Introduction
Lung cancer is one of the most common causes of
cancer-related mortality. To date, multiple genes have been found
to exhibit altered expression in the pathogenesis of lung cancer
(1,2).
Furthermore, the altered expression of these genes was found to be
closely associated with the mutation or methylation of promoter
sites of these genes (3,4). The overall 5-year survival rate of lung
cancer patients receiving traditional treatment remains poor
(5). Thus, it is crucial to identify
and characterize novel molecular markers and gene targets, as well
as investigate the mechanism underlying their altered expression,
in order to improve the accuracy of prognosis and develop optimal
targeted treatment strategies to improve the clinical outcome of
lung cancer patients.
Recently, a growing body of literature has
demonstrated that microRNAs (miRNAs), a class of non-coding RNA
molecules that regulate the expression of protein-coding genes
through binding to the 3′-untranslated regions of target mRNAs,
have emerged as important regulators in the development of lung
cancer and a novel potential target for the development of
therapeutic strategies for lung cancer patients (6,7). Various
specific miRNA molecules were reported to exhibit repressed
expression in lung cancer tissues and to be closely associated with
patient prognosis (8,9). However, the mechanism underlying the
altered expression of these specific miRNA molecules in lung
cancer, including mutation or methylation of their promoter
regions, remains largely unknown.
miRNA (miR)-7, a distinct member of the miRNA
family, was reported to be able to regulate the biology of various
tumor cells through repressing the expression of different target
molecules (10–13). miR-7 expression was found to be
repressed in lung cancer tissues (7,14).
Moreover, miR-7 was able to suppress the growth and metastatic
potential of human lung cancer cells in vitro (15). Our previous studies also demonstrated
that overexpression of miR-7 reduced the growth and metastasis of
human lung cancer cells in vivo (16,17). These
findings suggested that miR-7, as a tumor suppressor, may be a
potential candidate as a prognostic marker and a therapeutic target
in lung cancer. However, the presence of mutations in the miR-7
promoter and their possible association with the prognosis of lung
cancer remain to be elucidated.
The aim of this study was to investigate the
presence of promoter mutations of miR-7 in lung cancer and their
possible association with the prognosis of the patients.
Patients and methods
Patients and tissue samples
A total of 39 Chinese patients who were diagnosed
with lung cancer between 2007 and 2008 were included in the present
study. Clinical and pathological information, including age,
gender, smoking status, type of tumor and disease stage, were
collected. Paraffin blocks and fresh-frozen tumor specimens of
tumor samples from all 39 patients were prepared. In addition, 8
samples from normal tissues adjacent to the tumors were also
collected. All the patients were followed up until December, 2012.
This study was approved by the Ethics Committee of the First
Hospital of Zunyi Medical College (Guizhou, China) and written
informed consent was obtained from all the participants.
Sample preparation
Tissue sections (4–5 µm) were cut from the paraffin
blocks for pathological analysis. Total RNA was extracted from the
fresh-frozen tumor specimens for the detection of miR-7 expression
in lung cancer or normal lung tissues. Genomic DNA was isolated
from the frozen specimens using a NucleoSpin Tissue kit (Clontech
Laboratories Inc., Mountain View, CA, USA) according to the
manufacturer's instructions. The DNA samples were frozen at −70°C
until use.
Amplification of the 5′-flanking
region of the human miR-7 gene by polymerase chain reaction (PCR)
analysis
PCR primer sets were designed to amplify a 1.3-kb
product containing −1068 and +234 sites from the 5′-flanking region
of the human miR-7-2 gene. The sequences of the primer sets were as
follows: Sense (−1068 to −1051): 5′-AGCACCAATAGGGAAGGG-3′; and
antisense (+217 to +234): 5′-GAGTCTGCCGATGGGTGT-3′. PCR was
performed in 50 µl of reaction mixture containing 20 mmol/l
Tris-HCl, pH 8.8, 2 mmol/l MgSO4, 10 mmol/l KCl, 10
mmol/l (NH4)2SO4, 0.1% Triton
X-100, 1 mg/ml nuclease-free bovine serum albumin, 10 mmol/l each
of dATP, dCTP, dGTP and dTTP,, 0.2 pM of each primer, 6% dimethyl
sulfoxide, 1 µg genomic DNA and 2.5 U Pyrococcus furiosus
DNA polymerase. The thermal cycling settings for PCR included a
5-min initial denaturation at 95°C followed by 35 amplification
cycles (denaturation for 1 min at 94°C, annealing for 1 min at 58°C
and extension for 1.5 min at 72°C, with a final extension step at
72°C for 10 min). The PCR products were analyzed on a 1% agarose
gel in Tris-acetate buffer with ethidium bromide staining. The PCR
products were then purified from the agarose gels using the
GeneClean kit (Bio101 Inc., Vista, CA, USA) according to the
manufacturer's recommendations.
