Introduction
Cancer is among the leading causes of death
worldwide and colorectal cancer (CRC) is the third most frequently
diagnosed cancer type (1). Despite
the fact that some progress has been made in recent years,
colorectal cancer progression, metastasis and treatment still
constitute essential medical problems. Increasing evidence suggests
that CRC may develop according to the hierarchical model of
carcinogenesis which postulates existence of small sub-population
of poorly-differentiated, self-renewable cancer stem cells (CSCs),
also called tumor initiating cells (TICs) or cancer stem cell-like
cells (CSC-like cells) (2).
CSC-like cells have been induced in the SW480 colorectal cell line
by the transduction of OCT3/4, SOX2, KLF4 genes using retrovirus
vectors (3,4). Such a transformation may be caused
also by alterations in the cell microenvironment and may depend on
transforming growth factor-β (TGF-β) signaling (5). It was postulated that TGF-β through
activation of epithelial mesenchymal transition (EMT) may act by
retaining of dynamic equilibrium between CSC-like cells and
non-CSC-like cells within human colorectal and breast cancer lines
(6).
Lapidot et al (7) were the first to describe
characteristics of cells capable of developing acute myeloid
leukemia (AML) after transplantation into severe combined
immune-deficient (SCID) mice, which were suggested to be CSC-like
cells. Over the years, CSC-like cells were proven to exist in many
solid tumors including breast (8,9),
pancreas (10,11), skin (12), lung (13), glioblastoma (14,15),
prostate (16), and colon cancers
(17,18) as well as in brain and glioblastoma
(19,20). CSC-like cells are capable of
self-renewal and differentiation into non-tumorigenic cell progeny
and are resistant to conventional therapeutic procedures. Due to
their specific properties and insensitivity to chemo- and
radiotherapy, CSC-like cells are believed to be a source of cancer
recurrence (21-23). Therefore, targeting CSC-like cells
or their niches may lead to eradication of cancer cells, reduction
of tumor relapse risk and improved prognosis for CRC patients.
Isolation of the CSC-like cells from the total
cancer cell population is essential for circumstantial studies on
their participation in cancer progression, metastasis and drug
resistance. Their detection is based on cytological sorting,
morphological and biochemical features as well as
xenotransplantation assays (24).
The identification of specific surface biomarkers is one of the
most commonly used types of CSC-like cell analysis. To distinguish
colon CSC-like cell population from other tumor cells Ricci-Vitiani
et al (17) used as their
marker CD133 protein, known also as a prominin-1. CD133+
population was found to account for approximately 2.5% of all
cancer cells in CRC samples after tissue dissociation. Expression
of CD133 protein only in cell populations capable of sphere
formation, self-renewal and resistant to chemotherapeutics
confirmed its role as a putative CSC-like cell marker by other
groups (18,25,26).
They found expression of CD133 on the surface of CSC-like cells in
brain, pancreatic, gastric and gallbladder cancers. However,
Shmelkov et al (27)
reported that both, CD133+ and CD133−
metastatic colon subsets were capable of tumor initiation in the
mouse model. CD133− subset derived from metastatic CRC
was capable of long-term growth in a xenograft model within
NOD/SCID mice and appeared to be more aggressive in comparison to
CD133+ population. Moreover, the phenotypic analysis of
CD133− cells confirmed the expression of next CSC-like
marker, CD44 molecule, which correlated with the formation of more
aggressive tumors (27).
It has been proposed recently that the usage of
spherical cultures (SCs) presents a more adequate tool for
culturing and analysis of stem cells. This concept is based on
anchorage-independent properties of CSC-like cells which are
capable of surviving after being detached from niche elements and
forming next cellular aggregates floating freely in the serum-free
medium. That is contrary to the non-stem cancer cells population
which undergo anoikis under the same culture conditions (28-31).
These three-dimensional (3-D) models gained popularity in the field
of breast (32), lung (33), ovarian (34) and colon (29,31)
cancer research. In the two-dimensional (2-D) systems, cells are
grown as monolayer, lacking specific interactions that are present
in native tumors, thus, 2-D cultures poorly mirror the complexity
of cancer microenvironment. Hence, multicellular spheroid model
seems to partially simulate naturally-occurring heterogeneity in
regard to cellular morphology (35). Heterogeneous expo-sure to oxygen,
nutrients, physical and chemical stresses, and specific gene
expression (29,36-38).
SCs are more likely to be engaged in some functional assays since
they more adequately support CSC-like cell properties (25,30,31,38).
Although HCT116 and HT29 cancer cell lines belong to
the most often utilized CRC model cells in 2-D cultures, their
properties when cultured in adherent and spherical models have not
yet been fully compared. Therefore, we decided to investigate the
biological characteristics of colonospheres derived from HCT116 and
HT29 cell lines in vitro and verify the influence of the
culture conditions on cancer cell properties, and especially number
and features of CSC-like cells. Additionally, to fully evaluate the
features of SCs cells, we performed magnetic cell sorting based on
the presence of the CD133 protein on the cell surface. The
literature is lacking such comprehensive description thus we wish
to fulfill this field of knowledge.
Materials and methods
HT29 and HCT116 monolayer cell
culture
Both, HT29 and HCT116, human adenocarcinoma
colorectal cell lines were originally purchased from the American
Type Culture Collection (ATCC; Manassas, VA, USA). All experimental
chemicals were purchased from Sigma-Aldrich (Poznan, Poland). The
cells were cultured in medium recommended by the manufacturer,
McCoy's medium supplemented with 10% fetal bovine serum (FBS), 1%
penicillin-streptomycin and 2 mM L-glutamine and incubated at 37°C
under a humidified atmosphere of 5% CO2. The cells were
subcultured by trypsin-EDTA treatment when they achieved ~80%
confluency and the medium was renewed 3 times per week.
Colonosphere-derived HCT116 and HT29 cell
lines
All experimental chemicals were purchased from
Sigma-Aldrich, except for growth factors, which were purchased from
R&D Systems/Biokom (Warszawa, Poland). Cells were originally
cultivated in PCM, trypsinized, washed twice in phosphate-buffered
saline (PBS) and maintained in serum-free stem cell medium
containing Dulbecco's modified eagle's medium (DMEM)-F12
supplemented with ITS Liquid Media Complement, 5 mM HEPEs, 4 mg/ml
BSA, 2 nM L-glutamine, 3 mg/ml glucose, 20 ng/ml EGF, 20 ng/ml bFGF
and antibiotic antimycotic solution. This medium will be referred
to as stem cell medium (SCM). For characterization purposes, we
only used 5-6 passage SC within the present study.
Phenotypic flow cytometric analysis
Colonospheres were first washed in PBS-EDTA
(Sigma-Aldrich) medium for 5 min, to obtain single-cell suspension.
CRC line cells and cells from colonospheres were stained with the
following monoclonal antibodies: anti-CD29-APC (clone MAR4, IgG1,
κ), anti-CD44-FITC (clone, G44-26, IgG2b, κ), anti-EpCAM-FITC
(clone EBA-1, IgG1), and anti-LGR5-Biotin (clone 4D11F8, IgG2b, κ)
coupled with streptavidin-APC (BD Biosciences, San Jose, CA, USA).
Anti-CD133/2-pe (clone 293C3, IgG2b, κ) monoclonal antibody was
purchased from Miltenyi Biotec (Bergisch Gladbach, Germany). After
30 min of incubation in the dark in room temperature (RT) samples
were fixed and prepared for further analysis. Intracellular
staining was performer for anti-BMI-1-FITC (clone P51-311, IgG1, κ)
according to the manufacturer's instructions (BD biosciences).
Cells were mixed with warm fixation buffer in volume ratio 1:1 and
incubated for 10 min in 37°C. In the next steps, cells were
centrifuged at 250 × g for 10 min and the supernatant was
discarded. Pellet was washed with chilled stain buffer and
centrifuged under same conditions to remove supernatant. Cold Perm
buffer was added to the pellet while vortexing. Cells were
incubated on ice for 30 min, centrifuged and washed twice with
staining buffer. Cells (1×107) were suspended in 1 ml of
stain buffer, divided into separated test-tubes and stained with
anti-BMI-1. Next steps were analogical like in an extracellular
staining procedure presented above. Flow cytometric analysis was
performed using FACSCalibur flow cytometer (BD Biosciences).
Cell death assay
The 7-aminoactinomycin D (7-AAD) dye (BD
Biosciences), a compound binding to the DNA of impaired cells, was
used for death cell evaluation. After adding 10 µm of Via
Probe samples were incubated for 30 min, washed and re-suspended in
PBS prior to cytometric analysis.
Apoptosis assay
Analysis was based on changes of phosphatidylserine
localization within cell membrane using Annexin V-FITC apoptosis
detection kit I (BD Biosciences) according to the manufacturer's
instructions. Cells (1×106) were suspended in 1 ml of
binding buffer. A total of 5 µl Annexin V-FITC and 5
µl of PI were added, gently pipetted and incubated for 15
min in RT in the dark. After that time, 400 µl binding
buffer was added and flow cytometric analysis was performed within
1 h.
Magnetic cell sorting based on MACS
technology
The procedure was conducted according to the
manufacturer's recommendation (Miltenyi Biotec). In brief, 20
µl of Blocking Reagent and 20 µl of CD133 MicroBeads
Tumor Tissue were added to cells suspended in buffer composed of
PBS (Sigma-Aldrich), 0.5 M EDTA (Sigma-Aldrich) and 10% BSA
(Sigma-Aldrich) and incubated for 15 min. Cells were washed and 10
µl of Labeling Check Reagent-PE was added and was incubated
for 5 min. Labeled cells were washed and re-suspended in proper
amount of the buffer. Cell suspension was applied onto the MS
Column which was washed three times with buffer. Cells within
flow-through were collected. The purity of isolated subpopulation
was checked by addition of CD133/2 (293C3) antibody and flow
cytometric analysis.
