Introduction
Pancreatic cancer (PC) is an extremely aggressive
malignant tumor and the fourth leading cause of cancer-related
mortality in the USA, accounting for an estimated 57,600 new cases
and 47,050 related deaths each year (1). It has been also predicted to become
the second leading cause of cancer-related mortality in the ensuing
20-30 years (2). Moreover, the
incidence of PC tends to increase further as the population ages
(2). Due to its aggressive nature
and early metastatic behavior, the majority of patients with PC
exhibit a poor prognosis and resistance to most treatment
strategies, including chemotherapy, radiotherapy and immunotherapy.
Several studies have demonstrated a genomic, epigenetic and
metabolic remodeling that occurs during the onset and progression
of PC, leading to the complexities of tumor tissue, stromal tissue,
and the tumor-related immune system and their crosstalk (3,4).
Thus, an improved understanding of the underlying mechanisms
regulating PC tumorigenesis and tumor development would contribute
to the prevention of and therapeutic improvements for PC.
Long non-coding RNAs (lncRNAs) are a novel and
abundant type of transcripts, >200 nucleotides in length,
although with a limited protein-coding potential (5). lncRNAs are novel regulators of
oncogenesis, exerting oncogenic and tumor-suppressive effects in
various tumors (6-9). Acting as signals, decoys, guides, and
scaffolds, lncRNAs have been reported to form complex networks, in
order to modulate multiple cancer phenotypes. Furthermore, they may
regulate target gene expression and interact with various cellular
components, including DNAs, RNAs and proteins (10,11).
For instance, Gandhi et al (12) demonstrated that lncRNA lincNMR
regulated nucleotide metabolism through its binding to YBX1 protein
and affecting the key dNTP synthesizing enzymes RRM2, TYMS and TK1.
Additionally, the downregulation of lincNMR was found to decreased
dNTP levels and cell proliferation, and induce senescence in
multiple cancer types (12). In
another study, Hu et al (8)
revealed that the lncRNA XLOC-000647 downregulated the expression
of its adjacent genes associated with the NLRP3 inflammasome in
cis. High XLOC-000647 expression levels may markedly
attenuate PC cell proliferation. With advancements being made in
transcriptome-sequencing and functional genomics analysis, a number
of previously unsuspected lncRNAs have been identified in PC,
providing new perspectives towards the further elucidation of PC
pathogenesis (13). However, the
biological functions of the majority of lncRNAs remain largely
unknown.
Hence, the present study was designed to examine the
expression patterns and functions of lncRNAs in PC. Bioinformatics
analysis was first performed on publicly available genomic
databases to recognize lncRNAs aberrantly expressed in PC. It was
identified that linc01614 was upregulated in PC and thus it was
subjected to further investigations. Functionally, linc01614
significantly promoted the progression of PC in vitro and
in vivo. Moreover, RNA pull-down and co-immunoprecipitation
(Co-IP) assays indicated that linc01614 combined with GSK-3β to
perturb the interaction between GSK-3β and AXIN1, activated the
WNT/β-catenin signaling pathway and consequently enhanced β-catenin
protein expression levels. Thus, the results of the present study
revealed that linc01614 may be used as a candidate therapeutic
target for PC.
Materials and methods
Patients and samples
In total, 20 formalin-fixed and paraffin-embedded PC
and normal cancer-adjacent pancreatic tissues were retrieved
between September, 2020 to September, 2021 from the Pathological
Department of Renmin Hospital of Wuhan University for fluorescent
in situ hybridization (FISH) analysis. The median age of the
patients was 66.5 years (range, 49-79 years). The
clinicopathological parameters are summarized in Table SI. The present study was approved
by the Ethics Committee and the Human Research Review Committee of
Renmin Hospital of Wuhan University (approval no. WDRY2021-K188),
and written informed consent was obtained from all subjects.
Culture and transfection of cell
lines
The PC cell lines, Panc-1 (cat. no. CRL-1469),
SW1990 (cat. no. CRL-2172), BXPC-3 (cat. no. CRL-1687) and MIA-PaCa
(cat. no. CRL-1420), were purchased from the American Type Culture
Collection (ATCC). HPDE cells were obtained from Hunan Fenghui
Biotechnology Co., Ltd. (cat. no. CL0317). All the cells were
cultured according to the manufacturer's instructions. The cell
lines were authenticated by DNA fingerprinting and checked for
mycoplasma contamination (data not shown). The cells were cultured
in Dulbecco's modified Eagle's medium (DMEM; HyClone, Cytiva)
supplemented with 10% fetal bovine serum (Tianhang Biotechnology
Co., Ltd.). All cell lines were cultured in a 5% CO2 and
95% humidity incubator at 37°C.
Antisense oligonucleotides (ASOs) targeting
linc01614 and control ASO were obtained from RiboBio, Inc.
Transfection of ASOs was conducted using Neofect™
transfection Reagent (Neofect Biotech Co., Ltd.) with the final ASO
concentration of 70 nM. The ASO sequences are included in Table SII. To overexpress linc01614, the
cDNA was designed, synthesized and then cloned into GV658 vector
(Genchem, Inc.). 2 µg linc01614 plasmid, and corresponding
empty vector were transfected into HPDE cells using Neofect™
transfection Reagent (Neofect Biotech Co., Ltd.). The cells were
then cultured in a 5% CO2-humidified incubator at 37°C.
After 48 h, the cells were employed to perform subsequent
experimentation.
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from the cultured cells
using TRIzol reagent (Takara Bio, Inc.). All lncRNAs and mRNAs were
reverse transcribed with random and oligo dT primers using the
reverse transcription kit (cat. no. RR037A; Takara Bio, Inc.). The
reaction conditions were as follows: 37°C for 15 min and 85°C for 5
sec. Subsequently, qPCR was conducted using the TB green Premix Ex
Taq™ II (cat. no. RR820A; Takara Bio, Inc.) on a Bio-Rad 7500 PCR
system (Bio-Rad Laboratories, Inc.). The reaction conditions were
as follows: Initial denaturation at 95°C for 30 sec, followed by 40
cycles of amplification at 95°C for 5 sec and 60°C for 30 sec, and
melt curve analysis at 95°C for 10 sec and 65°C for 5 sec. GAPDH or
U6 were used as reference genes. The qPCR results were calculated
using the 2ΔΔCq method (14). The primers used are presented in
Table SIII. The experiments were
performed in triplicate.
RNA FISH
A fluorescent in situ hybridization kit
(RiboBio, Inc.) was used to conduct the FISH assay. Briefly, the
Panc-1 and SW1990 cells were fixed using 4% paraformaldehyde
(Biosharp, Inc.) and washed three times using phosphate-buffered
saline (PBS). The cells were then incubated with Cy3-labeled
linc01614 RNA probes (RiboBio, Inc.) in a hybridization buffer
overnight at 37°C. The nucleus of cells was stained with
4′,6-diamidino-2-phenylindole (DAPI) (RiboBio, Inc.) for 10 min at
room temperature. The stainings were observed using a confocal
microscope (Olympus FV1200; Olympus Corporation).
For PC tissues, lncRNA FISH on paraffin tissue
sections with linc01614 probes was also performed using the same
hybridization kit (RiboBio, Inc.). The sections were deparaffinized
and rehydrated using xylene and a graded alcohol solution series.
Following treatment with protease K (Biosharp, Inc.), the sections
were incubated with Cy3-labeled linc01614 probes in hybridization
buffer at 37°C overnight. Cell nuclei were stained with DAPI.
Images were acquired with the use of an Olympus fluorescence
microscope (Olympus IX71, Olympus Corporation).
Subcellular fractionation
Cytoplasmic and nuclear fractions of Panc-1 and
SW1990 cells were isolated using the PARIS Kit (Invitrogen; Thermo
Fischer Scientific, Inc.). The expression levels of mRNAs and
proteins isolated from cytoplasmic and nuclear fractions were
measured. GAPDH was used as the control for cytoplasmic expression
in RT-qPCR and western blot assays. U6 and LaminB were used as the
controls for nuclear expression in RT-qPCR and western blot
analysis, respectively.