Sequence analysis
To investigate mutations in the promoter region of
the human miR-7 gene, the above products of PCR templates were
prepared and the nucleotide sequence was determined on both strands
by Sanger's dideoxynucleotide chain-termination method with
Sequenase 2.0 (Amersham Pharmacia Biotech, Piscataway, NJ, USA).
Multiple overlapping fragments were sequenced at least twice in
each direction and the DNA sequence was analyzed using MacVector
software (Eastman Kodak Co., Rochester, NY, USA). DNA sequence
analysis was performed by a manual method (Thermo Sequenase Cycle
Sequencing kit; Amersham Pharmacia Biotech) and an automatic
sequence method (Applied Biosystems, Foster City, CA, USA)
according to the manufacturer's instructions.
Construction of the eukaryotic
vector
PCR primer sets were designed to amplify a 1.3-kb
product containing −1068 and +234 sites from the 5′-flanking region
of the human miR-7-2 gene. The sequences of the primer sets were as
follows: Sense (−1068 to −1051): 5′-CTA GCT AGC TAG AGC ACC AAT AGG
GAA GGG-3′; and antisense (+217 to +234): 5′-GAA GAT CTT CGA GTC
TGC CGA TGG GTG T-3′. The PCR products were amplified from DNA
derived from lung cancer tissues with or without miR-7 promoter
mutations, then subcloned into NheI and BglII sites
of the pGL3.0 basic vector (Invitrogen Corporation, San Diego, CA,
USA) to generate the pGL3.0-miR-7 expression plasmid [referred to
as p-wild-type (WT)-miR-7 or p-mutation (Mu)-miR-7, respectively].
For the construction of plasmids pGL-miR-7 promoter Luc, the
promoter region (−1068 to 0 bp) of miR-7 was amplified from DNA
derived from lung cancer tissues with or without miR-7 promoter
mutations using a forward primer (5′-CTA GCT AGC TAG AGC ACC AAT
AGG GAA GGG) and a reverse primer (5′-GAA GAT CTT CGC CAG TTC TGC
AAG GCG T) and subcloned into NheI and BglII sites of
the pGL basic vector (referred to as p-WT-promoter or
p-Mu-promoter, respectively). Clone identity was verified using
restriction digest analysis and plasmid DNA sequencing.
Endotoxin-free plasmids were obtained using the EndoFree Plasmid
Mega kit (Qiagen, Hilden, Germany). The plasmids were then
transiently transferred into the 95D human lung cancer cells using
Lipofectamine®-2000 (Invitrogen Corporation) in the following
experiments according to the manufacturer's instructions.
Luciferase reporter assay
The 95D cells were transiently co-transfected with
the p-WT-promoter or p-Mu-promoter and pCMV-lacZ plasmids using
Lipofectamine-2000 (Invitrogen Corporation) according to the
manufacturer's instructions and cultured in 37°C. After 24 h,
luciferase and β-galactosidase (β-gal) activity in 100 µl of cell
lysate were measured using the Luciferase Assay system and the
β-Galactosidase Enzyme Assay system (Promega Corporation, Madison,
WI, USA), respectively. Transfection efficiency was normalized
using β-gal activity.
Quantitative PCR (qPCR) assay
All the reagents, primers and probes were obtained
from Applied Biosystems. The relative expression of miR-7 was
determined as previously described (17). Briefly, a β-actin endogenous control
was used for normalization. Reverse transcription (RT) reactions
and qPCR were performed according to the manufacturer's protocols
(Applied Biosystems). RNA concentrations were determined with a
NanoDrop instrument (NanoDrop Technologies, Wilmington, DE, USA).
One nanogram of RNA per sample was used for the assays. All RT
reactions, including no-template controls and RT minus controls,
were run in triplicate in GeneAmp® PCR 9700 thermal cycler (Applied
Biosystems). The gene expression levels were quantified using the
ABI PRISM® 7900HT Sequence Detection system (Applied Biosystems).
Relative expression was calculated using the comparative threshold
cycle method.
Cell proliferation assays
95D cells transiently transfected with 10 nmol
p-WT-miR-7 or p-Mu-miR-7 vector using Lipofectamine-2000
(Invitrogen Corporation) were seeded at 3×103 cells per
well and incubated at 37°C in 5% CO2 in 96-well plates
for 72 h. Cell proliferation was measured in terms of optical
absorbance per well by a semi-automated tetrazolium-based
colorimetric assay using MTT.