Cell cycle analysis
Cells (1×107) after 7 days of culturing
were pipetted, washed twice in PBS (Sigma-Aldrich), fixed on 70%
ethanol at −20°C. Within 2 weeks, cells were centrifuged, suspended
in staining buffer composed of PBS, propidium iodide (pi) (50
µg/ml) and RNase (25 µg/ml) (both from Sigma-Aldrich)
for 30 min in the dark at 37°C. Samples were analyzed using
FACSCalibur flow cytometer (BD Biosciences).
Cell growth and proliferation assay
Adherent cells were seeded in cell culture flask
with vented cap in PCM and HCT116- and HT29-derived spheres in
24-well plates in SCM. After every passage, cells were dissociated
into single-cell suspension and counted.
Proliferation assay
Proliferation abilities were measured based on the
CellTrace™ Cell Proliferation kit (Invitrogen; Thermo Fisher
Scientific, inc., Carlsbad, CA, USA) according to the
manufacturer's protocol which differs among adherent and spherical
cells. Spherical cells in suspension were incu-bated with 5
µM CellTrace-CFSE Dye for 20 min at 37°C, protected from
light. Afterwards, free dye was removed by addition of culture
medium and centrifugation. Pellet cells were re-suspended in
complete SCM and proceeded with 7 days incubation and analysis.
Adherent cells were not trypsinized; old medium was removed and
replaced with 5 µM CellTrace-CFSE buffer and incubated in
the dark. Then, the solution was removed, the cells were washed
twice with PBS and replaced with fresh medium for 7 days. At day 0
and 7 cells were analyzed by the FACSCalibur flow cytometer (BD
Biosciences).
Dilution assay
Cells after magnetic sorting were seeded at
different density by using serial dilution method beginning with
2,500 cells/well and finishing with 10 cells/well. Cells were
cultured in 24-well plates, dedicated to non-adherent cells, in
SCM. After 7 days, cells were dissociated into single-cell
suspension, stained with Trypan blue (Sigma-Aldrich) and
counted.
3-D sphere invasion assay
After 7 days of culturing, formed spheres were taken
and the diameters were measured. Spheres were suspended in a
mixture of Matrigel™ Matrix growth factor reduced matrix (BD
biosciences) and DMeM/F12 medium (Sigma-Aldrich) in ratio 1:4 on
ice and transferred onto a 24-well plate. Invasion was monitored by
measuring the maximal outgrowth of the sphere diameter after 8
days. Photographs were taken with the use of inverted microscope
Olympus CKX53 coupled with digital camera Olympus SC50 (Olympus
Corp., Tokyo, Japan).
Statistical analysis
Data were computed using the GraphPad Prism ver.
6.05 (GraphPad Software, Inc., San Diego, CA, USA) and software
Statistica 12 (StatSoft, Kraków, Poland). Statistical significance
of differences between the mean values was based on non-parametric
tests and assessed by the Mann-Whitney U, the Kruskal-Wallis
analysis of variance (ANOVA) and the Spearman's rank correlation.
Values of P<0.001 or P<0.05 were considered as statistically
significant. Within the present study, data from at least 3
independent experiments have been analyzed to verify
reproducibility of the results. The data are presented as the means
± standard error of the mean (SEM).
Results
Establishing of spherical models derived
from HCT116 and HT29 cells
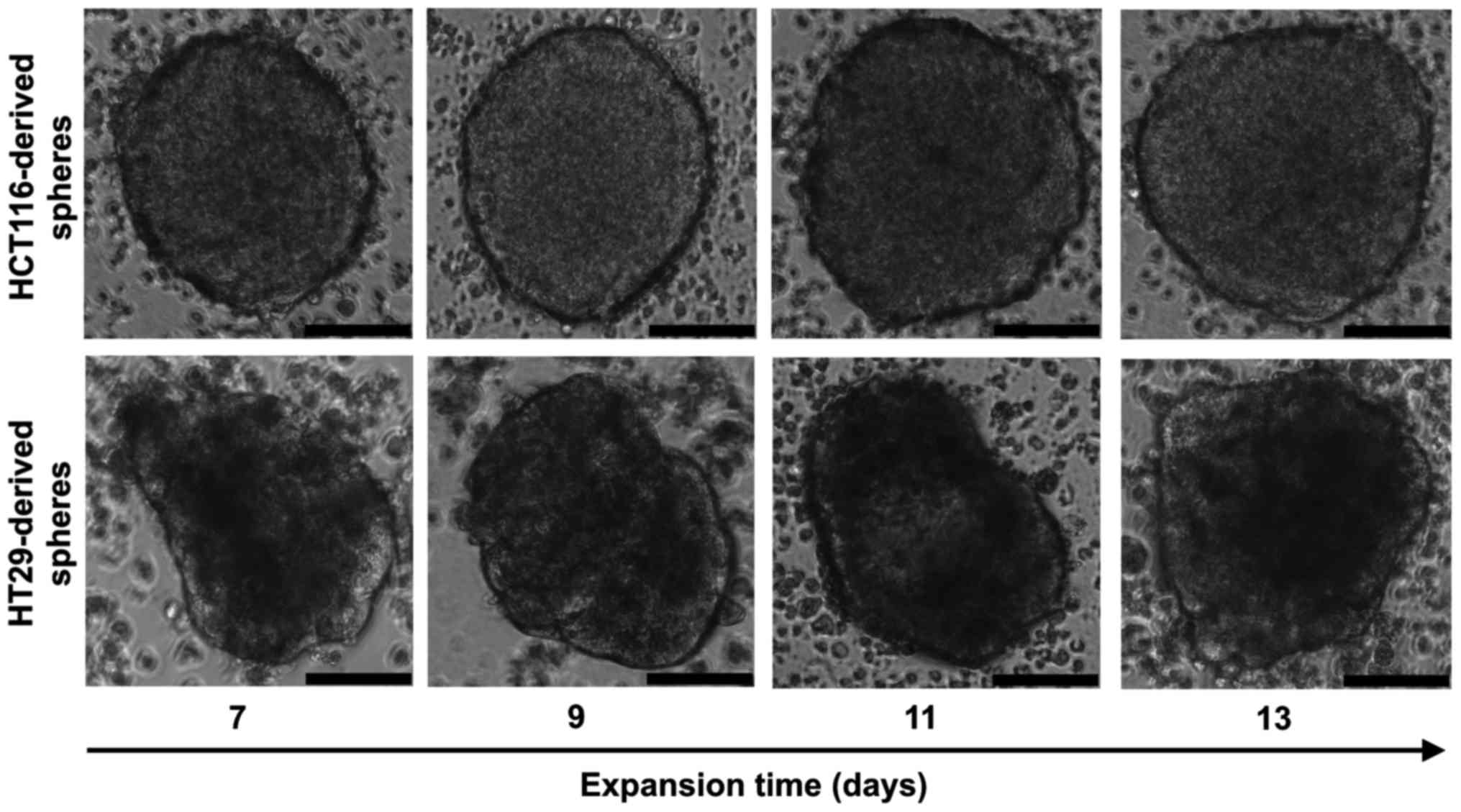
HCT116 and HT29 cell lines cultured in selective
conditions of stem cell medium (SCM) formed spheres (Fig. 1); however, colonospheres from both
lines displayed morphological differences. HCT116 cells formed
larger spheres with diameters varying from 150 up to 400 µm.
Shape of the HCT116-derived spheres was spherical with regular,
continuous and aquiline contour, which can be described as compact
tumor packaging. In comparison, the HT29-derived spheres were
smaller (100-300 µm), their outline was less regular and
cell aggregates presented various shapes. To calculate the sphere
size, the diameters of at least 5 representative spheres were
measured every second day and the average dimensions of HCT116- and
HT29-derived spheres are shown in Table I.
 | Table ISizes of the colonospheres. |
Table I
Sizes of the colonospheres.
| HCT116-derived
spheres | HT29-derived
spheres |
|---|
| Length
(µm) | 300.19±63.08 | 186.24±55.98 |
| Width
(µm) | 270.23±46.59 | 167.82±46.74 |
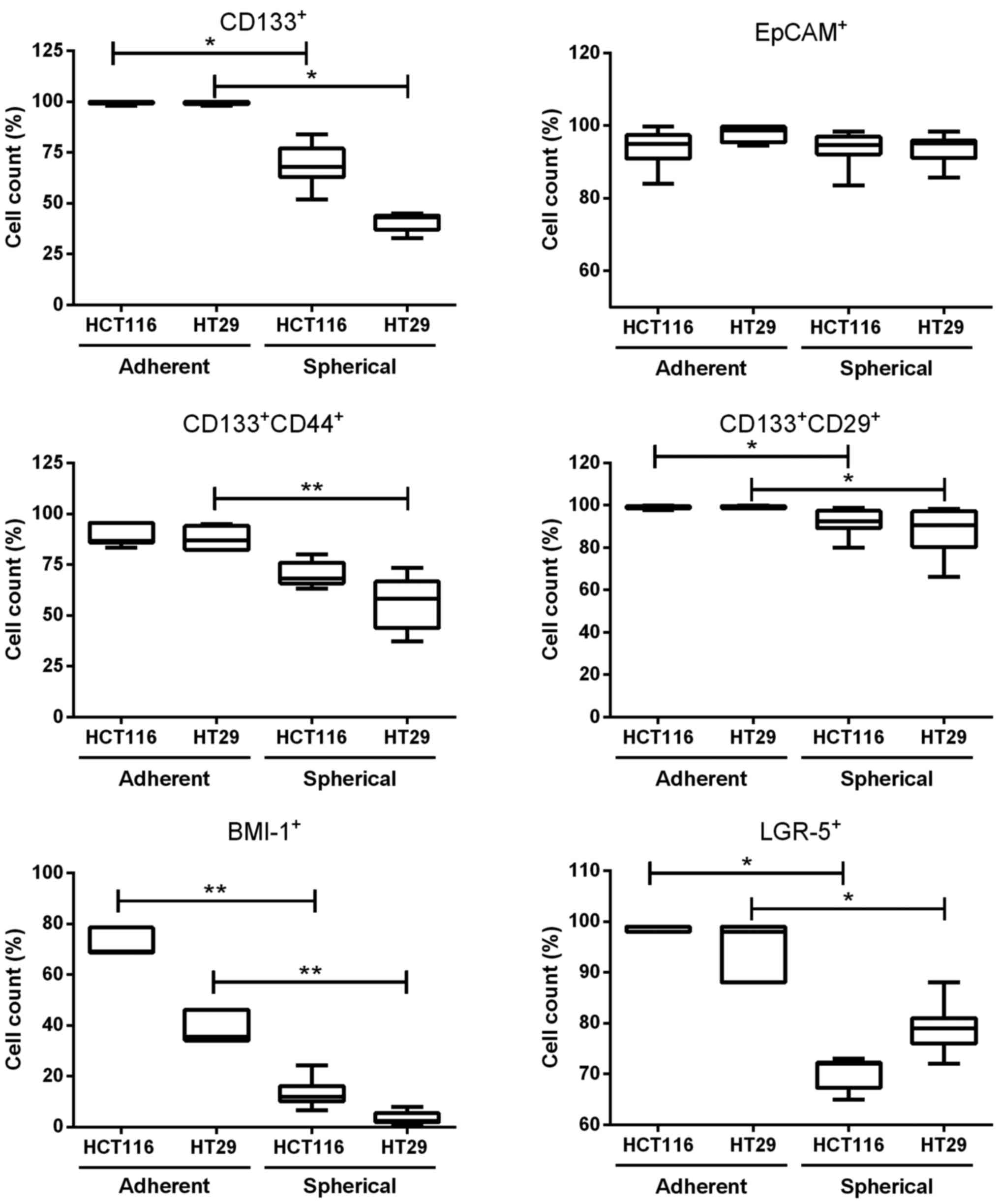
Different expression of CSC-like markers
on colonospheres as compared with adherent cell cultures
As CD133 and some other proteins, such as CD44,
CD29, BMI-1, LGR-5 and EpCAM, have been reported to be
characteristic for CRC-CSC-like cells, we determined their
expression by flow cytometry (Fig.
