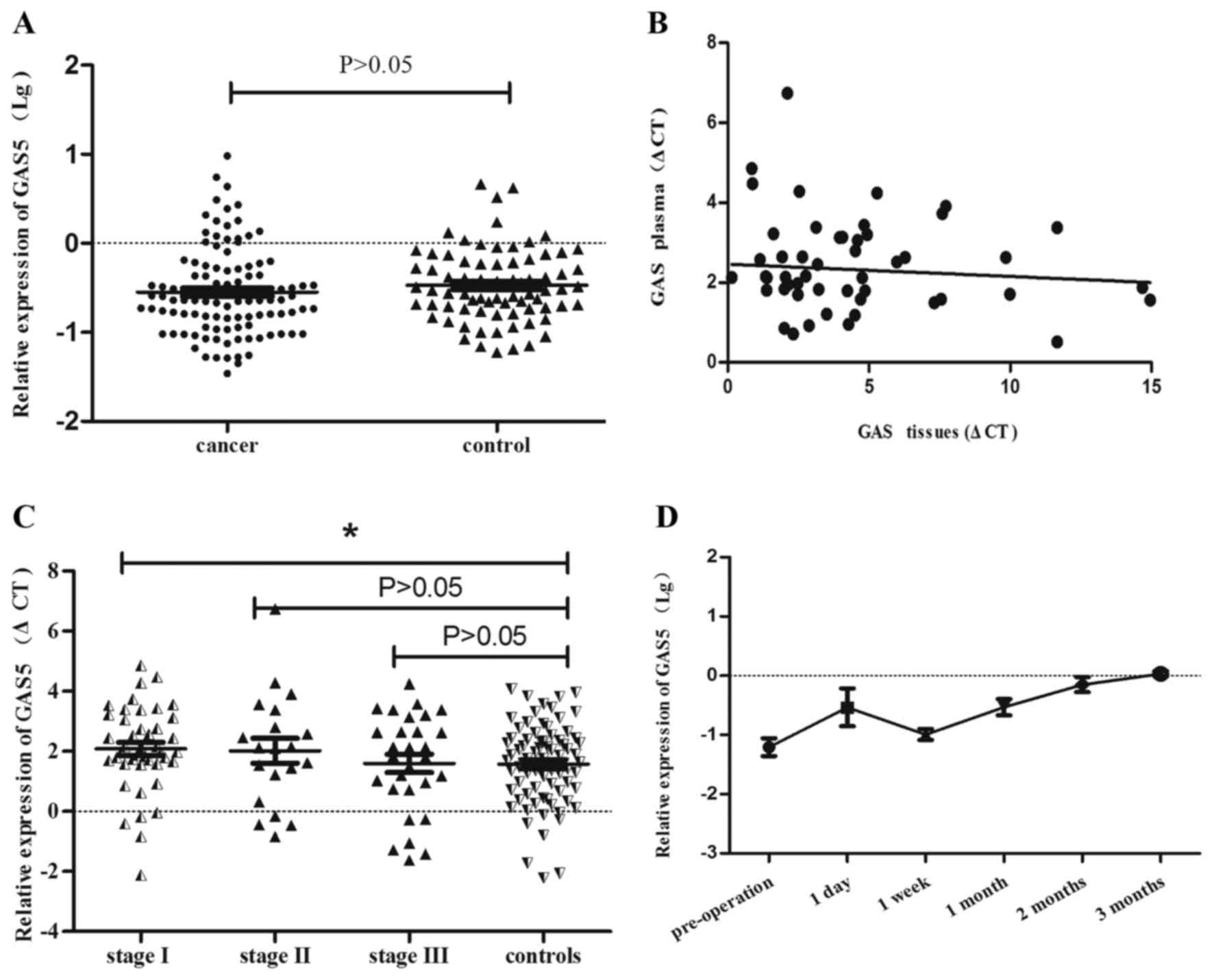
GAS5 expression level of plasma in NLCLC
patients
To further validate the contribution of GAS5 in
plasma of NSCLC patients, we compared the plasma GAS5 level between
111 NSCLC patients and 78 healthy controls using QRT-PCR.
Unexpectedly, no significant statistical differences were found
between NSCLC patients and controls (P>0.05) (Fig. 5A). We further assessed whether
there was a relationship of GAS5 expression between plasma and
cancer tissues by using liner correlation regression. As shown in
Fig. 5B, there was a weak
connection between 55 paired EDTA plasma samples and cancer tissues
obtained from the same individuals with NSCLC (P>0.05). However,
further data analysis showed that GAS5 expression was statistically
decreased by 29.94% in the group of stage I compared to controls
(P<0.05), while there was no statistical difference among other
stages of NSCLC and control group (P>0.05) (Fig. 5C, Tables II and III).
 | Table IIIThe plasma GAS5 level alteration
among stages of NSCLC and control group. |
Table III
The plasma GAS5 level alteration
among stages of NSCLC and control group.
| Group | −ΔΔCt | Percentage
(%)a |
|---|
| Stage I vs.