CCK-8 and EdU assay
Cell proliferation was assessed using a Cell
Counting Kit-8 (CCK-8; MCE, Inc.) and EdU Cell Proliferation Kit
(Epizyme, Inc.). In the CCK-8 assay, Panc-1 and SW1990 cells were
seeded on 96-well plates at a density of 2,000 cells/well, and were
then treated with 10 µl CCK-8 solution from day 1 to 5.
Following incubation for 2 h at 37°C, the absorbance was determined
at 450 nm using a microplate reader (HBS-1096A, DeTie Laboratory
Equipment Co., Ltd.). For the EdU assay, transfected cells were
seeded on 6-well plates, and 10 µM EdU were added to the
well. Following incubation for 2 h at 37°C, all cells were fixed
with 4% paraformaldehyde (Biosharp, Inc.), washed three times with
PBS containing 3% BSA, penetrated with PBS containing 0.3% Triton
X-100, washed three times with PBS containing 3% BSA, and 500
µl Click Additive Solution (Epizyme, Inc.) was then added
for 30 min at room temperature in the dark. Finally, the nucleus of
the cells was stained with Hoechst 33342 (Epizyme, Inc.) for 10 min
at room temperature in the dark. Images were acquired using an
Olympus fluorescence microscope (Olympus IX71, Olympus
Corporation).
Colony formation assay
Panc-1 and SW1990 cells were seeded in 6-well plates
at a concentration of 1,000 cells per well and cultured in DMEM
medium supplemented with 10% fetal bovine serum. Following a 2-week
incubation at 37°C, all cells were washed twice with PBS (Cienry,
Inc.), fixed with 4% paraformaldehyde (Biosharp, Inc.), and stained
with 0.1% crystal violet (Biosharp, Inc.) for 30 min. Finally, the
colonies were imaged using a camera (Redmi 10×4G, Xiaomi
Corporation) and counted using Image J software (National
Institutes of Health).
Transwell assay
Cell migration and invasion were assessed using the
Transwell assay. The upper chambers of the Transwells (Corning,
Inc.) were coated with 1:8 diluted Matrigel (BD Biosciences) or
without Matrigel for invasion and migration, respectively. A total
of 5×104 cells (Panc-1 and SW1990) was plated in the
upper chambers with 100 µl serum-free medium, while the
lower chambers were plated with 600 µl DMEM medium (HyClone,
Cytiva), containing 15% fetal bovine serum (Tianhang Biotechnology
Co., Ltd.). Following incubation for 24-48 h at 37°C, the cells on
the upper surface were cleared using a cotton swab, and the cells
on the lower surface were fixed with 4% paraformaldehyde, stained
with 0.1% crystal violet (Biosharp, Inc.) for 30 min. The results
were examined and photographed using an Olympus microscope (Olympus
IX71, Olympus Corporation).
Wound healing assay
Panc-1 and SW1990 cells were seeded in 6-well plates
and cultured in DMEM supplemented with 1.5% fetal bovine serum.
When the cells reached 90% confluency, the cell wound was then
generated utilizing a 10 µl pipette tip. The percentage of
wound closure [(original width-width after cell migration)/original
width] was examined and photographed at 0 and 48 h (Olympus IX71,
Olympus Corporation).
Flow cytometry
For the analysis of the cell cycle, Panc-1 and
SW1990 cells were harvested, washed with PBS, stained with binding
buffer containing 1 ml DNA staining solution and 10 µl
permeabilization solution, and incubated at room temperature for 30
min in the dark according to the manufacturer's instructions
(MultiSciences Biotech, Co., Ltd.). For the analysis of cell
apoptosis, cells were collected, washed in PBS, and resuspended in
a 500 µl mixture of 5 µl Annexin V-FITC (Annexin
V-PE) and 10 µl propidium iodide (7-AAD; MultiSciences
Biotech, Co., Ltd.). Modfit (Verity Software House, Inc.) and
FlowJo V10 (TreeStar, Inc.) software were employed to analyze the
cell cycle and apoptosis data, respectively.
Western blot analysis
Total protein was extracted from Panc-1 and SW1990
cells using RIPA lysis buffer (Beyotime Institute of
Biotechnology). Protein concentrations were determined using the
BCA Protein Assay kit (Beyotime Institute of Biotechnology).
Subsequently, 20 µg proteins from each sample were loaded,
separated by 10% SDS-PAGE gels, and then transferred onto a
polyvinylidene difluoride membrane (MilliporeSigma). After being
blocked with Protein Free Rapid Blocking Buffer (Epizyme, Inc.) for
30-60 min at room temperature, the membrane was incubated with the
following primary antibodies: Cyclin D1 (cat. no. WL01345a; 1:500),
CDK2 (cat. no. WL01543; 1:1,500), E-cadherin (cat. no. WL01482;
1:1,000), N-cadherin (cat. no. WL01047; 1:1,000), Snail (cat. no.
WL01863; 1:1,000), twist-related protein 1 (Twist1; cat. no.
WL00997; 1:500), β-catenin (cat. no. WL00962; 1:1,000), adenomatous
polyposis coli (APC; cat. no. WL02422; 1:500) and LaminB (cat. no.
WL01775; 1:1,000) (all from WanleiBio, Co., Ltd.); GSK-3β (cat. no.
22104-1-AP; 1:2,000) and AXIN1 (cat. no. 16541-1-AP; 1:500) from
ProteinTech Group, Inc.; active β-catenin (cat. no. 8814, 1:1,000)
from Cell Signaling Technology, Inc.; Vimentin (cat. no. GB111308;
1:1,000) and β-actin (cat. no. GB11001; 1:1,000) from Servicebio
Technology Co., Ltd.; and GAPDH (cat. no. ABPR001; 1:2,000) from
Goodhere, Inc. (https://www.goodhere.com/kysj/) at 4°C overnight. On
the second day, the membrane was washed three times using
Tris-buffered saline with 0.1% Tween (TBST) and incubated with a
goat anti-rabbit secondary antibody (cat. no. G1213; 1:3,000;
Servicebio Technology Co., Ltd.;) for 1 h at room temperature.
Finally, the signals of the immunoreactive protein bands were
detected with a Biosharp ECL detection kit (Biosharp, Inc.). The
WNT/β-catenin activator LiCl (Sigma-Aldrich; Merck KGaA) was used
to further investigate the association between linc01614 and
WNT/β-catenin pathway in PC cells. PC cells were transfected with
antisense oligonucleotide (ASO)-NC or ASO-1 for 24 h and then
treated with 20 mM LiCl for 24 h. Data were analyzed using ImageJ
software (v. 1.46; National Institutes of Health).
Xenograft tumors
A total of 10 male BALB/c-nu mice (aged 4 weeks)
were purchased from the Hunan SJA Laboratory Animal Center. All
mice were housed under specific-pathogen-free conditions
(temperature, 25°C; humidity, 50%; light/dark cycle, 12/12 h
cycle), with food and water ad libitum. Animal health and
behavior were monitored every day. Panc-1 tumor cells
(1×107) were suspended in 200 µl of sterile PBS
and subcutaneously injected into the right flanks of mice. After
the tumors were palpable, xenograft mice were grouped randomly as
negative control (n=5) and ASO-1 group (n=5). Then, 5 nM ASO-NC or
ASO-1 were injected intratumorally every 3 days for 4 times in
total. The tumor size was measured using a caliper every week.
Tumor volume was estimated as (length x width2)/2. After
3 weeks, the mice were euthanized using 1% pentobarbital sodium (50
mg/kg, intraperitoneal) followed by cervical dislocation. Death was
verified by the cessation of respiration and heartbeat, and the
absence of reflexes. Subsequently, the tumors were isolated and
maintained in liquid nitrogen for further analysis. All experiments
were conducted according to the relevant guidelines and regulations
and following the approval of the Institutional Animal Care
Committee of Renmin Hospital of Wuhan University (approval no.
WDRM20210606). The humane endpoints of the experiment were as
follows: i) A significant increase in tumor burden (≥10% body
weight); ii) weight loss ≥20% body weight; iii) occurrence of
ulceration or infection at the site of tumor and surrounding
tissue; iv) no movement for >24 h; v) no eating or drinking. In
this experiment, no animal reached the humane endpoints and none of
the mice were found dead.