2). We found that adherent forms of HCT116 and HT29 CRC cell
lines contained more CSC-like cells than their spherical
counterparts. Both parental HCT116 and HT29 cell lines contained
~90% of CD133+CD44+ and 100% of
CD133+CD29+ cells. On the contrary, SCs
derived from CRC cell lines presented significantly lower number of
CD133−, CD44− and CD29-positive cells
(Fig. 2). The expression of cell
adhesion molecule, EpCAM, remained at the same high level during
culture expansion time both in adhesive and spherical cultures
(Fig. 2). Transformation from
adherent to spherical forms of HCT116 and HT29 cell line culture
caused decreased expression of some other CSC-like cell markers
such as BMI-1 and LGR-5 in comparison to their adherent forms
(Fig. 2).
Viability of cells during adhering and
colonosphere-supporting culture
We evaluated the percentage of non-viable cells in
both cell culture systems by flow cytometric assays using 7-AAD dye
(Fig. 3), which is excluded by
living cells, but binds selectively to GC regions of DNA of damaged
cells. The spheres obtained from both studied CRC lines presented
significantly higher percentage of non-viable cells than their
parental (adherent) cell lines (Fig.
3). To evaluate these observations we analyzed the proportion
of 7-AAD+ cells labeled with Annexin V-FITC and PI by
cytometric analysis (data not shown). We confirmed the previous
observation that within adherent HCT116 and HT29 lines dead cells
constituted only a very minor percentage of the total population, 2
and 3%, respectively. HCT116-derived spheres displayed 64.5±9.1% of
7-AAD+ cells: among them 71.4±2.5% constituted
late-apoptotic cells, 19.7±4.2% presented early apoptotic features
and 9.0±6.1% were necrotic/dead cells (data not shown). In
comparison, 7-AAD+ cells from spherical model of HT29
were characterized by the presence of 74.7±5.5% of late apoptotic
and 14.3±4.9% of early apoptotic cells, the remaining subpopulation
was made up by necrotic/dead cells. The differences between HCT116-
and HT29-derived spheres in the proportions of early apoptotic,
late apoptotic and necrotic/ death cells were not significant.
Proliferative abilities of cells during
adhering and colo-nosphere-supporting culture
To estimate the proliferative abilities of SCs
cells, we calculated the proliferation rates of cells in both
culture systems during every passage (Fig. 4A). In HCT116- or HT29-derived
spheres the proliferation rate was 5- or 3-fold lower as compared
to the corresponding adherent cells, respectively (Fig. 4A).
The proliferative capacities of cells in adherent
and spherical models were also assessed by CFSE-based proliferation
assay. CFSE (carboxyfluoresceinsuccinimidyl ester), diffuses into
cells and after binding covalently to cellular amine residues,
emits fluorescence proportional to the number of stained cells.
Changes in CFSE median fluorescence intensity (MFI) after 7 days of
incubation compared to the MFI at day 0 represented fold decrease
indicative of the rate of proliferation (Fig. 4C). We found 1384±196-fold and
1339±30-fold MFI decrease in HCT116 and HT29 adherent cell lines,
respectively. SCs of the corresponding cell lines presented
significantly lower MFI fold decrease (34±5 for HCT116-spheres and
23±1 for HT29-spheres) between day 0 and 7, indicative of the lower
dilution of the CFSE dye and smaller number of cell divisions in
spheres as compared to adherent culture (Fig. 4C).
CFSE dye was also used to compare the proportion of
cells which underwent divisions to find out how many cells were
able to proliferate, thus, how many cells were quiescent. The
number of cells with decreased CFSE dye concentration after 7 days
of cultivation was estimated (Fig.
4B). Markedly, higher population of cells underwent divisions
in HCT116 and HT29 adherent models in comparison to their spherical
counterparts. Overall, we confirmed that the SCs had reduced number
of cells capable of dividing in comparison to adherent cells.
Cell cycle analysis of cells during
adhering and colonosphere-supporting culture
The cell cycle of the adherent- and sphere-forming
cells was analyzed by flow cytometry (Table II). Only viable cells were
considered since dead cells were gated and not analyzed. We
observed significant differences in the proportions of cells in
G0/G1 and S/G2/M phases between adherent HCT116 and HT29 cells and
their spherical counterparts.
 | Table IIDistribution of cells derived from
both culture systems in the cell cycle after incubation with pi and
flow cytometric analysis. |
Table II
Distribution of cells derived from
both culture systems in the cell cycle after incubation with pi and
flow cytometric analysis.
| G0/G1 | P-value | S/G2/M | P-value |
|---|
| HCT116 | 49.5±4.7% | <0.05a | 50.5±4.7% | <0.05a |
| HCT116-derived
spheres | 63.6±9.4% | | 36.4±9.4% | |
| HT29 | 44.9±12.9% | <0.001b | 55.1±12.9% | <0.001b |
| HT29-derived
spheres | 61.4±6.3% | | 38.6±6.3% | |
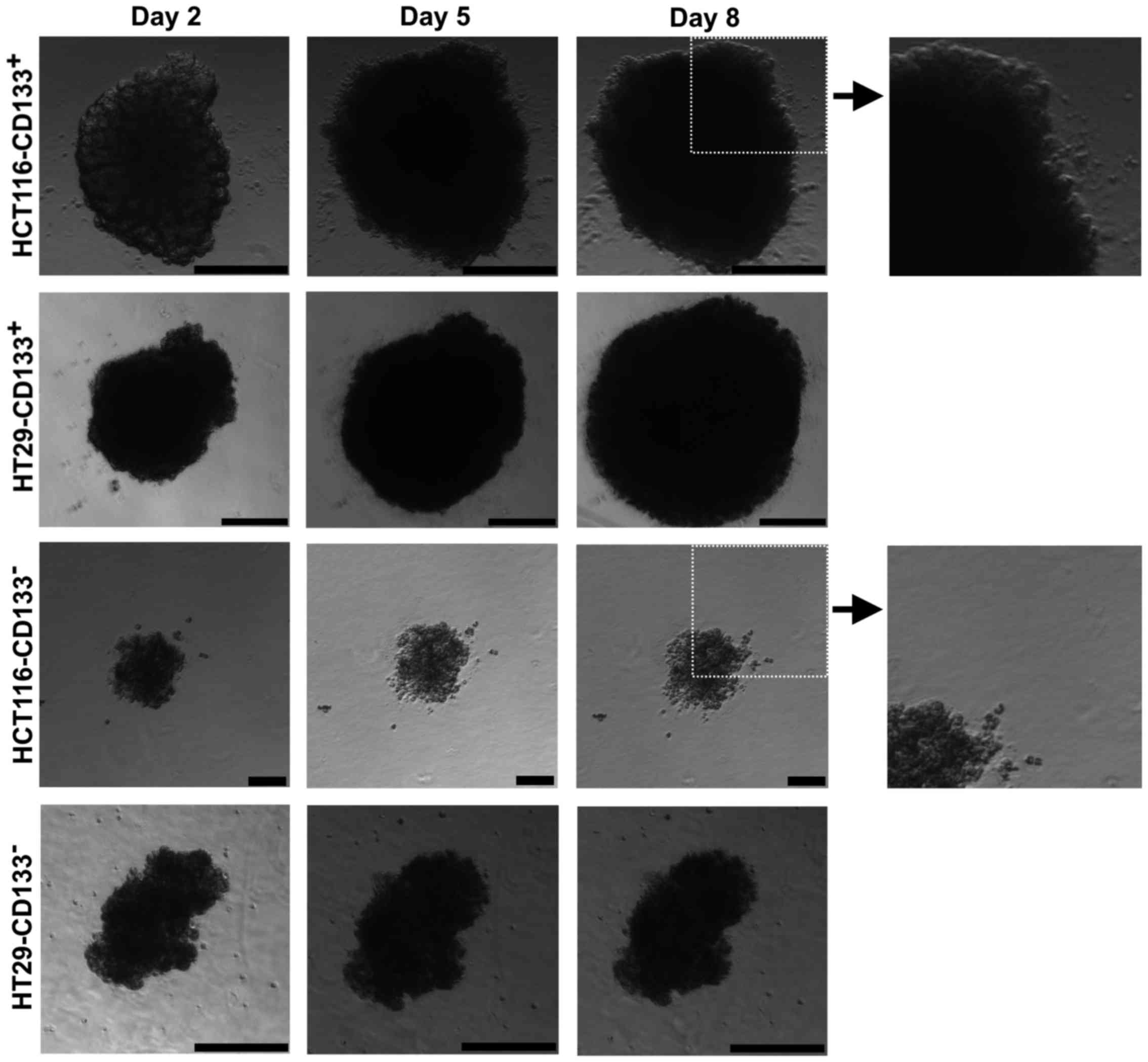
Morphological analysis of the obtained
CD133+ and CD133− cells after magnetic
separation
To further characterize the putative CSC-like cells
of the HCT116 and HT29 cell lines we performed the magnetic sorting
based on the expression of the CD133 molecule. Adherent cells from
CRC lines were separated into the following fractions:
HCT116-CD133+, HCT116-CD133−,
HT29-CD133+ and HT29-CD133−.
