Introduction
Merkel cell polyomavirus (MCPyV) was recently
discovered to be associated with Merkel cell carcinoma (MCC), a
rare and aggressive type of human skin cancer (1). MCPyV-induced oncogenesis is considered
to be involved in the transformative properties of the MCPyV large
T antigen. MCPyV has been detected in MCC patients and appears to
play a key role in tumourigenesis; ~80% of MCCs harbour MCPyV
(1). Using quantitative polymerase
chain reaction (qPCR), MCPyV has been detected in malignant and
benign tumours (1–16). Moreover, certain investigators have
reported that low viral loads of MCPyV have also been detected in
normal human tissue samples, including skin, liver and respiratory
secretions, suggesting that this virus is widespread in the human
body (7,17). However, previous studies on the
frequency of MCPyV infection in oral tumours or tumour-like lesions
are partial and incomplete (7,15). High
viral loads of MCPyV were detected in all saliva samples from 10
patients without cancer (7).
In previous studies conducted by Hashida et
al (16) and Pantulu et al
(18), MCPyV was found to be
associated with the pathogenesis of non-small-cell lung cancer and
chronic lymphocytic leukemia.
The aim of this study was to determine the
prevalence of MCPyV and identify new MCPyV-related tumours or
tumour-like lesions in the maxillofacial region, in addition to the
oral region. Thus, we measured the MCPyV DNA quantity using qPCR in
327 oral tumours or tumour-like lesions, 54 jaw tumours or cyst
lesions and 4 oral lesions from 4 immunosuppressed patients.
Materials and methods
Human tissue samples
The analysed samples comprised 385 of 404
formalin-fixed and paraffin-embedded (FFPE) samples, including 381
immunocompetent and 4 immunocompromised cases (Tables I and II). A total of 19 samples were excluded,
due to lack of detection of the protein tyrosine phosphatase
receptor type G. The 404 samples were obtained from 398 individuals
who underwent biopsy or surgical resection of tumours or
tumour-like lesions of the oral region at the Division of Oral and
Maxillofacial Biopathological Surgery and the Department of
Otolaryngology, Head and Neck Surgery of Tottori University
Hospital, Japan, between April, 2003 and March, 2013.
 | Table I.Merkel cell polyomavirus (MCPyV)
detection in tumours or tumour-like lesions from the oral cavity
and jaws of immunocompetent patients using quantitative polymerase
chain reaction (qPCR). |
Table I.
Merkel cell polyomavirus (MCPyV)
detection in tumours or tumour-like lesions from the oral cavity
and jaws of immunocompetent patients using quantitative polymerase
chain reaction (qPCR).
| Types of lesions | Case no. | Prevalence, no./total
(%) | MCPyV DNA load
(copies/cell) |
|---|
| Tumours or
tumour-like lesions of the oral cavity |
|
|
|
| Invasive
SCC | 176 | 7/176
(4.0) | 0.00038–0.00097 |
|
Common type | 155 | 7/155
(4.5) | 0.00038–0.00097 |
|
Tongue | 60 | 2/60
(3.33) | 0.00038–0.00097 |
|
Gingiva | 52 | 4/52 (7.7) | 0.00038–0.00092 |
|
Buccal
mucosa | 11 | 0/11 | - |
|
Floor
of the mouth | 19 | 1/19 (5.3) | 0.00078 |