Cb | 0.51332 | 29.94c |
| Stage II vs. C | 0.44632 | 26.61 |
| Stage III vs.
C | 0.02648 | 1.819 |
| Stage I+II vs.
C | 0.49205 | 28.90 |
| Stage II+III vs.
C | 0.19785 | 12.82 |
Expression alteration of GAS5 after
operation
We collected blood samples of 8 patients before
surgery and the corresponding blood specimens after resection
according to their post-operation time. Under the same treatment,
the GAS5 expression level for each plasma sample was analyzed by
QRT-PCR (Fig. 5D). The variation
of GAS5 expression was slightly increased after one day of
resection (P=0.1033), there was an abrupt decline after 7 days of
operation (P=0.2436). However, as seen from the figure along with
the changes of operation time, the GAS5 expression showed an
obvious elevated trend starting from the seventh day. There was a
statistical difference between pre-operation and post-operation
plasma after 1 month (P=0.0124) and 2 months P=0.001).
Surgical assessment of GAS5 in plasma of
NSCLC patients
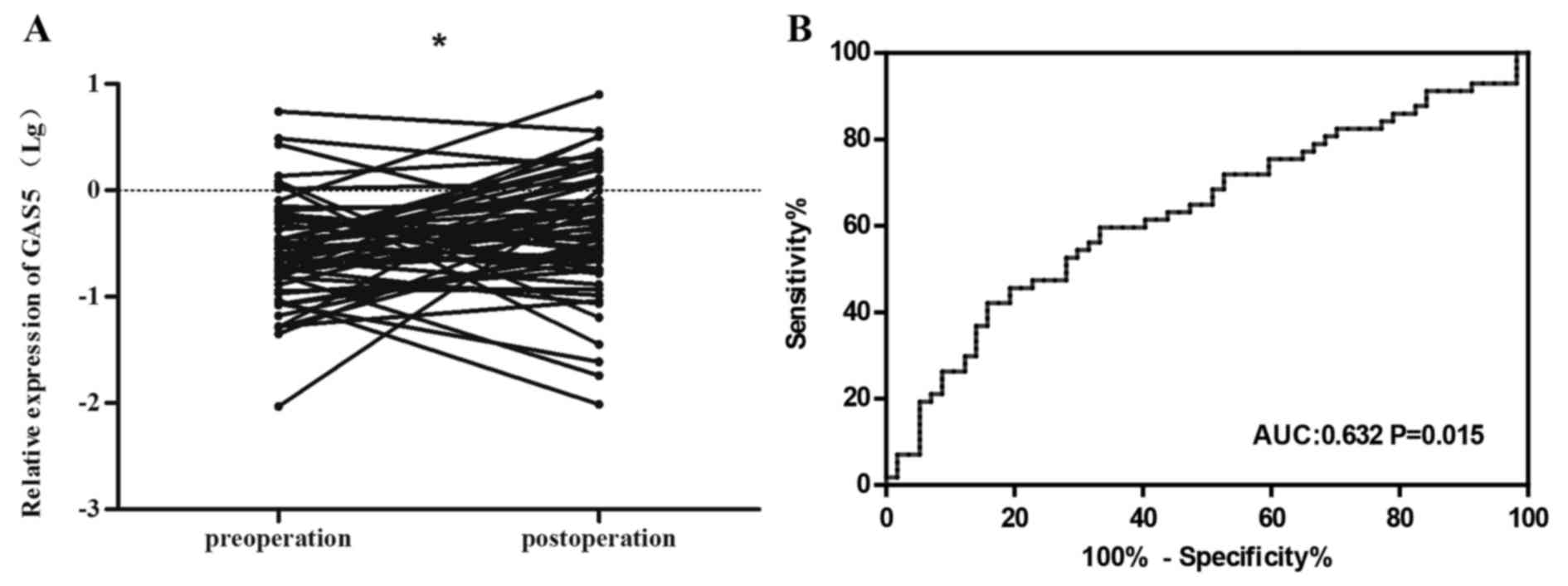
Our study also investigated the difference in
expression levels between 57 paired plasma specimens (pre-operation
and post-operation samples). We found that plasma levels of
circulating RNA GAS5 was significantly increased after surgical
treatment compared to pre-operation (P<0.05) (Fig. 6A). The result suggested that
circulating GAS5 in plasma could be acceptable for evaluation of
NSCLC surgical resection. Further analysis of the ROC curve for
post-operation was shown in Fig.
6B, with an area under curve (AUC) of 0.632 (95% CI,
0.529–0.735; P=0.015).