After magnetic separation the cell fractions were
cultured in the SCM for 14 days and analyzed (Fig. 5). Within the first week, we
observed morphological differences between isolated fractions from
both studied CRC cell lines. CD133+ subpopulations
formed big spheres with apparent contour, similar to their parental
floating counterparts, whereas CD133− cells presented
smaller cellular spheres. Additionally, cultures of
CD133− cells isolated from HCT116 and HT29 cell lines
presented prominent number of cells not involved in sphere
formation but just suspended in the medium. Parental spheres and
CD133+ cell-derived cultures had significantly larger
diameter in comparison to their CD133− analogues
(Table III).
 | Table IIISizes of spheres formed by separated
subsets from 3 independent experiments. |
Table III
Sizes of spheres formed by separated
subsets from 3 independent experiments.
|
HCT116-CD133+ |
HCT116-CD133− |
HT29-CD133+ | HT29-CD133- |
|---|
| Length
(µm) | 231.94±72.40 | 97.27±33.62 | 235.78±50.30 | 83.57±21.01 |
| Width
(µm) | 190.56±59.00 | 88.64±34.45 | 181.11±47.47 | 69.86±21.30 |
Phenotype of the obtained
CD133+ and CD133− cells after magnetic
separation
We found that the magnetically-separated
CD133+ fractions from both studied CRC cell lines were
enriched in double-positive cells for CD44+ and
CD29+, whereas CD133− cultures presented
significantly increased number of CD44−CD29+
cells (Fig. 6A and B). EpCAM
expression was similar within all subpopulations, whereas LGR-5
expression was markedly higher in both CD133+ and
CD133− subsets derived from HT29 cell line (Fig. 6C), similarly as in the parental
populations. However, HCT116-CD133+ and
HT29-CD133+ populations displayed higher proportion of
BMI-1+ cells when compared to their CD133-negative
analogues (Fig. 6C). The
expression of CD44 and CD29 on the cell surface correlated
positively with the sphere sizes (P<0.05, r=0.28, Spearman's
rank correlation coefficients).
Viability of the obtained
CD133+ and CD133− cells after magnetic
separation
The viability and apoptosis of the sorted
subpopulations was verified according to the same protocols as we
used for original HCT116 and HT29 cells; however, there were no
statistical differences between CD133+ and
CD133− fractions in studied cell lines (data not shown).
The detection of dying cells performed with the use of Annexin
V-FITC and PI staining did not reveal differences amongst
CD133+ and CD133− populations from HCT116 and
HT29 cell lines and confirmed similar viability of the studied
subsets of cells (data not shown).
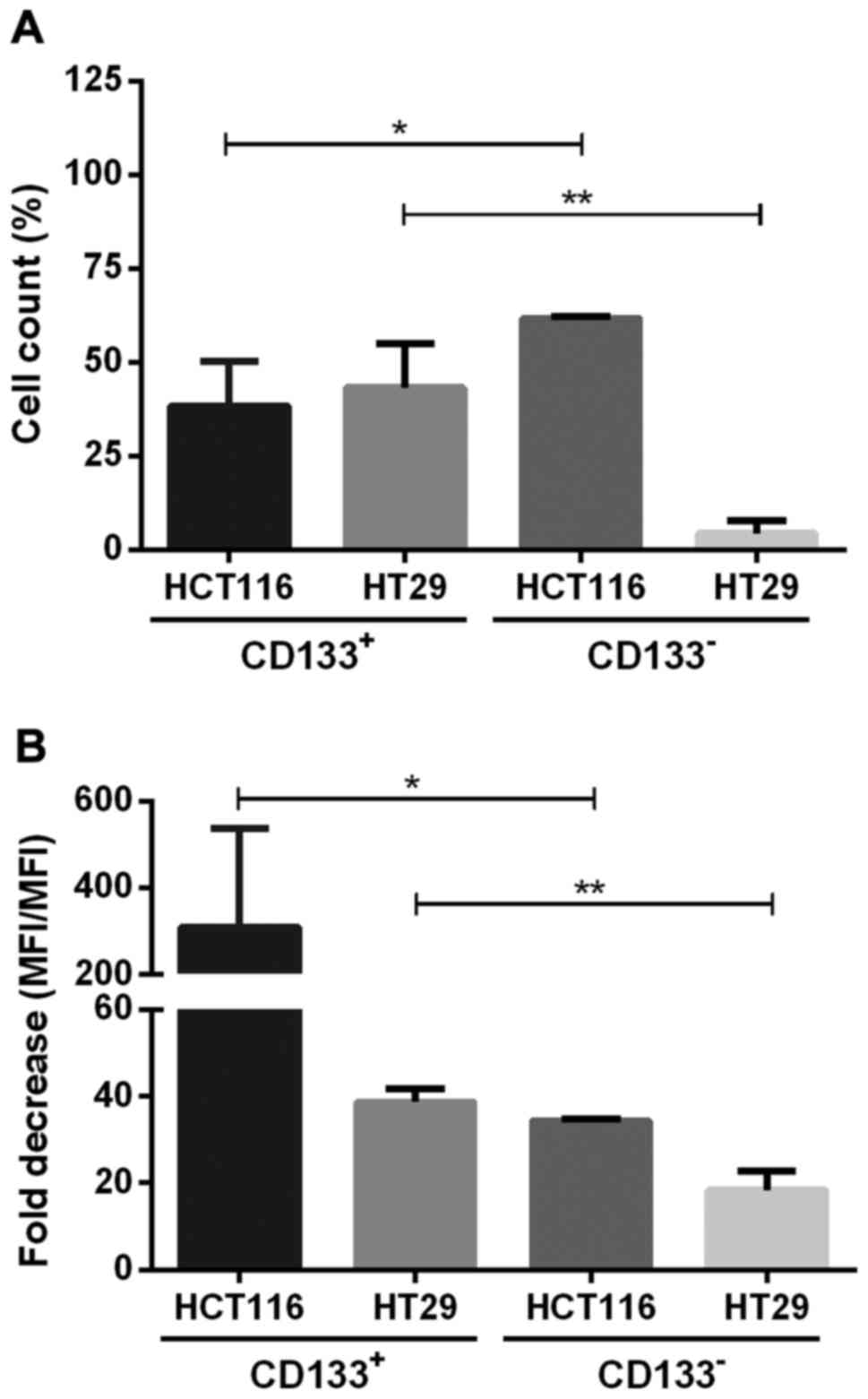
CD133+ and CD133−
cell fractions have different proliferation rates and dividing
potential
The proliferation of CD133+ and
CD133− subpopulations of the studied HC116 and HT29
cells cultured in adherent and spherical models was established by
the method of CFSE cell labeling (Fig.
7). HCT116-133− contained significantly higher
percentage of proliferating cells, with decreased amount of CFSE
dye in their cytoplasm, indicated as a population with lower MFI in
comparison to cells at day 0 (Fig.
7B). Fold decrease measured at day 0 and 7 was 309.3±119.7% for
HCT116-CD133+, whereas in HT29-CD133+
fraction this value was 38.7±2.9 (Fig.
7B). Based on the CFSE fluorescence we assessed the percentage
of cells which underwent divisions during expansion (Fig. 7A). Generally, we found that both
CRC lines represented different proliferative features because
HCT116-CD133+ fraction contained higher proportion of
actively proliferating cells in comparison to the
HCT116-CD133- cells, whereas HT29 cells induced the
opposite effects (Fig. 7A).
Cell cycle analysis of CD133+
and CD133− fractions
Staining with PI was performed to establish
distribution of sorted fractions of the studied cell lines in the
cell cycle, however, we did not observe any significant differences
amongst CD133+ and CD133− subpopulations
derived from the same CRC cell line. At the same time, we noted
changes between same type of fractions isolated from HCT116 and
HT29 cells. We observed markedly higher proportion of cells from
HCT116-CD133+ in G0/G1 phase in comparison to
HT29-CD133+ fraction, being 50.8 and 39.6%,
respectively. Furthermore, HT29-CD133+ possessed 60.4%
of actively dividing cells occurring in S/G2/M phase whereas this
percentage in CD133+ subset derived from HCT116
constituted <50%. When we compared the percentage of G0/G1 cells
amongst fractions from HCT116 and HT29 cell lines, we observed
slight increase of quiescent cells in HCT116-133− and
HT29-133− subpopulations (Table IV).
 | Table IVDistribution of CD133+ and
CD133− fractions derived from HCT116 and HT29 cell lines
in the cell cycle after incubation with pi and flow cytometric
analysis. |
Table IV
Distribution of CD133+ and
CD133− fractions derived from HCT116 and HT29 cell lines
in the cell cycle after incubation with pi and flow cytometric
analysis.
| G0/G1 | P-value | S/G2/M | P-value |
|---|
|
HCT116-CD133+ | 50.8±2.6% | <0.001a | 49.2±2.6% | <0.001a |
|
HT29-CD133+ | 39.6±5.6% | | 60.4±5.6% | |
|
HCT116-CD133− | 56.8±2.8% | <0.001a | 43.2±2.8% | <0.001a |
|
HT29-CD133− | 43.7±4.6% | | 56.3±4.6% | |
The ability to form colonospheres differs
between CD133+ and CD133− subpopulations
Cells from CD133-positive and CD133-negative
populations of HCT116 and HT29 cell lines were plated at different
cell densities, in range from 2.5×103 to 10 cells/well
by serial dilution method, in 24-well plates and cultured for 7
days to evaluate their sphere-formation abilities (spherogenicity).