Immunohistochemistry
Paraffin-embedded tissue sections were
deparaffinized and rehydrated using xylene and a graded alcohol
solution series as follows: Xylene I for 30 min, xylene II for 10
min, 100% ethanol I for 10 min, 100% ethanol II for 3 min, 90%
ethanol for 3 min, 70% ethanol for 3 min and H2O for 3
min. Subsequently, antigen retrieval was performed using microwave
heating in 10 mM citrate buffer (Servicebio Technology Co., Ltd.).
Endogenous peroxidase was blocked with 3%
H2O2. Subsequently, the sections were
incubated with primary antibodies specific for proliferating cell
nuclear antigen (PCNA; ProteinTech Group, Inc.; cat. no.
10205-2-AP; 1:200), Ki67 (ProteinTech Group, Inc.; cat. no.
27309-1-AP; 1:200) and β-catenin (Wanleibio, Co., Ltd.; cat.no.
WL00962; 1:50) at 4°C overnight. On the second day, the slides were
incubated with a goat anti-rabbit secondary antibody (1:200; cat.
no. G1213; Servicebio Technology Co., Ltd.) for 30 min at room
temperature. The results were assessed independently by two
pathologists, and photographed using an Olympus microscope (Olympus
BX51 TRF, Olympus Corporation). The staining intensity was scored
from 0 to 3, and the proportion of staining ranged from 0 to 4. The
immunoreactive score was obtained by multiplying the proportion and
intensity scores. A total score higher than 4 was considered a high
expression (15). ImageJ software
(v. 1.46; National Institutes of Health) was used to analyze the
relative protein expression.
TOP/FOP flash assay
In brief, the cells were added to a 24-well plate
and transfected with ASO-NC or ASO-linc01614 in the presence of TOP
Flash or FOP Flash plasmid and Renilla luciferase reporter
plasmid (Beyotime Institute of Biotechnology) using a Neofect™
transfection Reagent (Neofect Biotech Co., Ltd.). Following 48 h of
incubation at 37°C, luciferase intensity was detected utilizing a
Dual-Luciferase Reporter Assay System (GloMax™; Promega
Corporation). Finally, the relative ratio of TOP flash value/FOP
flash value was calculated and used as an indicator of the
activation of the WNT/β-catenin signaling pathway.
RNA pull-down assay
Biotin-labeled lncRNAs were transcribed and purified
using the T7 Biotin Labeling Transcription kit (Invitrogen; Thermo
Fischer Scientific, Inc.). Subsequently, biotinylated RNA was mixed
with the cell lysates for 4 h and incubated with streptavidin
agarose magnetic beads (cat. no. G1213; Thermo Fisher Scientific,
Inc.) for 1 h at room temperature. Finally, the purified
RNA-proteins complexes were washed, resolved and subjected to mass
spectrometry or western blot analysis. For mass spectrometry, the
gel piece was washed, in-gel digested, and the peptides were
extracted. The peptides were then selected using an autosampler and
transferred into a C18 analytical column for separation. For DDA
mode analysis, each scan cycle consisted of one full-scan mass
spectrum followed by 40 MS/MS events. Former target ion exclusion
was set for 12 sec. LC-MS/MS analysis was performed on a
quadrupole-TOF LC-MS/MS mass spectrometer (TripleTOF 5600 plus,
SCIEX) coupled with an Ekspert 400 MicroLC system. Raw data were
analyzed using MaxQuant (V1.6.6) using the Andromeda database
search algorithm.
RNA immunoprecipitation (RIP) assay
RIP assay was conducted using an RNA
Immunoprecipitation kit (Geneseed Biotech Co., Ltd.). Panc-1 and
SW1990 cells were harvested and lysed using RIP lysis buffer.
Subsequently, the whole lysates were incubated with magnetic beads
conjugated to 5 µg anti-GSK-3β antibodies (cat. no.
22104-1-AP; ProteinTech Group, Inc.), or IgG (GB111738; Servicebio
Technology Co., Ltd.) at 4°C overnight. The immunoprecipitated RNAs
were isolated and subjected to RT-qPCR analysis.
Co-IP assay
Co-IP assay was conducted using an
Immunoprecipitation kit (cat. no. ab206996; Abcam Co., Ltd.). PC
cells were harvested and lysed using immunoprecipitation lysis
buffer. Subsequently, cell extracts were incubated with the 2
µg indicated antibodies: GSK-3β (cat. no. 22104-1-AP;
ProteinTech Group, Inc.) and AXIN1 (cat. no. 2087S; Cell Signaling
Technology, Inc.) at 4°C overnight, followed by incubation with
Protein A/G magnetic beads for 4 h. Finally, the immunoprecipitates
were collected and examined by western blot analysis.
Bioinformatics and statistical
analysis
The expression and survival analyses for lncRNAs in
PC were obtained from The Cancer Genome Atlas (TCGA; https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga)
and Genotype-Tissue Expression project (GTEx; https://www.genome.gov/Funded-Programs-Projects/Genotype-Tissue-Expression-Project).
TGCA and GTEx datasets were analyzed using the R software. The
t-distributed stochastic neighbor embedding (t-SNE) and pan-cancer
analysis were conducted using an online tool Sangerbox (http://sangerbox.com). Gene set enrichment analysis
was performed using the GSEA software (v.4.0; Broad Institute,
Inc.). The RPISeq (http://pridb.gdcb.iastate.edu/RPISeq) and catRAPID
(http://service.tartaglialab.com)
databases were employed to predict the RNA-Protein interaction.
Experiments were performed in triplicate. The results were analyzed
using the SPSS 17.0 (SPSS, Inc.) or GraphPad Prism 8.0 (GraphPad
Software, Inc.) software and represented as percentages or mean ±
standard deviation. The Kaplan-Meier method was employed to
evaluate the overall survival (OS) and disease-free survival (DFS).
The two-tailed unpaired Student's t-test was employed to analyze
the means of two groups. One-way ANOVA was employed to evaluate the
differences among multiple groups. As a post hoc test following
ANOVA, Dunnett's multiple comparisons test was used for comparisons
with only the control group and Tukey's multiple comparisons test
was used for comparisons among multiple pairs of groups. The
association of linc01614 expression with the clinical parameters of
patients with PC was assessed using the Chi-squared test
(χ2). P<0.05 was considered to indicate a
statistically significant difference.
Results
Identification of linc01614 as a
PC-related lncRNA
To explore whether lncRNAs may be potentially
related to PC, a genomic analysis of gene expression data obtained
from the TCGA and GTEx database was conducted and 6,072 lncRNAs
were identified. Upon setting the thresholds to a fold change >2
and a false discovery rate (FDR) <0.01, 207 lncRNAs were found
to be dysregulated in PC, of which 104 were upregulated and 103
downregulated (Fig. 1A and B,
Table SIV). Hierarchical
clustering and t-SNE analysis revealed the notable differentiation
of these lncRNAs between the PC and non-cancerous pancreatic
tissues (Fig. 1C and D).
Subsequently, in order to determine to what extent these lncRNAs
have been previously studied in tumors, articles published in
PubMed up to January 8, 2022 were systematically retrieved. It was
observed that a number of lncRNAs, including UCA1, linc00460,
CASC9, MEG3, NEAT1 and ROCR, have already been studied in cancer
research (Fig. 1E).
linc01614 emerged as one of the prominently
differentially expressed lncRNAs in PC, as compared to
non-cancerous pancreatic tissues, with a log FC of 5.8 and FDR of
2.20E-52. As the authors wished to mainly focus on oncogenic
factors, the present study mainly focused on upregulated lncRNAs,
since oncogenic lncRNAs are preferable therapeutic targets and
diagnostic or prognostic biomarkers. The expression of linc01614
across different tumor types was evaluated. As demonstrated in
Fig. 1F, PC had the second highest
linc01614 expression among various tumor types. Subsequently, the
PubMed database was investigated and it was observed that the
function of linc01614 as a potential biomarker and oncogene in
gastric cancer (16), breast
cancer (17), lung cancer
(18,19), osteosarcoma (20) and esophageal squamous cell
carcinoma (21) has already been
studied. However, the biological roles and mechanisms of action of
linc01614 in PC remain relatively unknown. Additionally,
bioinformatics analysis of online databases predicted that
linc01614 may perform a regulatory function by interacting with
more RNAs and proteins (Table SV,
and Fig. 7B and C). linc01614 is
also closely associated with several signaling pathways, including
WNT/β-catenin, TGF-β, and focal adhesion kinase signaling, thus
suggesting that linc01614 is worthy of investigation in PC
(Fig. 6A). Thus, it was decided to
evaluate the biological functions and mechanisms of linc01614 in PC
with further experiments.
 | Figure 6linc01614 perturbs the WNT/β-catenin
signaling pathway in PC. (A) GSEA analysis suggesting a positive
association between linc01614 and the WNT/β-catenin signaling
pathway in PC. (B) TOP/FOP Flash reporter assay, for the
examination of the WNT signaling pathway activity in Panc-1 and
SW1990 cells following transfection with ASO-NC, ASO-1, or ASO-2.