|
Palate |
9 | 0/9 | - |
|
Lip |
4 | 0/4 | - |
|
Variant type | 21 | 0/21 | - |
|
Spindle
cell carcinoma |
3 | 0/3 | - |
|
Undifferentiated
carcinoma (lymphoepithelial carcinoma) | 18 | 0/18 | - |
| SCC in
situ |
3 | 0/3 | - |
| Dysplasia
(leukoplakia with atypia) | 10 | 1/10
(10.0) | 0.00066 |
|
Small-cell carcinoma |
1 | 0/1 | - |
|
Adenocarcinoma |
5 | 1/5 (20.0) | 0.00035 |
|
Mucoepidermoid carcinoma | 13 | 0/13 | - |
| Adenoid
cystic carcinoma | 13 | 2/13
(15.4) | 0.00040–0.00065 |
| Malignant
pleomorphic adenoma |
5 | 0/5 | - |
| Malignant
melanoma |
6 | 0/6 | - |
|
Non-Hodgkin's lymphoma | 10 | 1/10
(10.0) | 0.0021 |
|
Metastatic cancera |
6 | 0/6 | - |
|
Fibroma | 11 | 0/11 | - |
|
Papilloma | 12 | 0/12 | - |
|
Lipoma | 10 | 3/10
(30.0) | 0.00083–0.026 |
|
Neurofibroma |
5 | 3/5 (60.0) |
0.00034-0.0015 |
|
Schwannoma |
3 | 1/3 (33.3) | 0.00024 |
|
Pleomorphic adenoma | 14 | 0/14 | - |
| Warthin's
tumour | 12 | 2/12
(16.7) | 0.00030–0.0039 |
| Pyogenic
granuloma | 11 | 2/11
(18.2) |
0.000340–0.000345 |
| Epulis
in a patient with MCPyV+ MCC |
1 | 0/1 | - |
|
Total | 327 | 23/327 (7.0) | 0.00024-0.026 |
| Tumours or cysts of
the jaws |
|
|
|
|
Radicular cyst | 14 | 1/14 (7.1) | 0.00039 |
|
Dentigerous cyst | 10 | 0/10 | - |
|
Keratocystic odontogenic
tumour | 10 | 0/10 | - |
|
Ameloblastoma | 12 | 1/12 (8.3) | 0.00057 |
|
Ossifying fibroma |
3 | 0/3 | - |
| Fibrous
dysplasia |
5 | 0/5 | - |
|
Total | 54 | 2/54 (3.7) |
0.00039-0.00057 |
| Total | 381 | 25/381 (6.6) | 0.00024-0.026 |
 | Table II.MCPyV detection by qPCR in tumours or
tumour-like lesions from immunosuppressed patients. |
Table II.
MCPyV detection by qPCR in tumours or
tumour-like lesions from immunosuppressed patients.
| Tumour and
tumour-like lesions | No. of cases | Prevalence,
no./total (%) | MCPyV DNA load
(copies/cell) |
|---|
| Dysplasia from bone
marrow transplant patient | 1 | 0/1 | - |
| Papilloma from bone
marrow transplant patient | 1 | 1/1 (100.0) | 0.0003 |
| Ulcer from bone
marrow transplant patient | 1 | 0/1 | - |
| Ranula from
HIV+ patient | 1 | 0/1 | - |
| Total | 4 | 1/4 (25.0) | 0.0003 |
The 381 immunocompetent cases were as follows: 155
common-type invasive squamous cell carcinomas (SCCs) (60 of the
tongue, 52 of the gingiva, 11 of the buccal mucosa, 19 of the floor
of the mouth, 9 of the palate and 4 of the lip), 21 variant-type
invasive SCCs (3 spindle cell carcinomas and 18 undifferentiated
carcinomas), 3 SCCs in situ, 10 dysplasias, 1 small-cell
carcinoma, 5 adenocarcinomas, 13 mucoepidermoid carcinomas, 5
malignant pleomorphic adenomas, 6 malignant melanomas, 10
non-Hodgkin's lymphomas, 6 metastatic cancers (1 case each of
esophageal, gastric, renal and prostate cancer, and 2 cases of lung
cancer), 11 fibromas, 12 papillomas, 10 lipomas, 5 neurofibromas, 3
Schwannomas, 14 pleomorphic adenomas, 12 Warthin's tumours, 11
pyogenic granulomas, 1 epulis from an MCPyV-positive MCC patient,
14 radicular cysts, 10 dentigerous cysts, 10 keratocystic
odontogenic tumours, 12 ameloblastomas, 3 ossifying fibromas and 5
fibrous dysplasias.