Statistical analysis
Statistical evaluation was performed using one-way
analysis of variance (P<0.05). All comparisons between
categorical variables were performed by the Fisher's exact
Chi-square test. Relapse-free survival was calculated using the
Kaplan-Meier survival estimates and the log-rank test from the date
of diagnosis until the last contact or relapse. P<0.05 was
considered to indicate a statistically significant difference.
Statistical analyses were conducted using SPSS 13.0 software for
Windows (SPSS, Inc., Chicago, IL, USA).
Results
Promoter mutation of miR-7 in lung
cancer
To elucidate the mechanism underlying the reduced
expression of human miR-7 in lung cancer, we first searched for the
putative promoter region in the 5′-flanking region, which may alter
miR-7 gene transcription according to a previous report (13). Sequence analysis revealed putative
transcription factor binding sites for SRY, c-Myc, Gfi-1 and CdxA
in the 5′-flanking region, suggesting that the expression of the
human miR-7 gene may be controlled by these regulatory elements
(data not shown). Primer sets were used to amplify overlapping
fragments from the 5′-flanking region from −1068 bp upstream to
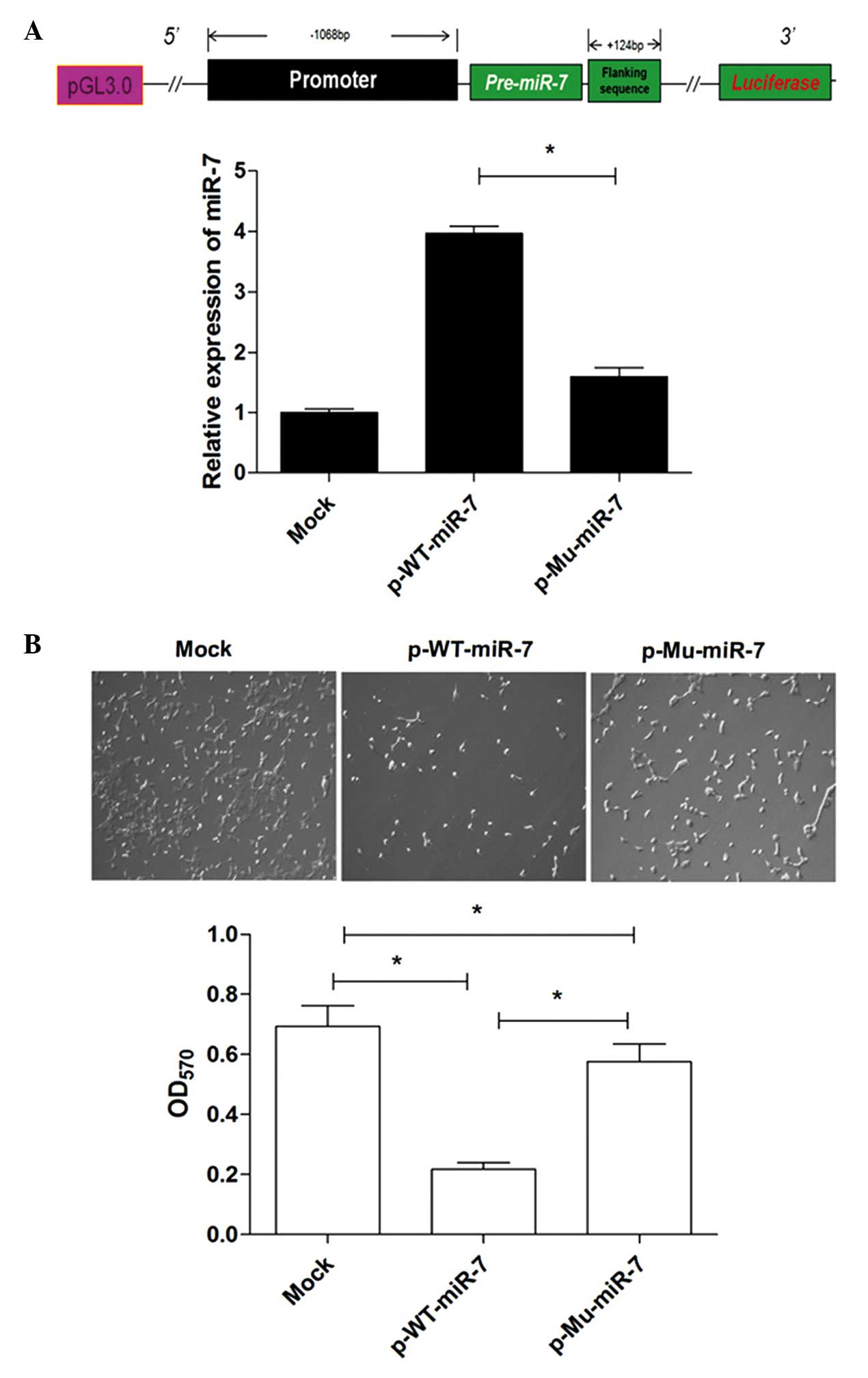
+234 bp downstream from the initiation site of pre-miR-7 (Fig. 1A) from the genomic DNA. The sizes of
PCR products amplified from the genomic DNA of lung cancer and
normal lung tissues were identical and corresponded to the expected
sizes, indicating that there were no major additions or deletions
in the 5′-flanking region of the human miR-7 gene in the lung
cancer tissues examined. Subsequently, the PCR products were
sequenced to assess the promoter mutation of miR-7 of lung cancer
tissues from 39 lung cancer patients and normal lung tissues from 8
healthy donors, respectively. Detailed DNA sequence analysis
identified two mutations in the PCR region in lung cancer tissues,
namely a G→C change at −617 and a A→G change at −604 (Fig. 1B). Moreover, as shown in Table I, the promoter mutation at the −617
site of miR-7 was detected in 25 patients (64.1%) and at the −604
site in 20 patients (51.3%). All A→G mutations were found in
combination with the G→C mutation; the A→G mutation alone was not
detected in any patients (0%), whereas the G→C mutation alone was
detected in 5 patients (12.8%). However, promoter mutations were
not detected in normal lung tissues (0%). We then investigated the
association between mutation sites of the miR-7 promoter and a
significant correlation was observed between the G→C and A→G sites
(P<0.05).
 | Table I.Mutation in the promoter region of the