Association of clinicopathological
factors and serological indicators with GAS5 expression level
To affirm the relationship of plasma GAS5 levels
with clinical parameters, NSCLC patients' pathologic information
including age, gender, smoking history, tumor size, TNM stage,
lymphatic metastasis, differentiation, and histological
classification were collected and analyzed by one-way ANOVA. As
shown in Table IV, the GAS5
expression levels showed huge variability between ADC and SCC
plasma samples (P<0.05), no differences were found among the
other characteristics.
 | Table IVCorrelation of plasma GAS5 levels
(ΔCt) among clinical parameters of NSCLC patients. |
Table IV
Correlation of plasma GAS5 levels
(ΔCt) among clinical parameters of NSCLC patients.
| Clinical
characteristic | No. of a cases (%) | Mean ± SEMb | P-valuec |
|---|
| Age (years) | | | 0.116 |
| ≤60 | 40 (36.0) | 2.21±0.29 | |
| >60 | 71 (64.0) | 1.69±0.18 | |
| Gender | | | 0.381 |
| Female | 25 (22.5) | 1.61±0.42 | |
| Male | 86 (77.5) | 1.94±0.17 | |
| Smoking
history | | | 0.520 |
| Never | 31 (28.4) | 1.70±0.35 | |
| Present | 78 (71.6) | 1.93±0.18 | |
| Histological
classification | | |
0.017d |
| ADC | 51 (48.1) | 1.41±0.21 | |
| SCC | 55 (51.9) | 2.18±0.24 | |
| Tumor size
(cm) | | | 0.920 |
| ≤3 | 22 (23.7) | 1.87±0.32 | |
| >3 | 71 (76.3) | 1.91±0.20 | |
| T stage | | | 0.545 |
| T1 | 22 (23.7) | 1.87±0.32 | |
| T2 | 48 (51.6) | 2.08±0.23 | |
| T3 | 16 (17.2) | 1.59±0.45 | |
| T4 | 7 (7.5) | 1.49±0.64 | |
| N stage | | | 0.457 |
| N0 | 61 (65.6) | 2.01±0.20 | |
| N1 | 8 (8.6) | 2.16±0.86 | |
| N2 | 24 (25.8) | 1.55±0.31 | |
| TNM stage | | | 0.344 |
| I | 44 (47.3) | 2.08±0.22 | |
| II | 20 (21.5) | 2.02±0.42 | |
| III | 29 (31.2) | 1.60±0.30 | |
|
Differentiation | | | 0.207 |
| High | 7 (8.0) | 1.06±0.56 | |
| High-moderate | 5 (5.7) | 2.21±0.73 | |
| Moderate | 47 (53.4) | 1.79±0.24 | |
| Moderate-low | 11 (12.5) | 1.19±0.49 | |
| Low | 18 (20.4) | 2.43±0.36 | |
| Ki67 proliferation
index (%) | | | 0.646 |
| ≤50 | 34 (50.0) | 1.94±0.24 | |
| >50 | 34 (50.0) | 1.77±0.28 | |
| EGFR | | | 0.084 |
| Normal | 16 (50.0) | 2.06±0.31 | |
| Abnormal | 16 (50.0) | 1.09±0.45 | |
Then, to explore whether there is a connection
between the patient serum index and GAS5 levels, traditional tumor
makers including CEA, NSE, SCC, CA211, CA199 and CA125 were
classified into normal and abnormal groups based on the criteria of
diagnosis in our hospital (Table
V). There were no differences among SCC, CA211 and NSE, but the
GAS5 expression level had a statistical relationship both in CEA
and CA199 groups (P<0.05 and P<0.01, respectively).
 | Table VRelationship of plasma GAS5 levels
(ΔCt) among serum index of NSCLC patients. |
Table V
Relationship of plasma GAS5 levels
(ΔCt) among serum index of NSCLC patients.
| Factors | No. of cases
(%)a | Mean ± SEMb | P-value |
|---|
| CEA
(µg/l) | | |
0.021c |
| ≤5.0 | 68 (73.9) | 2.19±0.18 | |
| >5.0 | 24 (26.1) | 1.27±0.39 | |