The 4 immunocompromised cases included 1 dysplasia
from a bone marrow transplant patient, 1 papilloma from a bone
marrow transplant patient, 1 ulcer from a bone marrow transplant
patient and 1 ranula from an HIV-positive patient.
The study protocol was approved by the Institutional
Review Board of the Faculty of Medicine, Tottori University.
qPCR
DNA was extracted from each sample. Tumour samples
sectioned into 10-µm slices were excised into three pieces. DNA was
extracted from each sample using the QIAamp DNA FFPE Tissue kit and
Mini kit (Qiagen, Hilden, Germany) according to the manufacturer's
instructions. The presence of adequate DNA in the samples was
confirmed by measuring the internal control DNA of RNase P. To
determine the MCPyV DNA quantity [relative ratio to MCPyV DNA (1.0
copy/cell) from the reference MCC] for each case, qPCR was
performed using an ABI PRISM 7900HT Sequence Detection system
(Applied Biosystems, Foster City, CA, USA). A total of 30 ng of
each DNA sample were amplified using 5 µl of Express qPCR Supermix
with Premixed ROX (Invitrogen, Carlsbad, CA, USA), 240 nmol/l
fluorescein-labelled locked nucleic acid hydrolysis probe 22
(5′-TGGTGGAG-3′) from a Universal Probe Library (Roche Diagnostics,
Basel, Switzerland) and 0.9 µmol/l primer in a final volume of 10
µl. The positive control was MCCs, whereas water was used as the
negative control. Thermal cycling consisted of incubation for 2 min
at 50°C, with initial denaturation for 10 min at 95°C, followed by
40 cycles of denaturation for 15 sec at 95°C and annealing for 1
min at 60°C, as previously described (19). The virus quantity was determined using
the virus signal in a positive MCC sample as reference. Thresholds
were plotted against each standard sample. All the reactions of
samples and controls were performed in triplicate, and the average
was described. The MCPyV DNA quantity in each sample was determined