(C and D) mRNA and protein levels of several vital proteins
involved in the WNT signaling pathway in ASO-NC-, ASO-1-, or
ASO-2-transfected PC cells. (E) Western blot analysis was used to
measure the nuclear and cytoplasmic levels of active β-catenin and
total β-catenin in PC cells transfected with ASO-NC, ASO-1, or
ASO-2. GAPDH and LaminB were used as internal cytoplasmic and
nuclear controls, respectively. (F) Western blot analysis of
β-catenin in normal PC cells or PC cells treated with ASO-NC,
ASO-1, LiCl, ASO-NC + LiCl, and ASO-1 + LiCl. Data are presented as
the mean ± SD. (ns, not significant). One-way ANOVA with Tukey's
test was employed to compare the expression of β-catenin in each
group. *P<0.05, **P<0.01 and
***P<0.001. p GSEA, Gene-set enrichment analysis; PC,
pancreatic cancer; ASO, antisense oligonucleotide; NC, negative
control; SD, standard deviation. |
linc01614 is upregulated in PC and is
associated with poor DFS
The expression pattern of linc01614 was first
investigated using the publicly available RNA-sequencing data from
the GTEx and TCGA databases. The level of linc01614 was higher in
PC tissues than non-tumor tissues (Fig. 2A). Moreover, linc01614 expression
was negatively associated with the DFS rather than OS (Fig. 2B and C). The association between
linc01614 expression and routine clinicopathological parameters was
also evaluated, based on TCGA database. The results did not reveal
any significant association between linc01614 level and the routine
clinicopathological parameters in patients with PC, including age,
sex, tumor size, tumor location, histological grade, tumor, node,
and metastasis (TNM) stage and the number of positive lymph nodes.
The results are summarized in Table
SVI. The expression of linc01614 was then analyzed in a cohort
of 20 cases of formalin-fixed paraffin-embedded PC tissues and
adjacent non-cancerous tissues using RNA FISH analysis. An
increased level of linc01614 in PC tissues was observed (Fig. 2D).
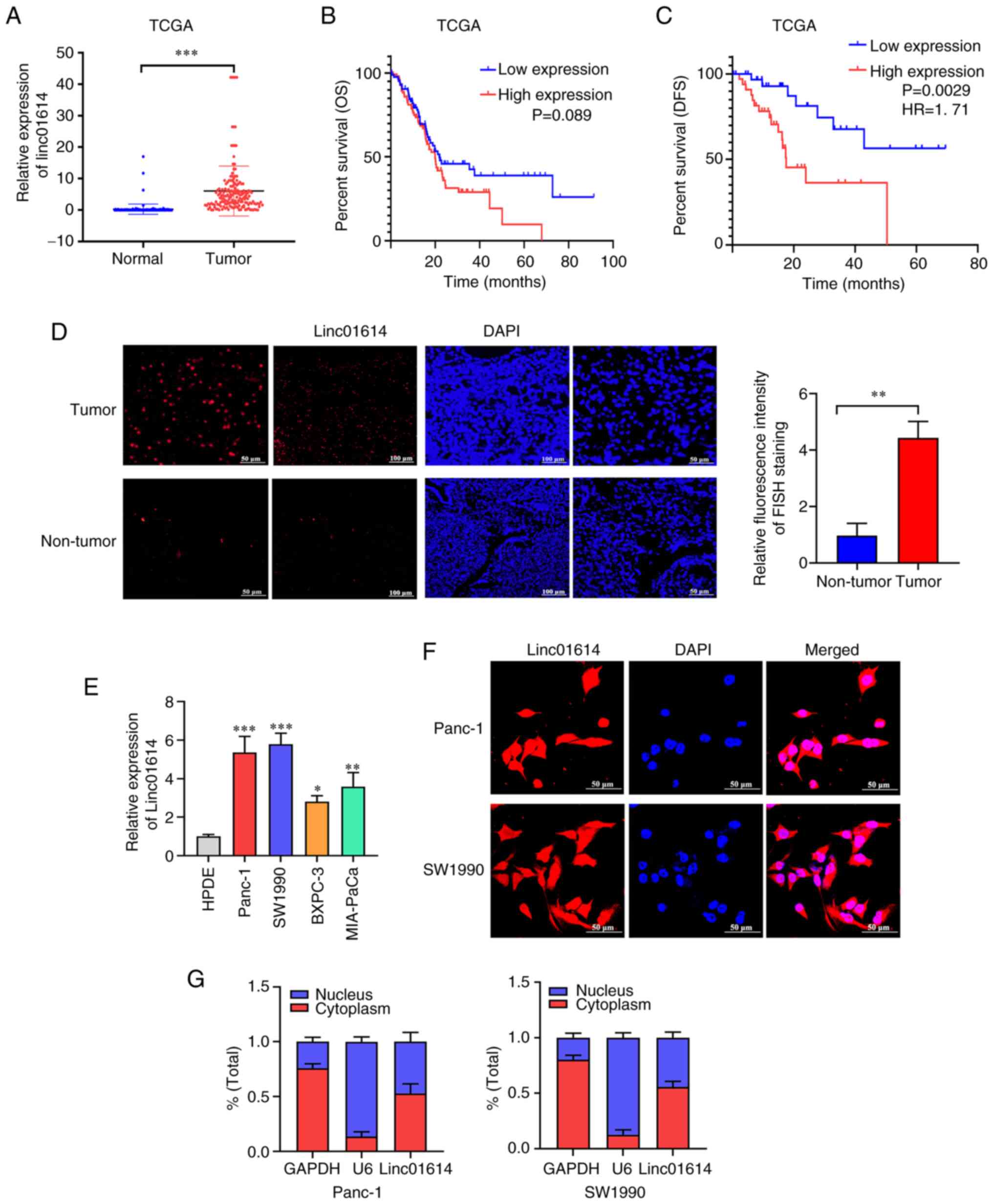 | Figure 2Upregulation of linc01614 in PC. (A)
Ectopic expression of linc01614 in PC tissues than in the normal
pancreatic tissues from TCGA and GTEx databases (N=171, T=178). (B
and C) Data from TCGA, indicating the association of the high
expression of linc01614 with poor DFS rather than OS. (D) RNA FISH
analysis of the expression of linc01614 in PC and normal pancreatic
cancer tissues. Magnification, ×200 and ×400. (E) Expression of
linc01614 in Panc-1, SW1990, BXPC-3 and MIA-PaCa cells vs. HPDE
cells. (F) RNA FISH analysis to determine the localization of
linc01614. Magnification, ×600. (G) Fractionation of PC cells
followed by reverse transcription-quantitative PCR, to confirm
linc01614 localization. Data are presented as the mean ± SD.
*P<0.05, **P<0.01,
***P<0.001 vs. the normal tissues or HPDE cells. PC,
pancreatic cancer; TCGA, The Cancer Genome Atlas; GTEx,
Genotype-Tissue Expression project; DFS, disease-free survival; OS,
overall survival; FISH, fluorescent in-situ hybridization; SD,
standard deviation. |
Subsequently, the linc01614 levels were investigated
in various PC cell lines (Panc-1, SW1990, BxPC-3 and Mia-PaCa)
using RT-qPCR. linc01614 expression in PC cell lines was also
significantly elevated in comparison with the human pancreatic duct
epithelium immortalized cells (HPDE; Fig. 2E). As the localization of lncRNA
significantly affects its function, the present study then aimed to
identify the localization of linc01614 in Panc-1 and SW1990 cells,
using the RNA FISH assay and subcellular fractionation. linc01614
was located in both the cytoplasm and nucleus, although its
abundance was slightly higher in the cytoplasm than nucleus
(Fig. 2F and G).
linc01614 promotes the proliferation of
PC cells in vitro
To determine the biological function of linc01614 in
PC cells, two independent ASOs against human linc01614 (ASO-1 and
ASO-2) were applied to knockdown linc01614 in Panc-1 and SW1990
cells, and the knockdown efficiency was examined using RT-qPCR
(Fig. 3A). The results of CCK-8
assay revealed that the knockdown of linc01614 suppressed the
proliferation of PC cells (Fig.