We noticed that both CD133+ and CD133−
subpopulations from HCT116 and HT29 lines created spheres at
various cell densities (Fig. 8B).
Cells after dilution assay were collected to assess their final
number in each well after 7 days of incubation (data not shown). We
observed rapid increase of cell numbers when they were plated in
reduced concentrations what suggested that the cell density
influences the cell proliferative capability. Increased number of
cells (2.5×103) seeded at the beginning of the
experiment exhibited inhibitory effect on cellular divisions and
their accumulation.
To verify the sphere forming abilities, we also
cultured both magnetically separated fractions in Matrigel™ Matrix
for 8 days and calculated their total outgrowth and morphology with
microscope equipped with digital camera. We measured the sizes of
the growing spheres and afterwards we calculated fold increase by
dividing the final size (at day 8) by the value at day 0 of the
incubation (Fig. 8A). The
trajectory of growth curves fluctuated until day 4,
HCT116-CD133+ and HT29-CD133+ increased
notably their sizes thereon.
Migration abilities of cells derived from
HCT116 cell line
Besides changes in sphere size, cultivating
CD133-positive and CD133-negative CRC cell subpopulations in
Matrigel™ Matrix for 8 days showed also structural differences in
colonospheres formed by cell fractions from both CRC lines. Cells
from all four fractions were able to degrade Matrigel Matrix and
enhanced their expansion (Fig. 9).
However, only HCT116-derived subpopulations,
HCT116-CD133+ and HCT116-CD133−, contained
cells with the ability to migrate from parental spheres resulting
in irregular outline of spheres and cell presence in distant
regions of Matrigel Matrix, suggestive of their invasive properties
(Fig. 9).
Discussion
It has been assumed that 3-D spherical model of the
expansion of cancer cells better corresponds with the natural tumor
microenvironment and presents properties which may be more relevant
to the in vivo tumor development than cancer cell lines
cultured in adherent forms (29,36-39).
The spherical cultures of cancer cells offer an appropriate model
for testing novel anticancer therapeutics, especially those which
would target CSC-like cells. It has been recently reported that
colonosphere cultures are enriched in CSC-like cells in a cell
line-dependent manner, thus, more data are necessary for precise
determination of the features of cells forming spherical cultures
in each particular cell line (40). Hierarchical model of tumor
development has been supported by data for various tumor types
including prostate, lung and colon cancers (13,16-18).
However, the increased utility of SCs in cancer studies provided
diver-gent results which obscure their real potential and enabled
the comparison of data obtained by various research groups.
Therefore, the present study aimed to explore the features of
colonospheres derived from two commonly used CRC cell lines, HCT116
and HT29, and compare them with the characteristics of the adherent
cell cultures. The obtained data present multiple evidence
suggesting advantageous use of 3-D cell cultures in the cancer
studies rather than traditional monolayer model.
Our results showed that cultivating human CRC cell
lines in spherical forms resulted in harvesting of cells
phenotypically different than their adherent counterparts, with an
important finding of more in vivo, like diversity creating
more reliably environment supporting CSC-like cells for
experimental purposes. Thus, colonospheres of the studied cell
lines represent more suitable tool for studying in vivo
tumor development than adherent cell cultures. Finding of adequate
biomarkers for the identification of CSC-like cells is a crucial
step to assess the proportion of CSC-like cells within the studied
populations. Therefore, apart from the CD133 molecule, which seems
to be controversial but still represents the most universal and the
most frequently used marker to identify CRC-CSC-like cells many
other markers such as CD44, CD166, CD29, CD24, epithelial-specific
antigen (ESA), leucine-rich repeat-containing G-protein coupled
receptor 5 (LGR-5), B lymphoma Mo-MLV insertion region 1 homolog
(BMI-1) have been used for the characterization of CSC-like cells
(22,24,39,41).
Our results indicated that both cell lines, HCT116 and HT29, when
cultured in a selecting, serum-free medium, were able to form
freely suspended spheres corroborating the results of earlier
studies on other CRC lines, including SW1222, Caco-2, SW480, DLD-1
and Sw620 (30,38,42,43).
However, the wide-ranged characterization and comparison of
spherical and adherent forms of the studied CrC cell lines was
accomplished first in the present study. Morphologically,
HCT116-derived spheres were almost ideally spherical, significantly
bigger and had more packed cells than HT29-drived spheres, which
presented less regular shapes and 'fuzzy' outlines in contrast to
the continuous contours of HCT116 spheres.
The important finding of the present study was the
observation of significantly lower ratio of CRC-CSC-like cells,
defined by the presence of the CD133, CD44, CD29, LGR-5 and BMI-1
molecules, in SCs of both studied cell lines rather than their
adherent counterparts. These results considerably differ from the
small fraction of CD133+ cells present in the primary
spherical cultures derived from CRC patients (17,18).
This discrepancy might have been caused by the use of fresh human
tissue resected from CRC patients (17,18),
whereas we studied SCs derived from the established CRC cell lines
commonly used in experimental studies. Furthermore, data concerning
CRC lines are very discrepant. CD133 expression in HCT116 line was
found to be greatly diversified. For instance Dittfeld et al
(44) showed that in parental
HCT116 CD133+ cells constituted 74% and in 3-D model 70%
of all cells, whereas Huang et al (45) reported that 2 and 65% of cells
expressed CD133 in monolayer and colonosphere model, respectively.
Some authors reported no differences in percentage of cells
positive for CD133 within HT29 CrC line cultured as monolayer and
in spherical form (6.25 and 5.6%, respectively) (46), but others (40) found ~15% increase of HT29
CD133+CD44+ cells when cultured under
CSC-selective conditions.
Notably, we found that the modification from
adherent into spherical form resulted in the major decrease of
CD29+ and CD44+ HCT116 and HT29 cells what we
associated with the weakening of the junctional complexes between
the cells and cell-matrix in spherical cultures (40). Similarly to other authors (47), we found that the expression of the
another adhesion molecule, EpCAM, did not differ between CRC cells
cultured as monolayer or spheres what suggests that EpCAM is rather
a surface marker of colon cancer cells, but not a marker of colon
CSC-like cells.
CRC cell lines were found to present rather high
expression of differentiation marker cytokeratin 20 (Ck20)
(48), which has been suggested to
be a useful diagnostic and prognostic marker in CRC patients
(49,50). In line with the idea that SCs
should maintain only primitive cells, a concomitant decrease in the
percentage of cells expressing Ck20 was observed in HCT116- and
HT29-derived colonospheres (data not shown) in comparison to their
adherence counterparts. Moreover, differences within the percentage
of Ck20-positive cells between CRC cell lines correspond with the
origin of cell lines, while HCT116-derived cells are less
differentiated and more immature, thus, have small fraction of
Ck20+ cells, whereas HT29 cell line is less aggressive
and possesses greater fraction of cells expressing Ck20.
LGR-5, a target of Wnt signaling, is a marker
protein for intestinal stem cells and CSC-like cells, hence it is
considered as a potential therapeutic target in colorectal cancer
(51-53). However, it has been recently
reported that selective LGr-5+ cell ablation curbs
primary CRC growth without total tumor regression due to the
proliferation of LGR-5− cells which refilled the
LGr-5+ pool (54). Our
finding of decreased proportion of LGR-5+ cells in
spherical cultures in comparison to their adherent counterparts
could be related to decreased cell-cell adhesion as it was found to
correlate with the expression of the LGR-5+ cells
(55). Moreover, this decline
might have been affected by the heterogeneity of spherical cultures
since they contained lower percentage of Ck20-expressing cells and
higher number of dying cells.
While BMI-1 was reported to play an important role
during the self-renewal and maintenance of many types of normal and
stem cells, its expression was also analyzed in different types of
cancers including leukemia, breast and colorectal (56-58).
BMI-1 is thought to protect tumor cells from apoptosis induced by
chemotherapeutics but the precise mechanism responsible for its
activity is not fully understood (59,60).
Although BMI-1 is highly enriched in many types of CSC-like cells,
however, not all BMI-1-positive cells are actual CSC-like cells
(59). The observations that
different subpopulations among CSC-like cells exhibit distinct rate
of growth, suggest that BMI-1 should be rather co-expressed with
other CSC-like cell markers, including CD133 or CD44 to induce such
pro-proliferating effect. The results of the present study seem to
support this notion since we found that the expression of BMI-1 was
positively correlated with the LGR-5 and CD133 expression in
colonospheres. Overall, the phenotypical analysis of the studied
CRC cell lines strongly suggests that SCs represented heterogeneous
cellular populations at the various development and differentiation
stages because of the varied expression of the LGR-5+
and BMI-1+ markers. BMI-1+ cells are rather
dormant type of CSC-like cells, which in case of eradication of
tumor bulk can be activated by the tumor microenvironment and
transform themselves into active, LGR-5+ cells, form
CSC-like cells with regenerative properties, to maintain the
balance between the CSC-like populations (5,60).