human microRNA-7 gene in lung cancer (n=39). |
Table I.
Mutation in the promoter region of the
human microRNA-7 gene in lung cancer (n=39).
| Mutation | Position | No. (%) |
|---|
| G→C | −617 | 25 (64.1) |
| A→G | −604 | 20 (51.3) |
| G→C and A→G | −617 and −604 | 25 (64.1) |
| No mutation |
| 14 (35.9) |
The mutation sites alter the activity
of the miR-7 promoter
To determine whether the mutations affected the
activity of the miR-7 promoter, PCR-amplified promoter fragments
(−1068 to 0), with or without the two mutations, were obtained from
lung cancer and normal lung tissues and subcloned into the
pGL3-basic vector (referred to as the p-Mu promoter and the p-WT
promoter, respectively). The promoter activities were then
evaluated by firefly luciferase reporter gene expression normalized
to a co-transfected pCMV-lacZ as a control for transfection
efficiency. The normalized luciferase reporter activities indicated
that the mutations in the promoter significantly reduced promoter
activity (Fig. 2, P<0.05).
Compared with normal promoter activity, the mutated promoter
activity decreased by >50% in the pGL3 constructs. These data
suggested that the mutation sites reduced the activity of the miR-7
promoter.
Promoter mutation reduces miR-7
expression in lung cancer cells
To further determine whether these mutations
affected the activity of the miR-7 promoter, subsequently altering
the expression of miR-7, the fragment containing the promoter
region and the pre-miR-7 region (−1068 to +234), with or without
the two mutations, from lung cancer and normal lung tissues, was
further amplified and subcloned into the pGL3-basic vector to
construct miR-7 expression vectors (referred to as p-Mu-miR-7 and
p-WT-miR-7, respectively). Human lung cancer cells were transiently
transfected with these constructed plasmids. The relative
expression level of miR-7 was then determined by qPCR analysis. The
data revealed that the expression level of miR-7 was significantly
decreased in the p-Mu-miR-7 vector transfection group compared with
that in the p-WT-miR-7 vector transfection group (Fig. 3A, P<0.05), indicating that the
mutation sites reduced the expression of miR-7. Consistently, the
growth of 95D cells in the p-Mu-miR-7 vector transfection group was
higher compared with that in the p-WT-miR-7 vector transfection
group (Fig. 3B, P<0.05), which was
consistent with our previous findings (17). Combining these data demonstrated that
the mutation sites significantly reduced the activity of the miR-7
promoter and altered the expression of miR-7, subsequently
affecting its biological activity.