based on the corresponding standard curves.
Results
MCPyV DNA detection in tumours and
tumour-like lesions in the oral cavity and jaws of immunocompetent
patients
MCPyV DNA was quantified from the FFPE samples of
381 immunocompetent and 4 immunocompromised cases and the data are
summarised in Tables I and II, respectively.
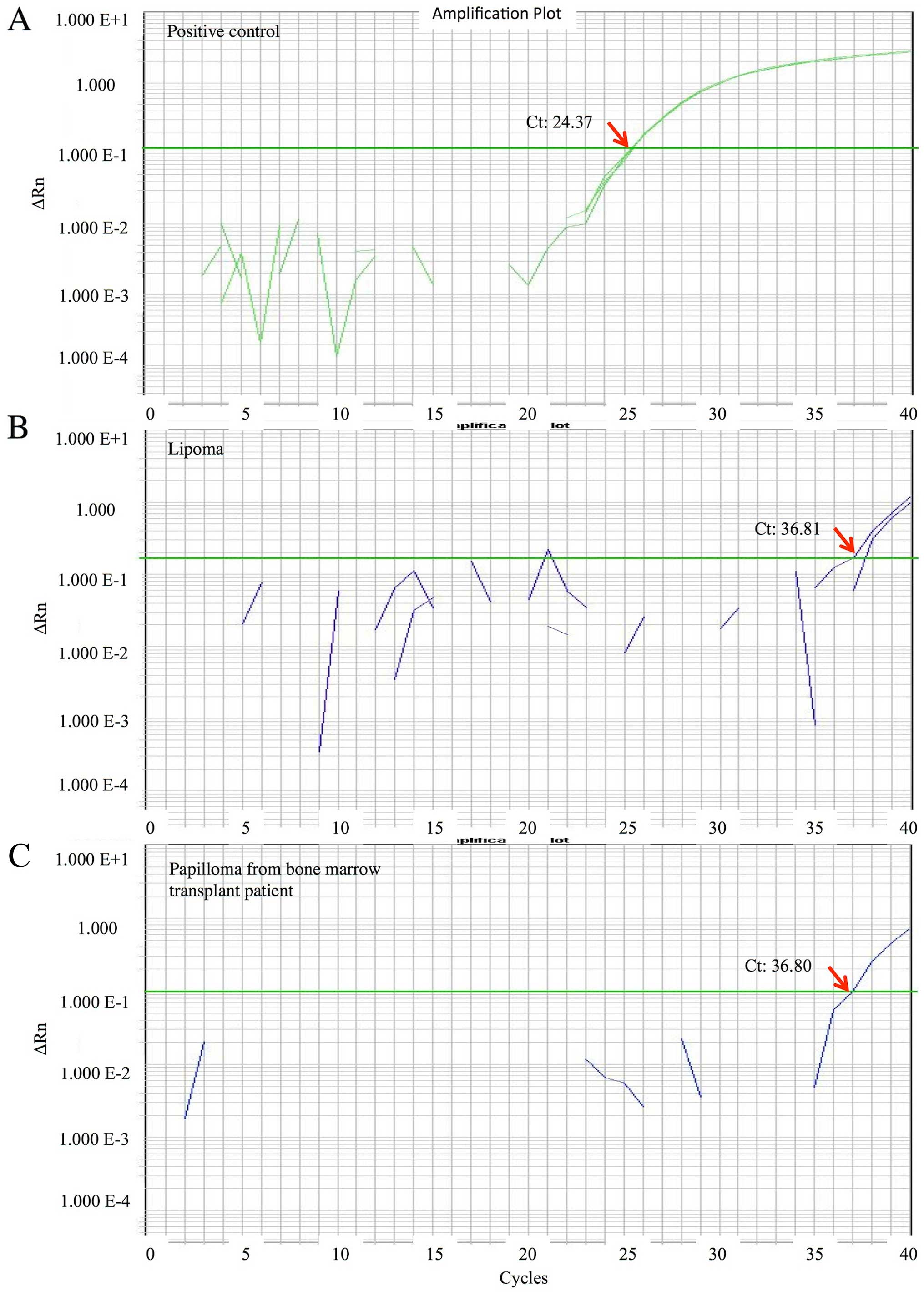
qPCR data for MCPyV DNA are shown for the positive
control (Fig. 1A) and 2
representative cases (Fig. 1B and C).
The range of the MCPyV DNA quantity was very low or low:
0.00024-0.026 copies/cell, with a median of 0.00053 copies/cell.
The overall prevalence of MCPyV in tumours or tumour-like lesions
of the oral cavity and jaws was 25/381 (6.6%).
Prevalence of MCPyV in oral tumour or
tumour-like lesions
The MCPyV prevalence in invasive SCCs was 7/176
(4.0%) [2/60 (3.33%) SCCs of the tongue, 4/52 (7.7%) SCCs of the
gingiva and 1/19 (5.3%) SCCs of the floor of the mouth], 1/10 (10%)
in dysplasias, 1/5 (20%) in adenocarcinomas, 2/13 (15.4%) in
adenoid cystic carcinomas, 1/10 (10%) in non-Hodgkin's lymphomas,
3/10 (30%) in lipomas, 3/5 (60%) in neurofibromas, 1/3 (33.3%) in
Schwannomas, 2/12 (16.7%) in Warthin's tumours and 2/11 (18.2%) in
pyogenic granulomas. The MCPyV DNA (0.00083–0.026) in a case of
lipoma was higher compared with that in other tumours or
tumour-like lesions.
Prevalence of MCPyV in jaw tumours or
cyst lesions
The MCPyV prevalence was 1/14 (7.1%) in radicular
cysts and 1/12 (8.3%) ameloblastomas. No significant difference was
observed between the MCPyV prevalence in tumours or tumour-like
lesions of the oral cavity and that in tumours or cysts of the jaws
(7.0 and 3.7%, respectively; P=0.5537).
MCPyV DNA detection in oral lesions
from immunosuppressed patients
MCPyV DNA was detected in one case of papilloma from
a bone marrow transplant patient but was not detected in the
remaining 3 cases (1/4, 25%) (Table
II).