Importantly, our observation that the viability of
cells in 3-D model was significantly lower than in adherent
cultures may have an implication for the in vitro studies of
cancer cells. According to analysis of phenotype, including LGR-5
and Ck20, and proliferative abilities of cells, we claim that
significant percentage of 7-AAD+ cells within SCs of all
studied cell populations (before and after magnetic separation) was
the result of cell differentiation and decreased cell-cell
interactions. This assumption is in agreement with the findings of
other authors who also revealed the ability of immature cells to
survive and grow in serum-free suspension, whereas more
differentiated cells undergo anoikis, a form of apoptosis triggered
by loss of anchorage to ECM elements (31,61,62).
What is more, the co-culture of CSC-like with non-CSC-like cells
derived from breast cancer lines revealed protective/supporting
effect of the latter one toward population with stem traits
(62). Despite the elevated
percentage of non-viable cells found in colonospheres, the overall
number of cells within them was constantly increasing, indicating
the presence of continually proliferating cells which prefer SCM
culture conditions.
The application of separation based on the CD133
molecule presence on the surface of CRC lines resulted in obtaining
cells which could be further cultured, similarly to that found in
other studies on primary cultures of CRC tumors (17,18,25,27,29)
or CRC cell lines (6,41,44,47).
Notably, besides changes in the presence of CD133 on the cell
surface, obtained fractions presented different proportions of
cells bearing other proteins important for the cell-cell and
cell-ECM interactions, i.e.
CD133+CD44+CD29+ or
CD133-CD44−CD29+ cells, that may
affect their sphere-formation ability. We found that spheres formed
by both CD133+ cell subsets were significantly smaller
than original colonospheres. This indicated that the expansion of
'parental' HCT116- and HT29-derived SCs depended not only on
CD133+ but also was influenced by CD133−
tumor cells. That is surprising because we noted positive
correlation between CD133 proportion and sphere sizes in SCs.
Additionally, analysis of dilution assay has shown that both
subpopulations from HCT116 and HT29 lines created spheres in SCM
independently of number of seeded cells, however, the bigger the
concentration of cells, the higher the proliferation rate was. We
suppose that sphere formation took place through proliferation and
partially also through following aggregation of cells in our
experimental settings.
It is widely known that HCT116 and HT29 represent
CRC cell lines that correspond to the more and less aggressive
forms of this cancer, respectively. Our findings support this
important distinction. The analysis of apoptosis based on Annexin
V-FITC and pi revealed significant proportion of cells in G0/G1
phase in HCT116-derived SCs indicating presence of dormant cells
within the culture, which might contain CSC-like cells, whereas SCs
originated from HT29 line presented more cells in active phases of
cell cycle, i.e. S/G2/M. This can be explained by differences in
the derivation of cell lines as HCT116 line represent
non-differentiated and highly aggressive cell line that corresponds
to the TNM 3 stage, so we assume that number of CSC-like cells
could be higher than in HT29 line, which is known as less invasive
(TNM 2). However, subsets obtained after magnetic isolation
possessed approximately similar percentage of cells in G0/G1 and
S/G2/M as their parental spherical counterparts, thus,
linage-dependent factors influence the cell cycle features of cells
during their expansion in culture, especially in spherical
forms.
The analysis of the proliferative potential of the
analyzed cell types revealed, as we expected, that adherent CRC
lines showed significantly higher proliferation rates than their
spherical counterparts. This can be associated with the
phenotypical heterogeneity of the culture forms of the studied CRC
cell lines. Since colonospheres presented lower proportion of
CSC-like cells, lower proliferative potential and higher proportion
of dead/dying cells in culture we suggests that SCs of cancer cells
better mimic the properties of primary CRC tumors. Therefore,
colonospheres should be more widely used for the studies of cancer,
e.g. such as ex vivo evaluation of chemotherapeutics.
The magnetic separation of cells from CRC cell
lines with different expression of CSC-like cell markers provides
another useful tool for the in vitro studies. In this
respect, the choice of CD133 molecule, a putative marker of
CSC-like cells, enabled us to isolate from colonospheres cell
populations with different proliferative capacities. For instance,
HCT116-CD133+ cells as compared with
HCT116-CD133− cells contained quite small fraction of
cells which underwent divisions during incubation time but
presented much higher proliferative potential, and these parameters
were even more obvious in HT29-CD133+. These
observations were confirmed by the dilution assays which showed
that CD133+ cell fractions derived from both CRC lines
proliferated more frequently and formed more prominent spheres than
CD133− cells. The finding that CD133+ cells
seem to possess high colony-formation ability should be used in the
future studies of colonospheres, especially those which investigate
targeted chemotherapy of CRC cells.
Overexpression of transcription factors inducing
EMT is associated with induction of stemness amongst cancer cells
suggesting the connection between EMT and CSC-like cells, thereby
revealing that those cells do not have to remain in their static
state but are dynamic instead (4,63).
Merlos-Suarez et al (64)
associated CSC-like cells with metastasis and suggested that
metastasizing cells may obtain CSC-like cell traits while expand
tumor region and invasion. The use of Matrigel Matrix, a natural
complex hydrogel of ECM proteins and associated components such as
laminin, type IV collagen, entactin, and heparin sulphate (65,66),
enabled us to assess the invasive capacities of CD133+
and CD133− derived from HT29 and HCT116 cell lines. We
observed large number of cells leaving spheres during expansion in
Matrigel Matrix from both HCT116-CD133+ and
HCT116-CD133− fractions proving their aggressive
properties and invasiveness. Notably, we are probably the first to
report that the cells of HT29-CD133+- and
HT29-CD133−-derived colonospheres did not present
invasive properties since they were not capable to leave the
abandon maternal sphere. This can be explained by the fact that
HT29 cells were originally isolated from primary, not metastatic
tumor and represent lower, ii stage, whereas HCT116 cells are
placed in iii class in the TnM classification. However, since the
HT29-CD133+ and HT29-CD133−-derived spheres
could increase in size, similarly to the HCT116 counterparts, we
assumed that they were able to degrade Matrigel Matrix. This
finding provided further evidence that the colonospheres present a
better model for the in vitro studies of cancer cells than
the adherent cell cultures.
It is noticeable that there are many epigenetic and
genetic features which differ in HCT116 and HT29 cells and might
have influenced the presented results. HCT116 cells were originally
isolated from primary tumor derived from colon ascendens of
48-year-old male (67,68), whereas HT29 cell lines represents
human colonic adenocarcinoma originating from 44-year-old female
(69). Ahmed et al
(70) explored the genetic and
epigenetic molecular phenotype of 24 colon cancer cell lines,
including HCT116 and HT29. Cells from these lines possessed
different status of one of the most commonly mutated genes in colon
cancer, KRAS. HT29 has wild-type of KRAS
(KRASWT), while HCT116 gained mutated
KRASG13D (70),
which resulted in a constitutive activation of KrAS signaling
pathway. In this pathway the RAS protein plays an important role as
central mediator downstream of growth factor receptors and
therefore, it is critical for cell proliferation, survival, and
differentiation involving mitogen-activated protein kinases (MApKs)
and phosphoinositide-3 kinase (pi3K) pathways (71,72).
Cells harboring KRAS mutations within codon 13 are reported
to possess high oncogenic potential and be very aggressive
(73). Additionally, KRAS
status has also biological relevance in terms of colorectal cancer
clinical outcome to anti-EGFR therapy (74) and senescence and/or apoptosis
mediated by Fasr/FasL (75,76).
Besides KRAS gene, HCT116 and HT29 cell lines represent
different mutation status of chromosomal instability (CIN)
phenotype and some cancer critical genes such as BRAF,
PIK3CA and TP53 genes (70). Some of these genes might be used
for prediction of clinical benefit from anti-EGFR treatment in
metastatic colorectal cancer (77), hence the usage of HCT116 and HT29
cell lines allows for comparison of broad spectrum of features
characteristic for these types of cells.
When it comes to the relations of our results to
the genetic status of both used CRC lines, we could only suspect
that the deregulation and constant activation of EGFR pathway (due
to mutated form of KRAS gene) may influence the proliferative
abilities, phenotype of cells and growth of spheres. However, if
such features are directly associated with one mutation or rather
with general cancer progression status remains unclear and requires
more studies.
Undeniably, 2-D immortalized CRC cell lines
represent elegant model to extend the knowledge concerning cell
trans-formation. However, they will never provide adequate culture
to analyze elaborated tumor biology in vivo, which seems to
be more reliably mimicked by the spherical culture system as was
postulated earlier by others (28-30,35,37)
and as we concluded from the presented study. Cells cultured under
CSC-supporting conditions seem to share more similarities with
original tumors, indicating that they provide a more biologically
relevant culture system when compared with widely used traditional
monolayer cultures, including the lower proportion of CSC-like
cells. Therefore, we claim that 3-D spherical model of CRC lines
should be considered as an important tool for the in vitro
studies of cancer, especially these which target non-differentiated
CSC-like cells and their microenvironment.
Acknowledgments
The present study was supported by a grant from the
Polish Ministry of Science and Higher education, contract grants
number: MN 01-0232/08/280 and N N402 684040.
Abbreviations:
|
7-AAD
|
7-aminoactinomycin D
|
|
BMI-1
|
B lymphoma Mo-MLV insertion region 1
homolog
|
|
CRC
|
colorectal cancer
|
|
CSC
|
cancer stem cell
|
|
EpCAM
|
epithelial cell adhesion molecule
|
|
LGR-5
|
leucine-rich repeat-containing
G-protein coupled receptor 5
|
|
MFI
|
median fluorescence intensify
|
|
SC
|
spherical culture
|
|
SCM
|
stem cell medium
|
References
|
1
|
Manhas J, Bhattacharya A, Agrawal SK,
Gupta B, Das P, Deo SV, Pal S and Sen S: Characterization of cancer
stem cells from different grades of human colorectal cancer. Tumour
Biol. 37:14069–14081. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zeuner A, Todaro M, Stassi G and De Maria
R: Colorectal cancer stem cells: From the crypt to the clinic. Cell
Stem Cell. 15:692–705. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Farhana L, Nangia-Makker P, Arbit E,
Shango K, Sarkar S, Mahmud H, Hadden T, YU Y and Majumdar AP: Bile
acid: A potential inducer of colon cancer stem cells. Stem Cell Res
Ther. 7:1812016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Oshima N, yamada Y, Nagayama S, Kawada K,
Hasegawa S, Okabe H, Sakai Y and Aoi T: induction of cancer stem
cell properties in colon cancer cells by defined factors. PLoS One.