3B). This finding was further verified by the EdU (Fig. 3C) and colony formation (Fig. 3D) assays. Cell cycle distribution
in the PC cells was also detected using flow cytometry. Notably,
the results revealed an increase in the number of PC cells in the
G1/G0 phase and a decrease in the number of PC cells in the S phase
following linc01614 knockdown compared with the control (Fig. 3E). To further verify the flow
cytometry data, western blot analysis was performed to detect the
G0/G1 phase regulating proteins cyclin D1 and CDK2. linc01614
knockdown decreased the expression of cyclin D1 and CDK2 in Panc-1
and SW1990 cells in comparison with the control group (Fig. 3F). Furthermore, flow cytometry
results revealed no significant difference in apoptosis between
linc01614 knockdown and control groups in both the Panc-1 and
SW1990 cells (Fig. S1).
Additionally, linc01614 overexpression was also
induced in HPDE cells by plasmid transfection. EdU and flow
cytometry assays were performed to investigate the effects of
linc01614 on cell proliferation and apoptosis in HPDE,
respectively. The results revealed that linc01614 overexpression
promoted HPDE cell proliferation, whereas no significant
differences in apoptosis were observed between the linc01614
upregulated and control HPDE cells (Fig. S2).
linc01614 promotes the migration and
invasion of PC cells in vitro
The effects of linc01614 on the migration and
invasion of PC cells were explored using wound healing and
Transwell assays, respectively. linc01614 knockdown markedly
decreased cell migration and invasion as compared to the control
(Fig. 4A and B).
 | Figure 4linc01614 facilitates the migration
and invasion of PC cells in vitro. (A) Transwell assays was
used, to assess the migration and invasion abilities of PC cells.
Magnification, ×100. (B) Wound healing assay, for the examination
of Panc-1 and SW1990 cells migration ability upon linc01614
knockdown. Magnification, ×40. (C) Western blot analysis was
performed for the measurement of EMT-related protein expression in
PC cells transfected with ASO-NC and ASO-1. Data are presented as
the mean ± SD. *P<0.05, **P<0.01 and
***P<0.001 vs. the ASO-NC group. PC, pancreatic
cancer; EMT, epithelial-mesenchymal transition; ASO, antisense
oligonucleotide; NC, negative control; SD, standard deviation. |
Accumulating evidence has emerged on the association
of invasion and metastasis with the epithelial-mesenchymal
transition (EMT) in PC (22-24).
Hence, the expression of EMT-related proteins was also evaluated
using western blot analysis. The expression of the EMT-related
epithelial marker, E-cadherin, markeldy increased, whereas the
expression of the mesenchymal markers, N-cadherin, vimentin, Snail
and Twist, decreased in the linc01614 knockdown groups (Fig. 4C). These results verified that
linc01614 promoted the migration, invasion and EMT of PC cells
in vitro.
linc01614 knockdown inhibits
tumorigenesis in vivo
To further validate the results of linc01614 in
vivo, a xenograft tumor model was established using the Panc-1
cell. It was found that the ASO-1-transfected group generated
smaller tumors in nude mice than the control group. The analysis of
tumor volume and weight also demonstrated that linc01614 knockdown
suppressed tumor growth in vivo (Fig. 5A-C). linc01614 expression was also
reduced in the xenograft tumors of the ASO-1-transfected group as
compared with the control group (Fig.
5D). Immunohistochemical staining revealed that the tumors
derived from the ASO-1 treated group exhibited a lower number of
PCNA-, Ki67- and β-catenin-positive cells compared with the control
group (Fig. 5E). In brief,
linc01614 inhibition by ASO-1 efficiently suppressed tumor growth
in vivo.
linc01614 modulates the WNT/β-catenin
signaling pathway in PC
To investigate the potential molecular mechanisms of
linc01614 in PC, GSEA analysis was performed. linc01614 was
significantly associated with several cancer-related signaling
pathways (Fig. 6A), including the
WNT/β-catenin signaling pathway, pathways in cancer, pancreatic
cancer pathway and TGF-β signaling pathway. The WNT/β-catenin
pathway is one of the most crucial signaling pathways in modulating
the occurrence of EMT in tumor cells (25-27).
In addition, several studies were detected regarding the activation
of WNT/β-catenin in PC (27-31).
Combined with the EMT results of the present study, it was
hypothesized that linc01614 may function as an oncogene partly by
modulating the WNT/β-catenin signaling pathway.
Furthermore, the TOP/FOP flash reporter assay was
performed to analyze the effects of linc01614 expression on the
WNT/β-catenin pathway in PC cells. It was demonstrated that
linc01614 knockdown significantly suppressed TOP-Flash reporter
activity in Panc-1 and SW1990 cells (Fig. 6B). The expression of several
proteins involved in the WNT signaling pathway was then also
measured. No significant differences were observed concerning WNT
pathway component mRNA levels between the linc01614 knockdown and
control groups (Fig. 6C). In the
western blot analysis, the expression of β-catenin significantly
decreased in the linc01614 knockdown group than in the control
group. However, no significant difference in the levels of other
proteins, including GSK-3β, APC, and AXIN1, between the linc01614
knockdown and control group were detected (Fig. 6D). Furthermore, the nuclear and
cytoplasmic levels of active and total β-catenin were decreased in
the linc01614 knockdown in comparison with the control group
(Fig. 6E). Hence, it was
hypothesized that linc01614 may influence β-catenin expression at
the post-transcriptional level.
To examine the association between linc01614 and
β-catenin in PC cells, WNT signaling was activated in Panc-1 and
SW1990 cells using the WNT signaling activator, LiCl. Western blot
analysis indicated that LiCl increased the expression of β-catenin
and partly offset the effect of linc01614 knockdown in PC cells
(Fig. 6F). Taken together, these
data demonstrated that the alteration of linc01614 levels
substantially affected β-catenin protein levels and WNT/β-catenin
signaling.
linc01614 combines with GSK-3β to
stabilize the level of β-catenin protein
To investigate the underlying mechanisms of
linc01614 in the regulation of WNT/β-catenin signaling, RNA-pull
down assays were conducted, followed by mass spectrometric
analysis, in order to determine potential linc01614-interacting
proteins in Panc-1 cells. A band between 43 and 55 kDa was
specifically enriched in the linc01614 pull-down proteins and was
subjected to mass spectrometry (Fig.
7A). GSK-3β, one of the key members of the WNT/β-catenin
pathway, was identified as a potential linc01614-interacting
protein (Fig. S3 and Table SVII). Bioinformatics analysis was
then performed, to verify the potential interaction between
linc01614 and GSK-3β in PC cells. By using the RPISeq database, it
was observed that GSK-3β possessed a high interaction probability
with linc01614 (Fig. 7B). The
catRAPID database was used to predict possible binding sites
between linc01614 and GSK-3β, and it was revealed that nucleotides
1471-1556 of the linc01614 sequence and amino acids 76-127 of the
GSK-3β sequence were the most probable binding sites (Fig. 7C). Furthermore, western blot
analysis revealed that GSK-3β was pulled down by biotin-labeled
linc01614 transcript and not by linc01614 antisense (Fig. 7D). GSK-3β RIP assays in Panc-1 and
SW1990 cells also revealed that linc01614 was markedly enriched by
the anti-GSK-3β antibody (Fig.
7E). These results validated that linc01614 could possibly bind
to GSK-3β.
Previous studies have demonstrated that AXIN1 can
recruit GSK-3β, APC and CK1 to form a multi-protein degradation
complex, which tightly regulates the degradation of β-catenin
(32,33). The results of the present study
also indicated that linc01614 can influence β-catenin expression at
the post-transcriptional level (Fig.
6C and D). In addition, lncRNAs can restrict protein
conformational shift or prevent the interaction among related
proteins through binding (34,35).