| NSE (ng/ml) | | | 0.934 |
| ≤18.3 | 72 (92.3) | 1.83±0.20 | |
| >18.3 | 6 (7.7) | 1.89±1.00 | |
| CA211 (ng/ml) | | | 0.657 |
| ≤3.3 | 14 (70.0) | 2.38±0.31 | |
| >3.3 | 6 (30.0) | 2.10±0.61 | |
| SCC
(µg/l) | | | 0.841 |
| ≤1.5 | 16 (84.2) | 2.26±0.35 | |
| >1.5 | 3 (15.8) | 2.42±0.14 | |
| CA125 (U/ml) | | | 0.096 |
| ≤35 | 79 (83.1) | 2.08±0.18 | |
| >35 | 16 (16.9) | 1.32±0.46 | |
| CA199 (U/ml) | | |
0.0008c |
| ≤37 | 82 (86.3) | 2.18±0.17 | |
| >37 | 13 (13.7) | 0.54±0.51 | |
Diagnostic potential of circulating GAS5
for early NSCLC
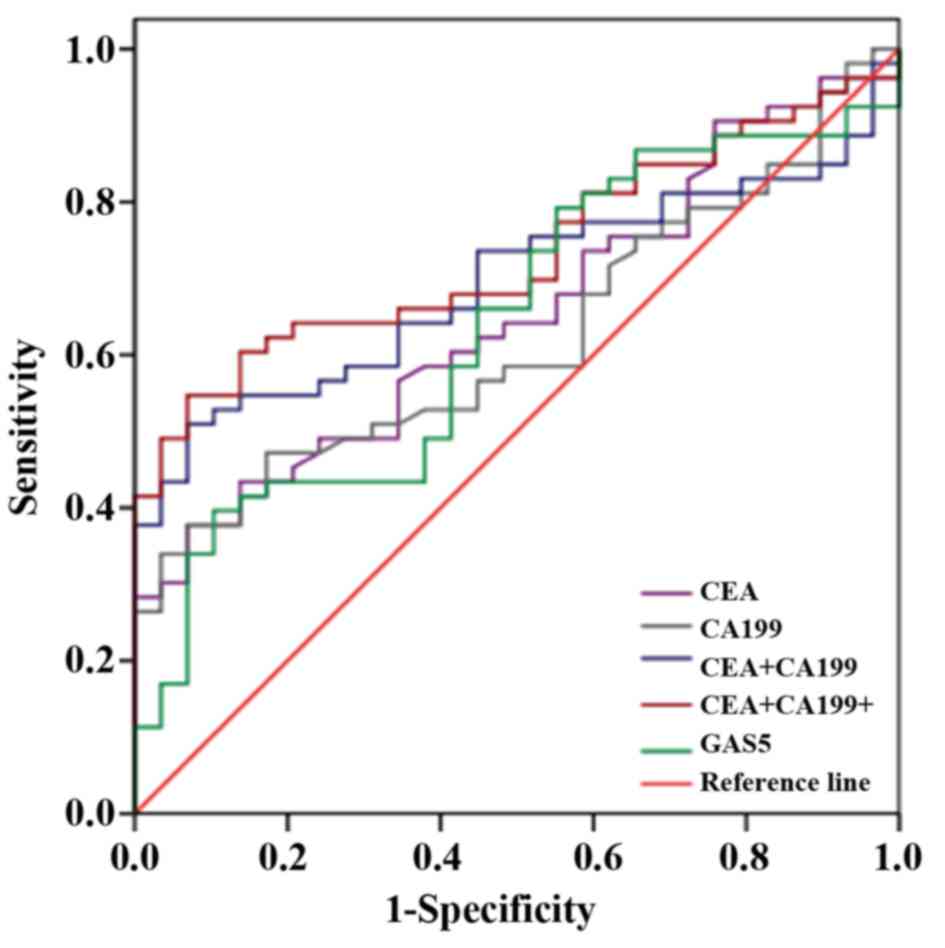
Based on our results, we further analyzed the ROC
curves of CEA, CA199 and GAS5 in early stage of NSCLC compared with
controls, and we found that the combination of the three indicators
possessed higher sensitivity and specificity when they were applied
alone as shown in Fig. 7 and
Table VI. The sensitivity and
specificity were ≤86.76% and 62.62%. Hence, circulating GAS5 might
serve as a combined diagnostic indicator for early diagnosis of
NSCLC.
 | Table VIAnalysis of ROC curves within
groups. |
Table VI
Analysis of ROC curves within
groups.
| Group | AUC | 95% CI | P-value | % Se | % Sp |
|---|
| CEA | 0.651 | 0.534–0.768 | 0.024 | 21.95 | 90.57 |
| CA199 | 0.622 | 0.503–0.742 | 0.068 | 6.897 | 98.11 |
| GAS5 | 0.638 | 0.515–0.760 | 0.223 | 42.31 | 77.78 |
| CEA+CA199 | 0.694 | 0.582–0.805 | 0.004 | 55.17 | 73.58 |
| CEA+CA199+GAS5 | 0.734 | 0.628–0.839 | <0.0005 | 86.76 | 62.62 |
Taken together, we have demonstrated that the lncRNA
GAS5 level was dysregulated in NSCLC patients in the present study.
Circulating GAS5 can potentially be useful for monitoring surgical
treatment in NSCLC patients and has a potential as a combined
biomarker for NSCLC screening. It provides a useful auxiliary
diagnostic method to screen for a large number of candidate
patients and increase our understanding of NSCLC pathology.
Discussion
It is well-known, early diagnosis is the main way to
improve the survival rate of lung cancer patients, whereas it is
still difficult at present. Single serological index including CEA,
SCC, CYFRA 21-1 and NSE for the diagnostic sensitivity and
specificity of lung cancer is often unsatisfactory, especially for
early clinical stages (28).
The lncRNAs have recently become a research focus
worldwide, as a group of RNA molecules that are longer than 200 nt
without protein-coding capacity (29). Increasing studies have revealed
that lncRNAs play fundamental roles in carcinomas such as breast
cancer (30), cervical cancer
(31) and gastric cancer (32), or diseases such as neurological
disorders (33), kidney and
cardiovascular diseases (34). Due
to the potential detection of lncRNAs in plasma or even urine of
cancer patients, they could be applied as biomarkers for the
earlier diagnosis of cancers. For example, dysregulated long
non-coding RNA prostate cancer antigen 3 (PCA3) promotes prostate
cancer cell proliferation possibly through the AR signal pathway.