Discussion
In this study, qPCR was used to investigate whether
MCPyV was present in oral and maxillofacial tumours and tumour-like
lesions. The possibility of contamination was unlikely, as
ultrapure water-negative controls were consistently negative. Loyo
et al (7) reported data on
MCPyV DNA prevalence in oral cavity fresh-frozen materials (51/67,
76%); the prevalence was 19/47 SCCs, 2/10 normal mucosal samples
and 10/10 saliva samples from patients without cancer. Oral cavity
samples had 0.026 copies/genome, while MCCs had an average of 10
copies/genome (range, 173–0.05 copies/genome) and saliva had an
average of 0.128 copies/genome (range, 5–0.01) (7). Matsushita et al (17) did not detect MCPyV DNA in
non-neoplastic tissues from 10 tongue samples and 2 salivary gland
samples from FFPE sections of autopsy cases. The MCPyV prevalence
in the present study was 7.0% (23/327). Matsushita et al
(17) and our study used FFPE samples
for MCPyV quantification, whereas Loyo et al (7) used fresh-frozen tissues. This is a major
reason for the differences between our data on prevalence and those
reported by Loyo et al (7).
Certain previous studies reported that MCPyV DNA detection in
fresh-frozen tissues is more reliable compared with detection in
FFPE samples (20–22).
In this study, low or very low levels of MCPyV DNA
were present in a wide variety of oral and maxillofacial tumours
and tumour-like lesions, while the reference MCC sample had 1 copy
of MCPyV/cell. The MCPyV DNA ratio in oral and maxillofacial
tumours and tumour-like lesions relative to the reference MCC
ranged between 0.00024 and 0.026, indicating that tumours and
tumour-like lesions positive for MCPyV DNA had lower viral loads
compared with those of the MCC sample. These very low MCPyV DNA
quantities detected in this study suggest the absence of
MCPyV-associated tumours or tumour-like lesions in the oral and
maxillofacial regions, as the low detected levels of MCPyV DNA may
be derived from background non-neoplastic tissues with mild MCPyV
infection, rather than from neoplastic tissues.
Cutaneous SCC is the second most frequent type of
skin cancer (23), and cervical SCC
is the major type of uterine cervical cancer (14). In SCC of the skin or uterine cervix,
absence or lower frequency of MCPyV has been detected compared with
MCCs (9–14). Dworkin et al reported that
MCPyV was present in 15% cases of cutaneous SCCs from
immunocompetent individuals; this indicates that the pathogenic
relevance of MCPyV in SCC is unknown (9). Murakami et al reported that MCPyV
was present in 13% cases of cutaneous SCCs from Japanese patients;
this indicates that cutaneous SCC in Japanese patients is
infrequently associated with MCPyV (13). Imajoh et al reported that MCPyV
was present in 19% of cervical SCCs from Japanese patients; this
suggested that MCPyV may be a cofactor of human papillomavirus for
tumour initiation and/or progression (14).
Oral SCC is the major type of oral cancer. Loyo
et al detected low levels of MCPyV DNA in 19/47 oral SCCs
and suggested that MCPyV was derived from background tissues and
was not associated with oral SCC.
In this study, the prevalence of MCPyV (23/327,
7.0%) in tumours or tumour-like lesions of the oral cavity was
higher compared with that in tumours or cysts of the jaws (2/54,
3.7%). However, the difference was not statistically significant
(P=0.5537, Fisher's exact test).
Baez et al (15) reported that MCPyV was present in 36.7%
samples of saliva and 21.4% oral tissue samples from
immunosuppressed patients. In this study, MCPyV was present in 25%
of immunosuppressed patients, which was compatible with the
prevalence data of Baez et al (15). It is evident that our estimated
prevalence (1/4, 25%) was higher compared with that for oral or jaw
lesions from immunocompetent patients (25/381, 6.6%; P=0.2448,
Fisher's exact test). In the present study, prevalence was
investigated in only 4 cases from immunosuppressed patients.