Hence, it was hypothesized that linc01614 perturbs the interaction
between GSK-3β and AXIN1 through its binding to GSK-3β, thereby
preventing the formation of the β-catenin degradation complex. In
the present study, the results of Co-IP results confirmed that
linc01614 knockdown enhanced the interaction between GSK-3β and
AXIN1 (Fig. 7F and G), indicating
that linc01614 negatively regulated the interaction between GSK-3β
and AXIN1. Thus, linc01614 stabilized the level of β-catenin
protein possibly through binding to GSK-3β, impeding the
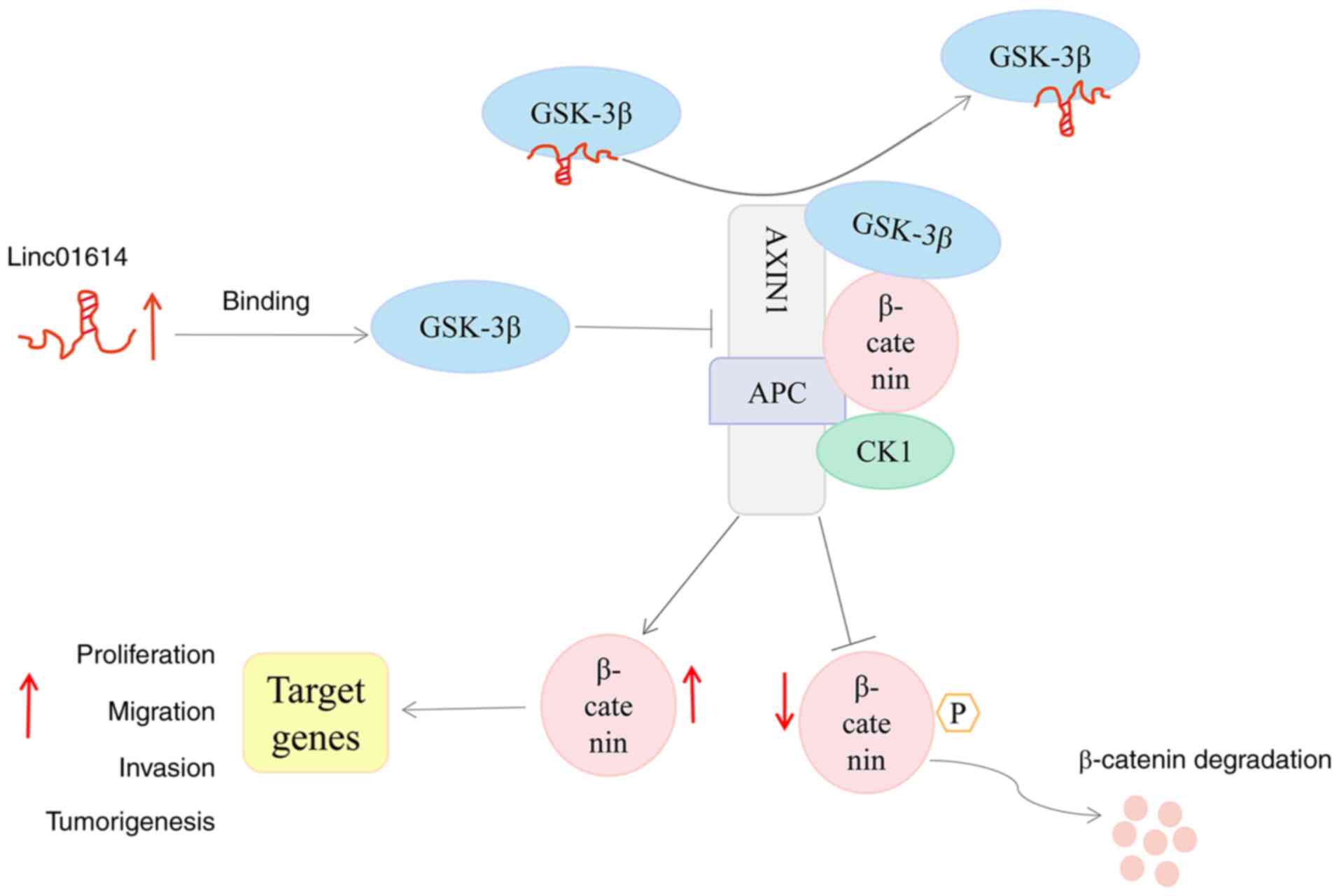
interaction between GSK-3β and AXIN1 (Fig. 8).
Discussion
In the present study, it was identified that
linc01614 was upregulated in PC. In PC cell lines, increased
linc01614 expression was positively associated with tumor growth,
invasiveness and migration. linc01614 functioned as a tumor
promoter and obstruct the interaction between GSK-3β and AXIN1 by
directly binding to GSK-3β, a vital protein of the WNT/β-catenin
signaling pathway, thereby preventing the formation of the
β-catenin degradation complex. Consequently, β-catenin may be
accumulated in the cytoplasm and nucleus of PC cells, and the
WNT/β-catenin pathway was activated. Therefore, the results
illustrated the role and mechanism of linc01614 as an oncogene,
activating the WNT/β-catenin pathway and promoting the progression
of PC.
linc01614 is a 2,180 nucleotide-long intergenic
non-protein coding RNA, encoded on the sense strand of chromosome
2. linc01614 is involved in the development of several malignancies
(16-21). These studies have mainly relied on
bioinformatics analysis of linc01614 or its interaction with miRNAs
as a competing endogenous RNA. However, whether linc01614 can
perform a regulatory function by interacting with other cellular
components (DNAs, RNAs and proteins) remains unclear. Additionally,
the subcellular localization of a lncRNA inside the cell is crucial
for understanding its functionality (36,37).
FISH and subcellular fractionation assays confirmed that linc01614
was enriched in both PC cell cytoplasm and nucleus. This indicates
that linc01614 may theoretically regulate fundamental cellular
processes at different levels, including epigenetic modification,
transcriptional, post-transcriptional, translational, and
post-translational regulation. Of note, RNA pull-down and RIP assay
demonstrated that linc01614 likely exerts its function by directly
binding to GSK-3β, a vital protein of the canonical WNT/β-catenin
pathway (38). Moreover, linc01614
disrupted the function of GSK-3β by affecting the interaction
between GSK-3β and AXIN1 rather than reducing the translational
production or increasing protein degradation. Hence, the present
findings illustrated the molecular mechanisms of linc01614 in the
progression of PC.
The canonical WNT signaling pathway controls the
expression of critical developmental genes by regulating the
accumulation of the transcriptional co-activator β-catenin.
Aberrant WNT/β-catenin signaling has been previously implicated in
PC (30,31,39).
Moreover, lncRNAs may function as important signal transduction
mediators in cancer signaling pathways, modifying cell fate and
function (40-43). Based on bioinformatics analysis and
experimental data, the present study successfully validated the
linc01614-mediated activation of the WNT/β-catenin signaling
pathway at the protein level in PC. Mechanistically, the
scaffolding AXIN1 protein interacts with GSK3, CK1α and β-catenin
at separate domains to form a β-catenin degradation complex. GSK-3β
can phosphorylate the amino-terminal region of β-catenin, leading
to subsequent β-catenin ubiquitination and proteasomal degradation
(44). Therefore, β-catenin levels
are possibly regulated by controlling GSK-3β activity in cells. It
was revealed that linc01614 could interact with GSK-3β in PC;
however, no significant differences in GSK-3β mRNA and protein
levels between the control and linc01614 knockdown group were
detected. However, the overexpression of linc01614 hindered the
interaction between GSK-3β and AXIN1, thereby preventing the
formation of the 'degradation complex' to degrade β-catenin.
Notably, GSK-3β itself participates in cell
proliferation, apoptosis, cell cycle progression and cell signal
transduction (45-47). As a serine-threonine kinase, GSK-3β
can exert tumor-promoter and tumor-suppressor effects by
phosphorylating a broad range of substrates (48,49).
The function of GSK-3β in the onset and progression of PC and the
acquisition of chemoresistance been previously established. For
instance, the overexpression of GSK-3β can promote the invasive
ability of Panc-1 cells by upregulating CXCR4 and MMP-2 expression
(50). GSK-3β can also regulate
cell cycle progression in cancer cells. In PC, GSK-3β can directly
modulate the phosphorylation status of several cell cycle
modulators, including cyclin D1 and p53 (51). Thus, GSK-3β may serve as a
candidate therapeutic target for PC. Consistently, the present
study revealed that LiCl, a GSK-3β inhibitor, upregulated the
β-catenin level and subsequently promoted the progression of PC.