Also, urine PCA3 can be used as a non-invasive method for early
diagnosis of prostate cancer (35,36).
Another study showed that plasma MALAT-1 was correlated with liver
damage and combination of MALAT1, AFP and PIVKAII can improve the
diagnostic value of HCC (37).
Moreover, according to the study of Li et al, linc00152
could be detected in plasma of GC patients as a result of exosome
protection (38). These findings
further confirmed that lncRNAs can serve as molecule biomarkers for
cancer diagnosis and prognosis.
Our target gene GAS5 was originally isolated from a
subtraction cDNA library and was expressed relatively higher in
growth arrest cells (39). It is
located at 1q25.1 containing 12 exons with only a short open
reading-frame that cannot encode a functional protein. The high
expression abundance of GAS5 may be explained by the interaction
between rapamycin (mTOR) pathway (40) and the nonsense-mediated decay (NMD)
pathway (41). Plenty of
experiments have demonstrated that overexpression of GAS5 can
induce apoptosis and reduce the rate of progression in S phase
(42). Because of the existence of
apoptosis in the physiological and pathological processes, the
molecular mechanisms of GAS5 in various cancers was uncovered in
different aspects. For instance, downregulated GAS5 in NSCLC
tissues is able to promote cell proliferation and reduce apoptosis
(25).
Why circulating lncRNAs could be detected in plasma
may be that they are packaged in some microparticles such as
exomes, microvesicles and apoptotic bodies (27). Long non-coding RNAs H19, HOTAIR and
MALAT1 were reported to stably exist in plasma (43). A recent study of our laboratory
showed that the detection level of GAS5 and GAPDH did not change
much when the whole blood samples were at 4°C for 24 h before
plasma pheresis. Also, plasma specimens can be stored at −80°C for
at least one month. Moreover, they are relatively stable when the
plasma samples were treated with freeze-thaw cycles (44). These findings agree with our
results to a certain degree.
How the circulating lncRNAs in blood are generated
and the mechanism for them to provide long-term circulation in the
bloodstream remain unclear. The most popular explanation is that,
as a kind of circulating nucleic acid (CNA), it may stem from the
self-secretion, cracking apoptosis and necrosis of tumor cells
(45,46). Under the speculation, our results
showed plasma GAS5 levels were significantly downregulated in early
stage of NSCLC patients compared with healthy controls
(P<0.0493), while tissue GAS5 levels were downregulated in
advanced stage, others studying other lncRNAs in gastric cancer
(GC) have found similar results. Shao et al demonstrated
that tissue lncRNA AA174084 levels were significantly downregulated
in both early and advanced stage of GC patients. However, the
plasma lncRNA AA174084 levels were downregulated in early stage of
GC compared to advanced stage. Also, they mentioned that the
increase of gastric juice AA174084 level in patients who had GC
(especially those who had early GC) did not indicate that the
AA174084 level in gastric juice will decrease along with the
increasing severity of GC (27).
Yang et al also found that tissue lncRNA
ABHD11-AS1 levels were upregulated in GC, while gastric
juice lncRNA ABHD11-AS1 levels were only significantly
upregulated in early gastric cancer (EGC) (47). All the results indicated that the
correlation of lncRNA levels between tissues and plasma might be
affected by other reasons. However, Chen and colleagues mentioned
that CNAs may be also correlated tightly with the immune response,
and it was not just a result of cancer but may actively contribute
to cancer defense (48). The
strong separation between LAD and SCC patient plasma (P=0.017)
could be explained by the difference mechanism of CNAs releasing to
bloodstream. However, further research is needed to explain this
phenomenon.
Many studies have presented that the expression
alteration of other lncRNAs of cancer patients who underwent
surgery could reflect surgical assessment and tumor dynamics. Han
et al discovered that lower plasma GAS5 level was observed
in breast cancer patients with a high Ki67 proliferation index
before surgery and patients with a positive lymph node metastasis
state after surgery (44). Yang
et al found that circulating H19 levels declined markedly in
gastric cancer patients after surgery and that lncRNAs may be
protected by exosomes (49). Zhang
et al found that the downregulation of circulating H19
levels in breast cancer patients after surgery can monitor the
patients condition (50). In our
study, the exploration of the alteration for GAS5 level over time
after surgery better confirmed our hypothesis that surgical
resection had important effects on GAS5 expression level. Our data
showed plasma GAS5 statistically elevated in postoperative NSCLC
patients after resection (P=0.026), indicating that lncRNA GAS5
might have the potential to assess the surgical effects for NSCLC
patient. Thus, the operation may stimulate cells to release an
amount of lncRNA to the circulation.
There commonly exist several EGFR mutations (~63.1%)
in Chinese NSCLC patients in the locations of 18, 19 and 21 exons.
It has been reported that EGFR mutation can lead to drug resistance
and poor prognosis of NSCLC patients (51,52).