9:e1017352014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pan T, Xu J and Zhu Y: Self-renewal
molecular mechanisms of colorectal cancer stem cells. Int J Mol
Med. 39:9–20. 2017. View Article : Google Scholar
|
|
6
|
Yang G, Quan Y, Wang W, Fu Q, Wu J, Mei T,
Li J, Tang Y, Luo C, Ouyang Q, et al: Dynamic equilibrium between
cancer stem cells and non-stem cancer cells in human SW620 and
MCF-7 cancer cell populations. Br J Cancer. 106:1512–1519. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lapidot T, Sirard C, Vormoor J, Murdoch B,
Hoang T, Caceres-Cortes J, Minden M, Paterson B, Caligiuri MA and
Dick JE: A cell initiating human acute myeloid leukaemia after
transplantation into SCID mice. Nature. 367:645–648. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Al-Hajj M, Wicha MS, Benito-Hernandez A,
Morrison SJ and Clarke MF: Prospective identification of
tumorigenic breast cancer cells. Proc Natl Acad Sci USA.
100:3983–3988. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Charafe-Jauffret E, Ginestier C, Iovino F,
Wicinski J, Cervera N, Finetti P, Hur MH, Diebel ME, Monville F,
Dutcher J, et al: Breast cancer cell lines contain functional
cancer stem cells with metastatic capacity and a distinct molecular
signature. Cancer Res. 69:1302–1313. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hermann PC, Huber SL, Herrler T, Aicher A,
Ellwart JW, Guba M, Bruns CJ and Heeschen C: Distinct populations
of cancer stem cells determine tumor growth and metastatic activity
in human pancreatic cancer. Cell Stem Cell. 1:313–323. 2007.
View Article : Google Scholar
|
|
11
|
Li C, Lee CJ and Simeone DM:
identification of human pancreatic cancer stem cells. Methods Mol
Biol. 568:161–173. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fang D, Nguyen TK, Leishear K, Finko R,
Kulp AN, Hotz S, Van Belle PA, Xu X, Elder DE and Herlyn M: A
tumorigenic subpopulation with stem cell properties in melanomas.
Cancer Res. 65:9328–9337. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kim CF, Jackson EL, Woolfenden AE,
Lawrence S, Babar I, Vogel S, Crowley D, Bronson RT and Jacks T:
Identification of bronchioalveolar stem cells in normal lung and
lung cancer. Cell. 121:823–835. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu G, Yuan X, Zeng Z, Tunici P, NG H,
Abdulkadir IR, Lu L, Irvin D, Black KL and Yu JS: Analysis of gene
expression and chemoresistance of CD133+ cancer stem
cells in glioblastoma. Mol Cancer. 5:672006. View Article : Google Scholar
|
|
15
|
Gilbertson RJ and Rich JN: Making a
tumour's bed: Glioblastoma stem cells and the vascular niche. Nat
Rev Cancer. 7:733–736. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Collins AT, Berry PA, Hyde C, Stower MJ
and Maitland NJ: prospective identification of tumorigenic prostate
cancer stem cells. Cancer Res. 65:10946–10951. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ricci-Vitiani L, Lombardi DG, Pilozzi E,
Biffoni M, Todaro M, Peschle C and De Maria R: Identification and
expansion of human colon-cancer-initiating cells. Nature.
445:111–115. 2007. View Article : Google Scholar
|
|
18
|
O'Brien CA, Pollett A, Gallinger S and
Dick JE: A human colon cancer cell capable of initiating tumour
growth in immunodeficient mice. Nature. 445:106–110. 2007.
View Article : Google Scholar
|
|
19
|
Singh SK, Clarke ID, Terasaki M, Bonn VE,
Hawkins C, Squire J and Dirks PB: identification of a cancer stem
cell in human brain tumors. Cancer Res. 63:5821–5828.
2003.PubMed/NCBI
|
|
20
|
Singh SK, Hawkins C, Clarke ID, Squire JA,
Bayani J, Hide T, Henkelman RM, Cusimano MD and Dirks PB:
identification of human brain tumour initiating cells. Nature.
432:396–401. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dragu DL, Necula LG, Bleotu C, Diaconu CC
and Chivu-Economescu M: Therapies targeting cancer stem cells:
Current trends and future challenges. World J Stem Cells.
7:1185–1201. 2015.PubMed/NCBI
|
|
22
|
Chen K, Huang YH and Chen JL:
Understanding and targeting cancer stem cells: Therapeutic
implications and challenges. Acta Pharmacol Sin. 34:732–740. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Touil Y, Igoudjil W, Corvaisier M, Dessein
AF, Vandomme J, Monté D, Stechly L, Skrypek N, Langlois C, Grard G,
et al: Colon cancer cells escape 5FU chemotherapy-induced cell
death by entering stemness and quiescence associated with the
c-yes/ YAP axis. Clin Cancer Res. 20:837–846. 2014. View Article : Google Scholar
|
|
24
|
Islam F, Gopalan V, Smith RA and Lam AK:
Translational potential of cancer stem cells: A review of the
detection of cancer stem cells and their roles in cancer recurrence
and cancer treatment. Exp Cell Res. 335:135–147. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fang DD, Kim YJ, Lee CN, Aggarwal S,
McKinnon K, Mesmer D, Norton J, Birse CE, He T, Ruben SM, et al:
Expansion of CD133+ colon cancer cultures retaining stem
cell properties to enable cancer stem cell target discovery. Br J
Cancer. 102:1265–1275. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li Z: CD133: A stem cell biomarker and
beyond. Exp Hematol Oncol. 2:172013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shmelkov SV, Butler JM, Hooper AT, Hormigo
A, Kushner J, Milde T, St Clair R, Baljevic M, White I, Jin DK, et
al: CD133 expression is not restricted to stem cells, and both
CD133+ and CD133− metastatic colon cancer
cells initiate tumors. J Clin Invest. 118:2111–2120.
2008.PubMed/NCBI
|
|
28
|
Weiswald LB, Bellet D and Dangles-Marie V:
Spherical cancer models in tumor biology. Neoplasia. 17:1–15. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Qureshi-Baig K, Ullmann P, Rodriguez F,
Frasquilho S, Nazarov PV, Haan S and Letellier E: What do we learn
from spheroidculture systems? Insights from tumorspheres derived
from primary colon cancer tissue. PLoS One. 11:e01460522016.
View Article : Google Scholar
|
|
30
|
Collura A, Marisa L, Trojan D, Buhard O,
Lagrange A, Saget A, Bombled M, Méchighel P, Ayadi M, Muleris M, et
al: Extensive characterization of sphere models established from
colorectal cancer cell lines. Cell Mol Life Sci. 70:729–742. 2013.
View Article : Google Scholar
|
|
31
|
Weiswald LB, Richon S, Massonnet G,
Guinebretière JM, Vacher S, Laurendeau I, Cottu P, Marangoni E,
Nemati F, Validire P, et al: A short-term colorectal cancer sphere
culture as a relevant tool for human cancer biology investigation.
Br J Cancer. 108:1720–1731. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chandrasekaran S, Marshall JR, Messing JA,
Hsu JW and King MR: TRAIL-mediated apoptosis in breast cancer cells
cultured as 3D spheroids. PLoS One. 9:e1114872014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Endo H, Okami J, Okuyama H, Kumagai T,
Uchida J, Kondo J, Takehara T, Nishizawa Y, Imamura F, Higashiyama
M, et al: Spheroid culture of primary lung cancer cells with
neuregulin 1/Her3 pathway activation. J Thorac Oncol. 8:131–139.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tong JG, Valdes YR, Barrett JW, Bell JC,
Stojdl D, McFadden G, McCart JA, DiMattia GE and Shepherd TG:
Evidence for differential viral oncolytic efficacy in an in vitro
model of epithelial ovarian cancer metastasis. Mol Ther Oncolytics.
2:150132015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hirschhaeuser F, Menne H, Dittfeld C, West
J, Mueller-Klieser W and Kunz-Schughart LA: Multicellular tumor
spheroids: An underestimated tool is catching up again. J
Biotechnol. 148:3–15. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Nath S and Devi GR: Three-dimensional
culture systems in cancer research: Focus on tumor spheroid model.
Pharmacol Ther. 163:94–108. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lee SH, Hong JH, Park HK, Park JS, Kim BK,
Lee JY, Jeong JY, Yoon GS, Inoue M, Choi GS, et al: Colorectal
cancer-derived tumor spheroids retain the characteristics of
original tumors. Cancer Lett. 367:34–42. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Stankevicius V, Kunigenas L, Stankunas E,
Kuodyte K, Strainiene E, Cicenas J, Samalavicius NE and Suziedelis
K: The expression of cancer stem cell markers in human colorectal
carcinoma cells in a microenvironment dependent manner. Biochem
Biophys Res Commun. 484:726–733. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Vermeulen L, Todaro M, de Sousa Mello F,
Sprick MR, Kemper K, perez Alea M, Richel DJ, Stassi G and Medema
JP: Single-cell cloning of colon cancer stem cells reveals a
multi-lineage differentiation capacity. Proc Natl Acad Sci USA.
105:13427–13432. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Calvet CY, André FM and Mir LM: The
culture of cancer cell lines as tumorspheres does not
systematically result in cancer stem cell enrichment. PLoS One.
9:e896442014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Botchkina IL, Rowehl RA, Rivadeneira DE,
Karpeh MS Jr, Crawford H, Dufour A, Ju J, Wang Y, Leyfman Y and
Botchkina GI: Phenotypic subpopulations of metastatic colon cancer
stem cells: Genomic analysis. Cancer Genomics Proteomics. 6:19–29.