Therefore, further studies using a large number of cases are
required.
In conclusion, the prevalence of MCPyV DNA in FFPE
samples of oral and maxillofacial tumours and tumour-like lesions
was estimated using qPCR, revealing a low MCPyV prevalence (25/381,
6.6%) with very low or low viral loads (0.00024-0.026 copies/cell)
in oral and maxillofacial tumours and tumour-like lesions from
immunocompetent patients, and also reconfirmed a high prevalence
(1/4, 25%) in oral lesions from immunosuppressed patients. To the
best of our knowledge, this study was the first to report
prevalence data on MCPyV DNA in tumours and cysts of the jaws
(2/54, 3.7%). These data suggest that the detected MCPyV DNA was
derived from non-neoplastic background tissues with widespread
low-level MCPyV infection, and the presence of MCPyV-related
tumourigenesis was not confirmed in oral and maxillofacial tumours
and tumour-like lesions. However, these prevalence study data may
provide valuable insights for further studies on MCPyV infection
and MCPyV-related diseases in the oral and maxillofacial
regions.
Acknowledgements
The present study was supported by the Japan Society
for the Promotion of Science (a Grant-in-Aid for Scientific
Research, no. 26460433). We would like to thank Professor H. Kitano
(Department of Otolaryngology, Head and Neck Surgery, Tottori
University) for kindly providing bioptic or surgically resected
samples. We would also like to thank Professor E. Nanba and all
other members of the Division of Functional Genomics, Research
Centre for Bioscience and Technology, Tottori University, for their
useful advice and excellent technical support.
References
|
1
|
Feng H, Shuda M, Chang Y and Moore PS:
Clonal integration of a polyomavirus in human Merkel cell
carcinoma. Science. 319:1096–1100. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gaynor AM, Nissen MD, Whiley DM, Mackay
IM, Lambert SB, Wu G, Brennan DC, Storch GA, Sloots TP and Wang D:
Identification of a novel polyomavirus from patients with acute
respiratory tract infections. PLoS Pathog. 3:e642007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bialasiewicz S, Lambert SB, Whiley DM,
Nissen MD and Sloots TP: Merkel cell polyomavirus DNA in
respiratory specimens from children and adults. Emerg Infect Dis.
15:492–494. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Goh S, Lindau C, Tiveljung-Lindell A and
Allander T: Merkel cell polyomavirus in respiratory tract
secretions. Emerg Infect Dis. 15:489–491. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Katano H, Ito H, Suzuki Y, Nakamura T,
Sato Y, Tsuji T, Matsuo K, Nakagawa H and Sata T: Detection of
Merkel cell polyomavirus in Merkel cell carcinoma and Kaposi's
sarcoma. J Med Virol. 81:1951–1958. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Foulongne V, Kluger N, Dereure O, Mercier
G, Molès JP, Guillot B and Segondy M: Merkel cell polyomavirus in
cutaneous swabs. Emerg Infect Dis. 16:685–687. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Loyo M, Guerrero-Preston R, Brait M, Hoque
MO, Chuang A, Kim MS, Sharma R, Liégeois NJ, Koch WM, Califano JA,
et al: Quantitative detection of Merkel cell virus in human tissues
and possible mode of transmission. Int J Cancer. 126:2991–2996.
2010.PubMed/NCBI
|
|
8
|
Teman CJ, Tripp SR, Perkins SL and
Duncavage EJ: Merkel cell polyomavirus (MCPyV) in chronic
lymphocytic leukemia/small lymphocytic lymphoma. Leuk Res.
35:689–692. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dworkin AM, Tseng SY, Allain DC, Iwenofu
OH, Peters SB and Toland AE: Merkel cell polyomavirus in cutaneous
squamous cell carcinoma of immunocompetent individuals. J Invest
Dermatol. 129:2868–2874. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kassem A, Technau K, Kurz AK, Pantulu D,
Löning M, Kayser G, Stickeler E, Weyers W, Diaz C, Werner M, et al:
Merkel cell polyomavirus sequences are frequently detected in
nonmelanoma skin cancer of immunosuppressed patients. Int J Cancer.