Recent studies explored a series of lncRNAs dysregulated in EGFR
exon deletion in lung adenocarcinoma that played critical roles by
regulating neighboring molecules associated with EGFR-TKIs
resistance (53,54). Our result found that a low level of
plasma GAS5 has a tendency for patients with EGFR mutation
(P=0.084), which was consistent with the outcomes in EGFR-TKI
sensitive and resistant cell lines discovered by Dong et al
(55).
We further found that GAS5 level has tight
relationship with CEA and CA199 (P=0.017 and P=0.001 respectively).
Further analysis by ROC curves showed that the combination of
circulating GAS5 levels with CEA and CA199 is considered to be more
advantageous for the diagnosis of NSCLC (Table IV).
However, there are some research limitations in our
study, including insufficient sample size for advanced stage, the
missing data of some patients, failure in further mechanism
research. Therefore, more studies should be carried out for the
biological role of circulating lncRNAs.
Acknowledgments
This study was supported by the National Basic
Research Program of China (973 Program) (2012CB720605).
References
|
1
|
Haaland B, Tan PS, de Castro G Jr and
Lopes G: Meta-analysis of first-line therapies in advanced
non-small-cell lung cancer harboring EGFR-activating mutations. J
Thorac Oncol. 9:805–811. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ettinger DS, Akerley W, Bepler G, Blum MG,
Chang A, Cheney RT, Chirieac LR, D'Amico TA, Demmy TL, Ganti AK, et
al NCCN Non-Small Cell Lung Cancer Panel Members: Non-small cell
lung cancer. J Natl Compr Canc Netw. 8:740–801. 2010.PubMed/NCBI
|
|
4
|
Wu Y, Liu H, Shi X, Yao Y, Yang W and Song
Y: The long non-coding RNA HNF1A-AS1 regulates proliferation and
metastasis in lung adenocarcinoma. Oncotarget. 6:9160–9172. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Koudelakova V, Kneblova M, Trojanec R,
Drabek J and Hajduch M: Non-small cell lung cancer - genetic
predictors. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub.
157:125–136. 2013.PubMed/NCBI
|
|
6
|
Han Li C and Chen Y: Small and long
non-coding RNAs: Novel targets in perspective cancer therapy. Curr
Genomics. 16:319–326. 2015. View Article : Google Scholar
|
|
7
|
Zhang A, Zhang J, Kaipainen A, Lucas JM
and Yang H: Long non-coding RNA: A newly deciphered 'code' in
prostate cancer. Cancer Lett. 375:323–330. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jin Z, Guan L, Song Y, Xiang GM, Chen SX
and Gao B: MicroRNA-138 regulates chemoresistance in human
non-small cell lung cancer via epithelial mesenchymal transition.
Eur Rev Med Pharmacol Sci. 20:1080–1086. 2016.PubMed/NCBI
|
|
10
|
Zhu K, Ding H, Wang W, Liao Z, Fu Z, Hong
Y, Zhou Y, Zhang CY and Chen X: Tumor-suppressive miR-218-5p
inhibits cancer cell proliferation and migration via EGFR in
non-small cell lung cancer. Oncotarget. 7:28075–28085.
2016.PubMed/NCBI
|
|
11
|
Liu H: MicroRNAs in breast cancer
initiation and progression. Cell Mol Life Sci. 69:3587–3599. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Schmitz SU, Grote P and Herrmann BG:
Mechanisms of long noncoding RNA function in development and
disease. Cell Mol Life Sci. 73:2491–2509. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tantai J, Hu D, Yang Y and Geng J:
Combined identification of long non-coding RNA XIST and HIF1A-AS1
in serum as an effective screening for non-small cell lung cancer.
Int J Clin Exp Pathol. 8:7887–7895. 2015.PubMed/NCBI
|
|
14
|
Esteller M: Non-coding RNAs in human
disease. Nat Rev Genet. 12:861–874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gibb EA, Brown CJ and Lam WL: The
functional role of long non-coding RNA in human carcinomas. Mol
Cancer. 10:382011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kaplan R, Luettich K, Heguy A, Hackett NR,
Harvey BG and Crystal RG: Monoallelic up-regulation of the
imprinted H19 gene in airway epithelium of phenotypically normal
cigarette smokers. Cancer Res. 63:1475–1482. 2003.PubMed/NCBI
|
|
17
|
Ricciuti B, Mencaroni C, Paglialunga L,
Paciullo F, Crinò L, Chiari R and Metro G: Long noncoding RNAs: New
insights into non-small cell lung cancer biology, diagnosis and
therapy. Med Oncol. 33:182016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Prensner JR and Chinnaiyan AM: The
emergence of lncRNAs in cancer biology. Cancer Discov. 1:391–407.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jin C, Peng X, Xie T, Lu X, Liu F, Wu H,
Yang Z, Wang J, Cheng L and Wu N: Detection of the long noncoding
RNAs nuclear-enriched autosomal transcript 1 (NEAT1) and metastasis
associated lung adenocarcinoma transcript 1 in the peripheral blood
of HIV-1-infected patients. HIV Med. 17:68–72. 2016. View Article : Google Scholar
|
|
20
|
Yang Y, Cai Y, Wu G, Chen X, Liu Y, Wang
X, Yu J, Li C, Chen X, Jose PA, et al: Plasma long non-coding RNA,
CoroMarker, a novel biomarker for diagnosis of coronary artery
disease. Clin Sci. 129:675–685. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhou X, Yin C, Dang Y, Ye F and Zhang G:
Identification of the long non-coding RNA H19 in plasma as a novel
biomarker for diagnosis of gastric cancer. Sci Rep. 5:115162015.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zheng K, Dou Y, He L, Li H, Zhang Z, Chen
Y, Ye A, Liu W and Kong L: Improved sensitivity and specificity for
prostate cancer diagnosis based on the urine PCA3/PSA ratio
acquired by sequence-specific RNA capture. Oncol Rep. 34:2439–2444.