2009.PubMed/NCBI
|
|
42
|
Shaheen S, Ahmed M, Lorenzi F and Nateri
AS: Spheroid-formation (Colonosphere) assay for in vitro sssessment
and expansion of stem cells in colon cancer. Stem Cell Rev.
12:492–499. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yeung TM, Gandhi SC, Wilding JL, Muschel R
and Bodmer WF: Cancer stem cells from colorectal cancer-derived
cell lines. Proc Natl Acad Sci USA. 107:3722–3727. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Dittfeld C, Dietrich A, Peickert S, Hering
S, Baumann M, Grade M, Ried T and Kunz-Schughart LA: CD133
expression is not selective for tumor-initiating or radioresistant
cell popu-lations in the CrC cell lines HCT-116. Radiother Oncol.
92:353–361. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Huang R, Wang G, Song Y, Tang Q, You Q,
Liu Z, Chen Y, Zhang Q, Li J, Muhammand S, et al: Colorectal cancer
stem cell and chemoresistant colorectal cancer cell phenotypes and
increased sensitivity to Notch pathway inhibitor. Mol Med Rep.
12:2417–2424. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Fan X, Ouyang N, Teng H and Yao H:
isolation and characterization of spheroid cells from the HT29
colon cancer cell line. Int J Colorectal Dis. 26:1279–1285. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Pang C, Xie J, Guo J, Manning HC, Gore JC
and Guo N: Evaluation of CD44 and CD133 as cancer stem cell markers
for colorectal cancer. Oncol Rep. 28:1301–1308. 2012. View Article : Google Scholar
|
|
48
|
Chan CW, Wong NA, Liu Y, Bicknell D,
Turley H, Hollins L, Miller CJ, Wilding JL and Bodmer WF:
Gastrointestinal differentiation marker Cytokeratin 20 is regulated
by homeobox gene CDX1. Proc Natl Acad Sci USA. 106:1936–1941. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Ning Y, Hanna DL, Zhang W, Mendez A, Yang
D, El-Khoueiry R, Matsusaka S, Sunakawa Y, Stremitzer S, Parekh A,
et al: Cytokeratin-20 and survivin-expressing circulating tumor
cells predict survival in metastatic colorectal cancer patients by
a combined immunomagnetic qRT-PCR approach. Mol Cancer Ther.
14:2401–2408. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Imai Y, Yamagishi H, Fukuda K, Okamura T,
Ono Y, Ban S, Inoue T and Ueda Y: Expression of cytokeratin 20
indicates invasive histological phenotype in poorly differentiated
colorectal adenocarcinoma. Anticancer Res. 34:159–167.
2014.PubMed/NCBI
|
|
51
|
Yanai H, Atsumi N, Tanaka T, nakamura N,
Komai Y, Omachi T, Tanaka K, ishigaki K, Saiga K, Ohsugi H, et al:
Intestinal cancer stem cells marked by Bmi1 or Lgr5 expression
contribute to tumor propagation via clonal expansion. Sci Rep.
7:418382017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Shimokawa M, Ohta Y, Nishikori S, Matano
M, Takano A, Fujii M, Date S, Sugimoto S, Kanai T and Sato T:
visualization and targeting of LGR5+ human colon cancer
stem cells. Nature. 545:187–192. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Hirsch D, Barker N, Mcneil N, Hu Y, Camps
J, McKinnon K, Clevers H, ried T and Gaiser T: LGr5 positivity
defines stem-like cells in colorectal cancer. Carcinogenesis.
35:849–858. 2014. View Article : Google Scholar :
|
|
54
|
de Sousa E, Melo F, Kurtova AV, Harnoss
JM, Kljavin N, Hoeck JD, Hung J, Anderson je, Storm EE, Modrusan Z,
Koeppen H, et al: A distinct role for LGR5+ stem cells in primary
and metastatic colon cancer. Nature. 543:676–680. 2017. View Article : Google Scholar
|
|
55
|
Walker F, Zhang HH, Odorizzi A and Burgess
AW: LGR5 is a negative regulator of tumourigenicity, antagonizes
Wnt signal-ling and regulates cell adhesion in colorectal cancer
cell lines. PLoS One. 6:e227332011. View Article : Google Scholar
|
|
56
|
Parvathi MV, Murthy PB, Vennila M and
Suresh BV: Regulation of BMI1 Polycomb gene expression in
histological grades of invasive ductal breast carcinomas and its
correlation with hormone receptor status. Tumour Biol.
34:3807–3815. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Peng HX, Liu XD, Luo ZY, Zhang XH, Luo XQ,
Chen X, Jiang H and Xu L: Upregulation of the proto-oncogene bmi-1
predicts a poor prognosis in pediatric acute lymphoblastic
leukemia. BMC Cancer. 17:762017. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Li X, Zheng X, Xu B, Zhang D, Xu Y, Xie Q,
Hu W, Zheng Z, Shao Y, Wu J, et al: Lower Bmi-1 expression may
predict longer survival of colon cancer patients. Cell Physiol
Biochem. 39:2421–2426. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Siddique HR and Saleem M: role of bMi1, a
stem cell factor, in cancer recurrence and chemoresistance:
Preclinical and clinical evidences. Stem Cells. 30:372–378. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Yan KS, Chia LA, Li X, Ootani A, Su J, Lee
JY, Su N, Luo Y, Heilshorn SC, Amieva MR, et al: The intestinal
stem cell markers Bmi1 and Lgr5 identify two functionally distinct
populations. Proc Natl Acad Sci USA. 109:466–471. 2012. View Article : Google Scholar :
|
|
61
|
Charafe-Jauffret E, Monville F, Ginestier
C, Dontu G, Birnbaum D and Wicha MS: Cancer stem cells in breast:
Current opinion and future challenges. Pathobiology. 75:75–84.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Kim SY, Hong SH, Basse PH, WU C, Bartlett
DL, Kwon YT and Lee YJ: Cancer stem cells protect non-stem cells
from anoikis: bystander effects. J Cell Biochem. 117:2289–2301.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Sato R, Semba T, Saya H and Arima Y:
Concise review: Stem cells and epithelial-mesenchymal transition in
cancer: Biological implications and therapeutic targets. Stem
Cells. 34:1997–2007. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Merlos-Suárez A, Barriga FM, Jung P,
Iglesias M, Céspedes MV, Rossell D, Sevillano M, Hernando-Momblona
X, da Silva-Diz V, Muñoz P, et al: The intestinal stem cell
signature identifies colorectal cancer stem cells and predicts
disease relapse. Cell Stem Cell. 8:511–524. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Albini A and Noonan DM: The
'chemoinvasion' assay, 25 years and still going strong: The use of
reconstituted basement membranes to study cell invasion and
angiogenesis. Curr Opin Cell Biol. 22:677–689. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Benton G, Arnaoutova I, George J, Kleinman
HK and Koblinski J: Matrigel: From discovery and ECM mimicry to
assays and models for cancer research. Adv Drug Deliv Rev. 79–80.
3–18. 2014.
|
|
67
|
Brattain MG, brattain DE, Fine WD, Khaled
FM, Marks ME, Kimball PM, Arcolano LA and Danbury BH: initiation
and characterization of cultures of human colonic carcinoma with
different biological characteristics utilizing feeder layers of
confluent fibroblasts. Oncodev Biol Med. 2:355–366. 1981.PubMed/NCBI
|
|
68
|
Brattain MG, Fine WD, Khaled FM, Thompson
J and brattain DE: Heterogeneity of malignant cells from a human
colonic carcinoma. Cancer Res. 41:1751–1756. 1981.PubMed/NCBI
|
|
69
|
Fogh J and Trempe G: New human tumor cell
lines. Human Tumor Cells in Vitro. Fogh J: Springer; US, Boston,
MA: pp. 115–159. 1975, View Article : Google Scholar
|
|
70
|
Ahmed D, Eide PW, Eilertsen IA, Danielsen
SA, Eknæs M, Hektoen M, Lind GE and Lothe RA: epigenetic and
genetic features of 24 colon cancer cell lines. Oncogenesis.
2:e712013. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Aksamitiene E, Kiyatkin A and Kholodenko
BN: Cross-talk between mitogenic RAS/MAPK and survival PI3K/Akt
pathways: A fine balance. Biochem Soc Trans. 40:139–146. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Zenonos K and Kyprianou K: RAS signaling
pathways, mutations and their role in colorectal cancer. World J
Gastrointest Oncol. 5:97–101. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Bazan V, Migliavacca M, Zanna I, Tubiolo
C, Grassi N, Latteri MA, La Farina M, Albanese I, Dardanoni G,
Salerno S, et al: Specific codon 13 K-ras mutations are predictive
of clinical outcome in colorectal cancer patients, whereas codon 12
K-ras mutations are associated with mucinous histotype. Ann Oncol.
13:1438–1446. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Knickelbein K and Zhang L: Mutant KRAS as
a critical deter-minant of the therapeutic response of colorectal
cancer. Genes Dis. 2:4–12. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Raats DA, Frenkel N, van Schelven SJ,
Rinkes IH, Laoukili J and Kranenburg O: CD95 ligand induces
senescence in mismatch repair-deficient human colon cancer via
chronic caspase-medi-ated induction of DNA damage. Cell Death Dis.
8:e26692017. View Article : Google Scholar
|
|
76
|
Szarynska M, Olejniczak A, Wierzbicki P,
Kobiela J, Laski D, Sledzinski Z, Adrych K, Guzek M and Kmiec Z:
Fasr and FasL in colorectal cancer. Int J Oncol. 51:975–986.
2017.PubMed/NCBI
|
|
77
|
Therkildsen C, Bergmann TK,
Henrichsen-Schnack T, Ladelund S and Nilbert M: The predictive
value of KRAS, NRAS, BRAF, PIK3CA and PTEN for anti-EGFR treatment
in metastatic colorectal cancer: A systematic review and
meta-analysis. Acta Oncol. 53:852–864. 2014. View Article : Google Scholar : PubMed/NCBI
|