125:356–361. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Reisinger DM, Shiffer JD, Cognetta AB, Jr,
Chang Y and Moore PS: Lack of evidence for basal or squamous cell
carcinoma infection with Merkel cell polyomavirus in
immunocompetent patients with Merkel cell carcinoma. J Am Acad
Dermatol. 63:400–403. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ridd K, Yu S and Bastian BC: The presence
of polyomavirus in non-melanoma skin cancer in organ transplant
recipients is rare. J Invest Dermatol. 129:250–252. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Murakami M, Imajoh M, Ikawa T, Nakajima H,
Kamioka M, Nemoto Y, Ujihara T, Uchiyama J, Matsuzaki S, Sano S, et
al: Presence of Merkel cell polyomavirus in Japanese cutaneous
squamous cell carcinoma. J Clin Virol. 50:37–41. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Imajoh M, Hashida Y, Nemoto Y, Oguri H,
Maeda N, Furihata M, Fukaya T and Daibata M: Detection of Merkel
cell polyomavirus in cervical squamous cell carcinomas and
adenocarcinomas from Japanese patients. Virol J. 9:1542012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Baez CF, Guimarães MA, Martins RA, Zalona
AC, Cossatis JJ, Zalis MG, Cavalcanti SM and Varella RB: Detection
of Merkel cell polyomavirus in oral samples of renal transplant
recipients without Merkel cell carcinoma. J Med Virol.
85:2016–2019. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hashida Y, Imajoh M, Nemoto Y, Kamioka M,
Taniguchi A, Taguchi T, Kume M, Orihashi K and Daibata M: Detection
of Merkel cell polyomavirus with a tumour-specific signature in
non-small cell lung cancer. Br J Cancer. 108:629–637. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Matsushita M, Kuwamoto S, Iwasaki T,
Higaki-Mori H, Yashima S, Kato M, Murakami I, Horie Y, Kitamura Y
and Hayashi K: Detection of Merkel cell polyomavirus in the human
tissues from 41 Japanese autopsy cases using polymerase chain
reaction. Intervirology. 56:1–5. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pantulu ND, Pallasch CP, Kurz AK, Kassem
A, Frenzel L, Sodenkamp S, Kvasnicka HM, Wendtner CM, Zur H and
Ausen A: Detection of a novel truncating Merkel cell polyomavirus
large T antigen deletion in chronic lymphocytic leukemia cells.
Blood. 116:5280–5284. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kuwamoto S, Higaki H, Kanai K, Iwasaki T,
Sano H, Nagata K, Kato K, Kato M, Murakami I, Horie Y, et al:
Association of Merkel cell polyomavirus infection with morphologic
differences in Merkel cell carcinoma. Hum Pathol. 42:632–640. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Foulongne V, Dereure O, Kluger N, Molès
JP, Guillot B and Segondy M: Merkel cell polyomavirus DNA detection
in lesional and nonlesional skin from patients with Merkel cell
carcinoma or other skin diseases. Br J Dermatol. 162:59–63. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Touzé A, Gaitan J, Maruani A, Le Bidre E,
Doussinaud A, Clavel C, Durlach A, Aubin F, Guyétant S, Lorette G,
et al: Merkel cell polyomavirus strains in patients with merkel
cell carcinoma. Emerg Infect Dis. 15:960–962. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Martel-Jantin C, Filippone C, Cassar O,
Peter M, Tomasic G, Vielh P, Brière J, Petrella T, Aubriot-Lorton
MH, Mortier L, et al: Genetic variability and integration of Merkel
cell polyomavirus in Merkel cell carcinoma. Virology. 426:134–142.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Boukamp P: UV-induced skin cancer:
Similarities - variations. J Dtsch Dermatol Ges. 3:493–503. 2005.
View Article : Google Scholar : PubMed/NCBI
|