2015.PubMed/NCBI
|
|
23
|
Sun M, Song J, Zhou Z, Zhu R, Jin H, Ji Y,
Lu Q and Ju H: Comparison of serum microRNA21 and tumor markers in
diagnosis of early non-small cell lung cancer. Dis Markers.
2016:38231212016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen F, Li WM, Wang DM, Gao SS, Bao Y,
Chen WB and Liu D: Clinical value of combined detection of serum
tumor markers in lung cancer diagnosis. Sichuan Da Xue Xue Bao Yi
Xue Ban. 39:832–835. 2008.In Chinese. PubMed/NCBI
|
|
25
|
Shi X, Sun M, Liu H, Yao Y, Kong R, Chen F
and Song Y: A critical role for the long non-coding RNA GAS5 in
proliferation and apoptosis in non-small-cell lung cancer. Mol
Carcinog. 54(Suppl 1): E1–E12. 2015. View
Article : Google Scholar
|
|
26
|
Tong YS, Wang XW, Zhou XL, Liu ZH, Yang
TX, Shi WH, Xie HW, Lv J, Wu QQ and Cao XF: Identification of the
long non-coding RNA POU3F3 in plasma as a novel biomarker for
diagnosis of esophageal squamous cell carcinoma. Mol Cancer.
14:32015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shao Y, Ye M, Jiang X, Sun W, Ding X, Liu
Z, Ye G, Zhang X, Xiao B and Guo J: Gastric juice long noncoding
RNA used as a tumor marker for screening gastric cancer. Cancer.
120:3320–3328. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yang-Chun F, Min F, Di Z and Yan-Chun H:
Retrospective study to determine diagnostic utility of 6 commonly
used lung cancer biomarkers among Han and Uygur population in
Xinjiang Uygur Autonomous Region of People's Republic of China.
Medicine (Baltimore). 95:e35682016. View Article : Google Scholar
|
|
29
|
Ma L, Bajic VB and Zhang Z: On the
classification of long non-coding RNAs. RNA Biol. 10:925–933. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Malih S, Saidijam M and Malih N: A brief
review on long noncoding RNAs: a new paradigm in breast cancer
pathogenesis, diagnosis and therapy. Tumour Biol. 37:1479–1485.
2016. View Article : Google Scholar
|
|
31
|
Peng L, Yuan X, Jiang B, Tang Z and Li GC:
LncRNAs: key players and novel insights into cervical cancer.
Tumour Biol. 37:2779–2788. 2016. View Article : Google Scholar
|
|
32
|
Yang Q, Zhang RW, Sui PC, He HT and Ding
L: Dysregulation of non-coding RNAs in gastric cancer. World J
Gastroenterol. 21:10956–10981. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Roberts TC, Morris KV and Wood MJ: The
role of long non-coding RNAs in neurodevelopment, brain function
and neurological disease. Philos Trans R Soc Lond B Biol Sci.
369:3692014. View Article : Google Scholar
|
|
34
|
Lorenzen JM and Thum T: Long noncoding
RNAs in kidney and cardiovascular diseases. Nat Rev Nephrol.
12:360–373. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ferreira LB, Palumbo A, de Mello KD,
Sternberg C, Caetano MS, de Oliveira FL, Neves AF, Nasciutti LE,
Goulart LR and Gimba ER: PCA3 noncoding RNA is involved in the
control of prostate-cancer cell survival and modulates androgen
receptor signaling. BMC Cancer. 12:5072012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cui Y, Cao W, Li Q, Shen H, Liu C, Deng J,
Xu J and Shao Q: Evaluation of prostate cancer antigen 3 for
detecting prostate cancer: A systematic review and meta-analysis.
Sci Rep. 6:257762016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Konishi H, Ichikawa D, Yamamoto Y, Arita
T, Shoda K, Hiramoto H, Hamada J, Itoh H, Fujita Y, Komatsu S, et
al: Plasma level of metastasis-associated lung adenocarcinoma
transcript 1 is associated with liver damage and predicts
development of hepatocellular carcinoma. Cancer Sci. 107:149–154.