Several lncRNAs, including MIR22HG and linc01197, have been
reported to regulate the WNT/β-catenin signaling (40,52),
whereas none of these lncRNAs bind to GSK-3β. However, considering
the multiple biological functions of GSK-3β, the net effect of
GSK-3β and the WNT/β-catenin pathway in PC still needs further
analysis.
In conclusion, the present study demonstrated that
linc01614 functioned as an oncogene and promoted the progression of
PC. By binding to GSK-3β, linc01614 may stabilize the level of
β-catenin protein to hyperactivate WNT/β-catenin signaling activity
in PC. The current findings provide new insight into the underlying
mechanisms of PC progression and a potential future therapeutic
target for PC.
Supplementary Data
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZGT and LJC conceived and designed the study. LJC
and LW conducted most of the experiments with the assistance of WW
and FX. LLZ and WBL analyzed the data. LJC wrote the first draft of
the manuscript. ZGT performed critical revision and supervised all
phases of the study. ZGT and LJC confirm the authenticity of all
the raw data. All authors have read and approved the final
manuscript.
Ethical approval and consent to
participate
The present study was performed in accordance with
the Declaration of Helsinki and approved by the Ethics Committee of
Renmin Hospital of Wuhan University (approval no. WDRY2021-K188),
and written informed consent was obtained from all subjects. All
experiments were conducted according to the relevant guidelines and
regulations and following the approval of the Institutional Animal
Care Committee of Renmin Hospital of Wuhan University (approval no.
WDRM20210606).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
The present study was supported by the Foundation of Health
Commission of Hubei Province (grant nos. WJ2021M063,
WJ2021F052).
References
|
1
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer statistics, 2021. CA Cancer J Clin. 71:7–33. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
GBD 2017 Pancreatic Cancer Collaborators:
The global, regional, and national burden of pancreatic cancer and
its attributable risk factors in 195 countries and territories,
1990-2017: A systematic analysis for the global burden of disease
study 2017. Lancet Gastroenterol Hepatol. 4:934–947. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Herting CJ, Karpovsky I and Lesinski GB:
The tumor micro-environment in pancreatic ductal adenocarcinoma:
Current perspectives and future directions. Cancer Metastasis Rev.
40:675–689. 2021. View Article : Google Scholar
|
|
4
|
Ho WJ, Jaffee EM and Zheng L: The tumour
microenvironment in pancreatic cancer-clinical challenges and
opportunities. Nat Rev Clin Oncol. 17:527–540. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jarroux J, Morillon A and Pinskaya M:
History, discovery, and classification of lncRNAs. Adv Exp Med
Biol. 1008:1–46. 2017. View Article : Google Scholar
|
|
6
|
Liu S, Zhan N, Gao C, Xu P, Wang H, Wang
S, Piao S and Jing S: Long noncoding RNA CBR3-AS1 mediates
tumorigenesis and radiosensitivity of non-small cell lung cancer
through redox and DNA repair by CBR3-AS1/miR-409-3p/SOD1 axis.
Cancer Lett. 526:1–11. 2022. View Article : Google Scholar
|
|
7
|
Sun Y, Tian Y, He J, Tian Y, Zhang G, Zhao
R, Zhu WJ and Gao P: Linc01133 contributes to gastric cancer growth
by enhancing YES1-dependent YAP1 nuclear translocation via sponging
miR-145-5p. Cell Death Dis. 13:512022. View Article : Google Scholar
|
|
8
|
Hu H, Wang Y, Ding X, He Y, Lu Z, Wu P,
Tian L, Yuan H, Liu D, Shi G, et al: Long non-coding RNA
XLOC_000647 suppresses progression of pancreatic cancer and
decreases epithelial-mesenchymal transition-induced cell invasion
by down-regulating NLRP3. Mol Cancer. 17:182018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Singh N, Ramnarine VR, Song JH, Pandey R,
Padi SK, Nouri M, Olive V, Kobelev M, Okumura K, Mccarthy D, et al:
The long noncoding RNA H19 regulates tumor plasticity in
neuroendocrine prostate cancer. Nat Commun. 12:73492021. View Article : Google Scholar
|
|
10
|
Kopp F and Mendell JT: Functional
classification and experimental dissection of long noncoding RNAs.
Cell. 172:393–407. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gandhi M, Groß M, Holler JM, Coggins SA,
Patil N, Leupold JH, Munschauer M, Schenone M, Hartigan CR,
Allgayer H, et al: The lncRNA lincNMR regulates nucleotide
metabolism via a YBX1-RRM2 axis in cancer. Nat Commun. 11:32142020.
View Article : Google Scholar
|
|
13
|
Iyer MK, Niknafs YS, Malik R, Singhal U,
Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, et
al: The landscape of long noncoding RNAs in the human
transcriptome. Nat Genet. 47:199–208. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
15
|
Pan S, Deng Y, Fu J, Zhang Y, Zhang Z and
Qin X: N6-methyladenosine upregulates miR181d5p in exosomes derived
from cancer associated fibroblasts to inhibit 5FU sensitivity by
targeting NCALD in colorectal cancer. Int J Oncol. 60:142022.
View Article : Google Scholar
|
|
16
|
Chen Y, Cheng WY, Shi H, Huang S, Chen H,
Liu D, Xu W, Yu J and Wang J: Classifying gastric cancer using
FLORA reveals clinically relevant molecular subtypes and highlights
linc01614 as a biomarker for patient prognosis. Oncogene.
40:2898–2909. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Vishnubalaji R, Shaath H, Elkord E and
Alajez NM: Long non-coding RNA (lncRNA) transcriptional landscape
in breast cancer identifies linc01614 as non-favorable prognostic
biomarker regulated by TGFβ and focal adhesion kinase (FAK)
signaling. Cell Death Discov. 5:1092019. View Article : Google Scholar
|
|
18
|
Liu AN, Qu HJ, Yu CY and Sun P: Knockdown
of linc01614 inhibits lung adenocarcinoma cell progression by
up-regulating miR-217 and down-regulating FOXP1. J Cell Mol Med.
22:4034–4044. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sun Y and Ling C: Analysis of the long
non-coding RNA linc01614 in non-small cell lung cancer. Medicine
(Baltimore). 98:e164372019. View Article : Google Scholar
|
|
20
|
Cai Q, Zhao X, Wang Y, Li S, Wang J, Xin Z
and Li F: linc01614 promotes osteosarcoma progression via
miR-520a-3p/SNX3 axis. Cell Signal. 83:1099852021. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tang L, Chen Y, Peng X, Zhou Y, Jiang H,
Wang G and Zhuang W: Identification and validation of potential
pathogenic genes and prognostic markers in ESCC by integrated
bioinformatics analysis. Front Genet. 11:5210042020. View Article : Google Scholar
|
|
22
|
Aiello NM, Brabletz T, Kang Y, Nieto MA,
Weinberg RA and Stanger BZ: Upholding a role for EMT in pancreatic
cancer metastasis. Nature. 547:E7–E8. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Thiery JP: Epithelial-mesenchymal
transitions in tumour progression. Nat Rev Cancer. 2:442–454. 2002.
View Article : Google Scholar
|
|
24
|
Wang L, Wu H, Wang L, Zhang H, Lu J, Liang
Z and Liu T: Asporin promotes pancreatic cancer cell invasion and
migration by regulating the epithelial-to-mesenchymal transition
(EMT) through both autocrine and paracrine mechanisms. Cancer Lett.
398:24–36. 2017. View Article : Google Scholar
|
|
25
|
Liu Y, Tang T, Yang X, Qin P, Wang P,
Zhang H, Bai M, Wu R and Li F: Tumor-derived exosomal long
noncoding RNA LINC01133, regulated by Periostin, contributes to
pancreatic ductal adenocarcinoma epithelial-mesenchymal transition
through the Wnt/β-catenin pathway by silencing AXIN2. Oncogene.