2016. View Article : Google Scholar :
|
|
38
|
Li Q, Shao Y, Zhang X, Zheng T, Miao M,
Qin L, Wang B, Ye G, Xiao B and Guo J: Plasma long noncoding RNA
protected by exosomes as a potential stable biomarker for gastric
cancer. Tumour Biol. 36:2007–2012. 2015. View Article : Google Scholar
|
|
39
|
Schneider C, King RM and Philipson L:
Genes specifically expressed at growth arrest of mammalian cells.
Cell. 54:787–793. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Mourtada-Maarabouni M, Hasan AM, Farzaneh
F and Williams GT: Inhibition of human T-cell proliferation by
mammalian target of rapamycin (mTOR) antagonists requires noncoding
RNA growth-arrest-specific transcript 5 (GAS5). Mol Pharmacol.
78:19–28. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Tani H, Torimura M and Akimitsu N: The RNA
degradation pathway regulates the function of GAS5 a non-coding RNA
in mammalian cells. PLoS One. 8:e556842013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Mourtada-Maarabouni M, Hedge VL, Kirkham
L, Farzaneh F and Williams GT: Growth arrest in human T-cells is
controlled by the non-coding RNA growth-arrest-specific transcript
5 (GAS5). J Cell Sci. 121:939–946. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ren S, Wang F, Shen J, Sun Y, Xu W, Lu J,
Wei M, Xu C, Wu C, 'Zhang Z, et al: Long non-coding RNA metastasis
associated in lung adenocarcinoma transcript 1 derived miniRNA as a
novel plasma-based biomarker for diagnosing prostate cancer. Eur J
Cancer (Oxford). 49:2949–2959. 2013. View Article : Google Scholar
|
|
44
|
Han L, Ma P, Liu SM and Zhou X:
Circulating long noncoding RNA GAS5 as a potential biomarker in
breast cancer for assessing the surgical effects. Tumour Biol.
37:6847–6854. 2016. View Article : Google Scholar
|
|
45
|
Schwarzenbach H, Hoon DS and Pantel K:
Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev
Cancer. 11:426–437. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Stroun M, Maurice P, Vasioukhin V, Lyautey
J, Lederrey C, Lefort F, Rossier A, Chen XQ and Anker P: The origin
and mechanism of circulating DNA. Ann NY Acad Sci. 906:161–168.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yang Y, Shao Y, Zhu M, L Q, Yang F, Lu X,
Xu C, Xiao B, Sun Y and Guo J: Using gastric juice
lncRNA-ABHD11-AS1 as a novel type of biomarker in the
screening of gastric cancer. Tumour Biol. 37:1183–1188. 2016.
View Article : Google Scholar
|
|
48
|
Chen G, Wang J and Cui Q: Could
circulating miRNAs contribute to cancer therapy? Trends Mol Med.
19:71–73. 2013. View Article : Google Scholar
|
|
49
|
Yang T, Zeng H, Chen W, Zheng R, Zhang Y,
Li Z, Qi J, Wang M, Chen T, Lou J, et al: Helicobacter pylori
infection, H19 and LINC00152 expression in serum and risk of
gastric cancer in a Chinese population. Cancer Epidemiol.
44:147–153. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhang K, Luo Z, Zhang Y, Zhang L, Wu L,
Liu L, Yang J, Song X and Liu J: Circulating lncRNA H19 in plasma
as a novel biomarker for breast cancer. Cancer Biomark. 17:187–194.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhang M, Guo H, Zhao S and Wang Y, Yang M,
Yu J, Yan Y and Wang Y: Efficacy of epidermal growth factor
receptor inhibitors in combination with chemotherapy in advanced
non-small cell lung cancer: A meta-analysis of randomized
controlled trials. Oncotarget. 7:39823–39833. 2016.PubMed/NCBI
|
|
52
|
Greenhalgh J, Dwan K, Boland A, Bates V,
Vecchio F, Dundar Y, Jain P and Green JA: First-line treatment of
advanced epidermal growth factor receptor (EGFR) mutation positive
non-squamous non-small cell lung cancer. Cochrane Database Syst
Rev. 5:CD0103832016.
|
|
53
|
Cheng N, Li X, Zhao C, Ren S, Chen X, Cai
W, Zhao M, Zhang Y, Li J, Wang Q, et al: Microarray expression
profile of long non-coding RNAs in EGFR-TKIs resistance of human
non-small cell lung cancer. Oncol Rep. 33:833–839. 2015.
|
|
54
|
Wang Y, Chen W, Chen J, Pan Q and Pan J:
LncRNA expression profiles of EGFR exon 19 deletions in lung
adenocarcinoma ascertained by using microarray analysis. Med Oncol.
31:1372014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Dong S, Qu X, Li W, Zhong X, Li P, Yang S,
Chen X, Shao M and Zhang L: The long non-coding RNA, GAS5, enhances
gefitinib-induced cell death in innate EGFR tyrosine kinase
inhibitor-resistant lung adenocarcinoma cells with wide-type EGFR
via downregulation of the IGF-1R expression. J Hematol Oncol.
8:432015. View Article : Google Scholar : PubMed/NCBI
|