40:3164–3179. 2021. View Article : Google Scholar :
|
|
26
|
Zhang J, Cai H, Sun L, Zhan P, Chen M,
Zhang F, Ran Y and Wan J: LGR5, a novel functional glioma stem cell
marker, promotes EMT by activating the Wnt/β-catenin pathway and
predicts poor survival of glioma patients. J Exp Clin Cancer Res.
37:2252018. View Article : Google Scholar
|
|
27
|
Liu SL, Cai C, Yang ZY, Wu ZY, Wu XS, Wang
XF, Dong P and Gong W: DGCR5 is activated by PAX5 and promotes
pancreatic cancer via targeting miR-3163/TOP2A and activating
Wnt/β-catenin pathway. Int J Biol Sci. 17:498–513. 2021. View Article : Google Scholar :
|
|
28
|
Ram Makena M, Gatla H, Verlekar D,
Sukhavasi S, K Pandey M and C Pramanik K: Wnt/β-Catenin signaling:
The culprit in pancreatic carcinogenesis and therapeutic
resistance. Int J Mol Sci. 20:42422019. View Article : Google Scholar
|
|
29
|
Tang N, Xu S, Song T, Qiu Y, He J and Fu
X: Zinc finger protein 91 accelerates tumour progression by
activating β-catenin signalling in pancreatic cancer. Cell Prolif.
54:e130312021. View Article : Google Scholar
|
|
30
|
Wang L, Heidt DG, Lee CJ, Yang H, Logsdon
CD, Zhang L, Fearon ER, Ljungman M and Simeone DM: Oncogenic
function of ATDC in pancreatic cancer through Wnt pathway
activation and beta-catenin stabilization. Cancer Cell. 15:207–219.
2009. View Article : Google Scholar :
|
|
31
|
Zhou C, Liang Y, Zhou L, Yan Y, Liu N,
Zhang R, Huang Y, Wang M, Tang Y, Ali DW, et al: TSPAN1 promotes
autophagy flux and mediates cooperation between WNT-CTNNB1
signaling and autophagy via the MIR454-FAM83A-TSPAN1 axis in
pancreatic cancer. Autophagy. 17:3175–3195. 2021. View Article : Google Scholar :
|
|
32
|
Kishida S, Yamamoto H, Ikeda S, Kishida M,
Sakamoto I, Koyama S and Kikuchi A: Axin, a negative regulator of
the wnt signaling pathway, directly interacts with adenomatous
polyposis coli and regulates the stabilization of beta-catenin. J
Biol Chem. 273:10823–10826. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Seeling JM, Miller JR, Gil R, Moon RT,
White R and Virshup DM: Regulation of beta-catenin signaling by the
B56 subunit of protein phosphatase 2A. Science. 283:2089–2091.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jiang M, Zhang S, Yang Z, Lin H, Zhu J,
Liu L, Wang W, Liu S, Liu W, Ma Y, et al: Self-recognition of an
inducible host lncRNA by RIG-I feedback restricts innate immune
response. Cell. 173:906–919. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xu J, Shao T, Song M, Xie Y, Zhou J, Yin
J, Ding N, Zou H, Li Y and Zhang J: MIR22HG acts as a tumor
suppressor via TGFβ/SMAD signaling and facilitates immunotherapy in
colorectal cancer. Mol Cancer. 19:512020. View Article : Google Scholar
|
|
36
|
Cabili MN, Dunagin MC, Mcclanahan PD,
Biaesch A, Padovan-Merhar O, Regev A, Rinn JL and Raj A:
Localization and abundance analysis of human lncRNAs at single-cell
and single-molecule resolution. Genome Biol. 16:202015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
van Heesch S, van Iterson M, Jacobi J,
Boymans S, Essers PB, De Bruijn E, Hao W, Macinnes AW, Cuppen E and
Simonis M: Extensive localization of long noncoding RNAs to the
cytosol and mono- and polyribosomal complexes. Genome Biol.
15:R62014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li Q, Sun M, Wang M, Feng M, Yang F, Li L,
Zhao J, Chang C, Dong H, Xie T, et al: Dysregulation of
Wnt/β-catenin signaling by protein kinases in hepatocellular
carcinoma and its therapeutic application. Cancer Sci.
112:1695–1706. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Morris JP IV, Wang SC and Hebrok M: KRAS,
Hedgehog, Wnt and the twisted developmental biology of pancreatic
ductal adenocarcinoma. Nat Rev Cancer. 10:683–695. 2010. View Article : Google Scholar
|
|
40
|
Han M, Wang S, Fritah S, Wang X, Zhou W,
Yang N, Ni S, Huang B, Chen A, Li G, et al: Interfering with long
non-coding RNA MIR22HG processing inhibits glioblastoma progression
through suppression of Wnt/β-catenin signalling. Brain.
143:512–530. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Nyati KK, Hashimoto S, Singh SK, Tekguc M,
Metwally H, Liu YC, Okuzaki D, Gemechu Y, Kang S and Kishimoto T:
The novel long noncoding RNA AU021063, induced by IL-6/Arid5a
signaling, exacerbates breast cancer invasion and metastasis by
stabilizing Trib3 and activating the Mek/Erk pathway. Cancer Lett.
520:295–306. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wu N, Jiang M, Liu H, Chu Y, Wang D, Cao
J, Wang Z, Xie X, Han Y and Xu B: LINC00941 promotes CRC metastasis
through preventing SMAD4 protein degradation and activating the
TGF-β/SMAD2/3 signaling pathway. Cell Death Differ. 28:219–232.
2021. View Article : Google Scholar
|
|
43
|
Xu M, Cui R, Ye L, Wang Y, Wang X, Zhang
Q, Wang K, Dong C, Le W and Chen B: LINC00941 promotes glycolysis
in pancreatic cancer by modulating the Hippo pathway. Mol Ther
Nucleic Acids. 26:280–294. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kimelman D and Xu W: beta-catenin
destruction complex: Insights and questions from a structural
perspective. Oncogene. 25:7482–7491. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ryu HY, Kim LE, Jeong H, Yeo BK, Lee JW,
Nam H, Ha S, An HK, Park H, Jung S, et al: GSK3B induces autophagy
by phosphorylating ULK1. Exp Mol Med. 53:369–383. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Park R, Coveler AL, Cavalcante L and Saeed
A: GSK-3β in pancreatic cancer: Spotlight on 9-ING-41, its
therapeutic potential and immune modulatory properties. Biology
(Basel). 10:6102021.
|
|
47
|
Zhang Z, Gao Q and Wang S: Kinase GSK3β
functions as a suppressor in colorectal carcinoma through the
FTO-mediated MZF1/c-Myc axis. J Cell Mol Med. 25:2655–2665. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Duda P, Akula SM, Abrams SL, Steelman LS,
Martelli AM, Cocco L, Ratti S, Candido S, Libra M, Montalto G, et
al: Targeting GSK3 and associated signaling pathways involved in
cancer. Cells. 9:11102020. View Article : Google Scholar :
|
|
49
|
Pecoraro C, Faggion B, Balboni B, Carbone
D, Peters GJ, Diana P, Assaraf YG and Giovannetti E: GSK3β as a
novel promising target to overcome chemoresistance in pancreatic
cancer. Drug Resist Updat. 58:1007792021. View Article : Google Scholar
|
|
50
|
Ying X, Jing L, Ma S, Li Q, Luo X, Pan Z,
Feng Y and Feng P: GSK3β mediates pancreatic cancer cell invasion
in vitro via the CXCR4/MMP-2 pathway. Cancer Cell Int. 15:702015.
View Article : Google Scholar
|
|
51
|
Mccubrey JA, Rakus D, Gizak A, Steelman
LS, Abrams SL, Lertpiriyapong K, Fitzgerald TL, Yang LV, Montalto
G, Cervello M, et al: Effects of mutations in Wnt/β-catenin,
hedgehog, Notch and PI3K pathways on GSK-3 activity-diverse effects
on cell growth, metabolism and cancer. Biochim Biophys Acta.
1863:2942–2976. 2016. View Article : Google Scholar
|
|
52
|
Ling J, Wang F, Liu C, Dong X, Xue Y, Jia
X, Song W and Li Q: FOXO1-regulated lncRNA LINC01197 inhibits
pancreatic adenocarcinoma cell proliferation by restraining
Wnt/β-catenin signaling. J Exp Clin Cancer Res. 38:1792019.
View Article : Google Scholar
|