Promoter mutation is associated with
repressed expression of miR-7 in lung cancer tissues
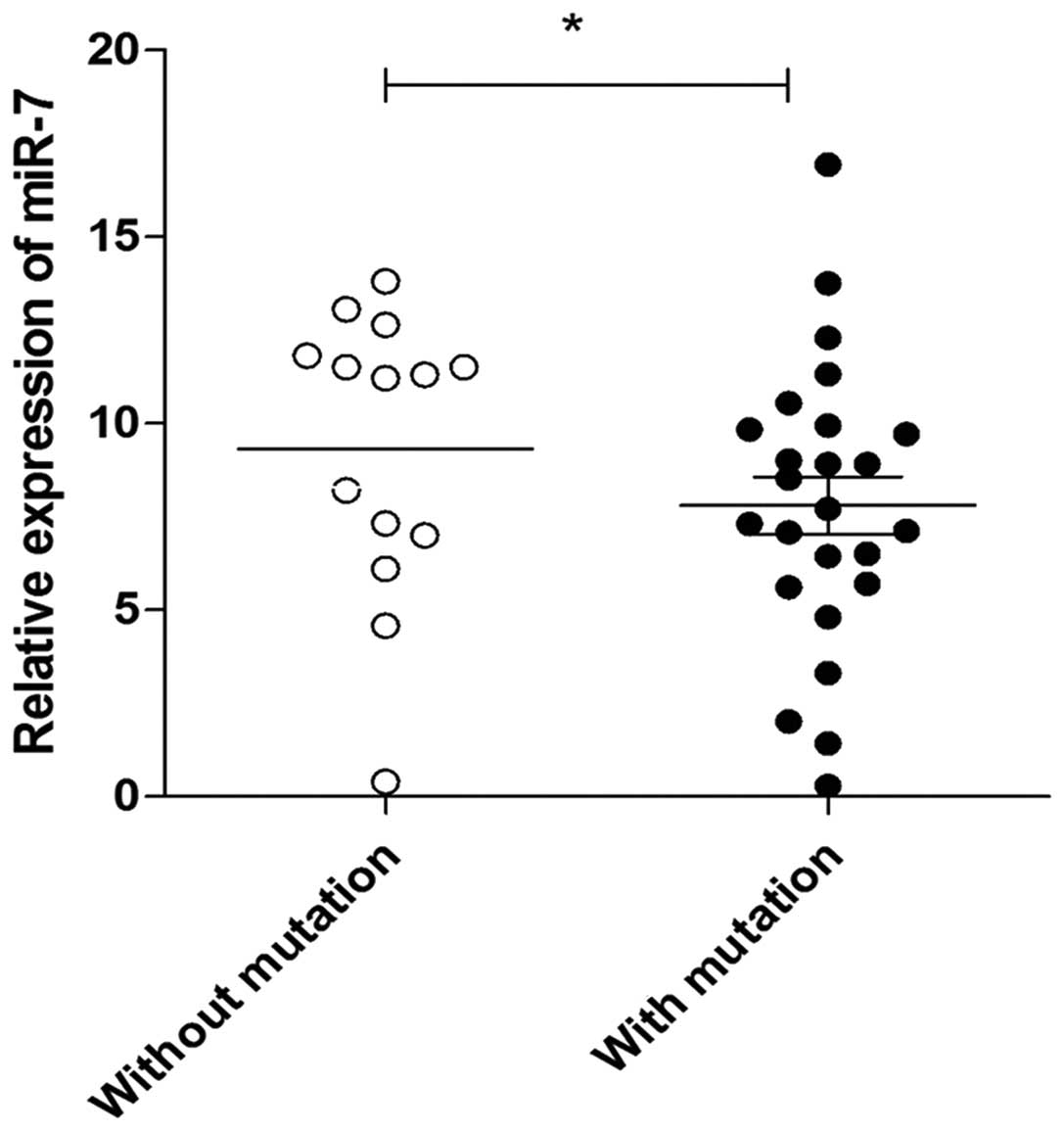
We next sought to determine whether the mutation of
the miR-7 promoter was associated with the expression of miR-7 in
lung cancer tissues. The relative expression of miR-7 was also
assessed by qPCR in 39 lung cancer and 8 normal lung tissue
specimens. Consistent with previous findings (14), the relative expression of miR-7
decreased significantly in lung cancer tissues compared with that
in normal lung tissues (data not shown). Notably, we found that the
expression level of miR-7 in lung cancer tissues with mutation
sites was lower compared with that in lung cancer tissues without
mutation sites (Fig. 4, P<0.05),
indicating that the promoter mutation is closely associated with
the repressed expression of miR-7 in lung cancer tissues.
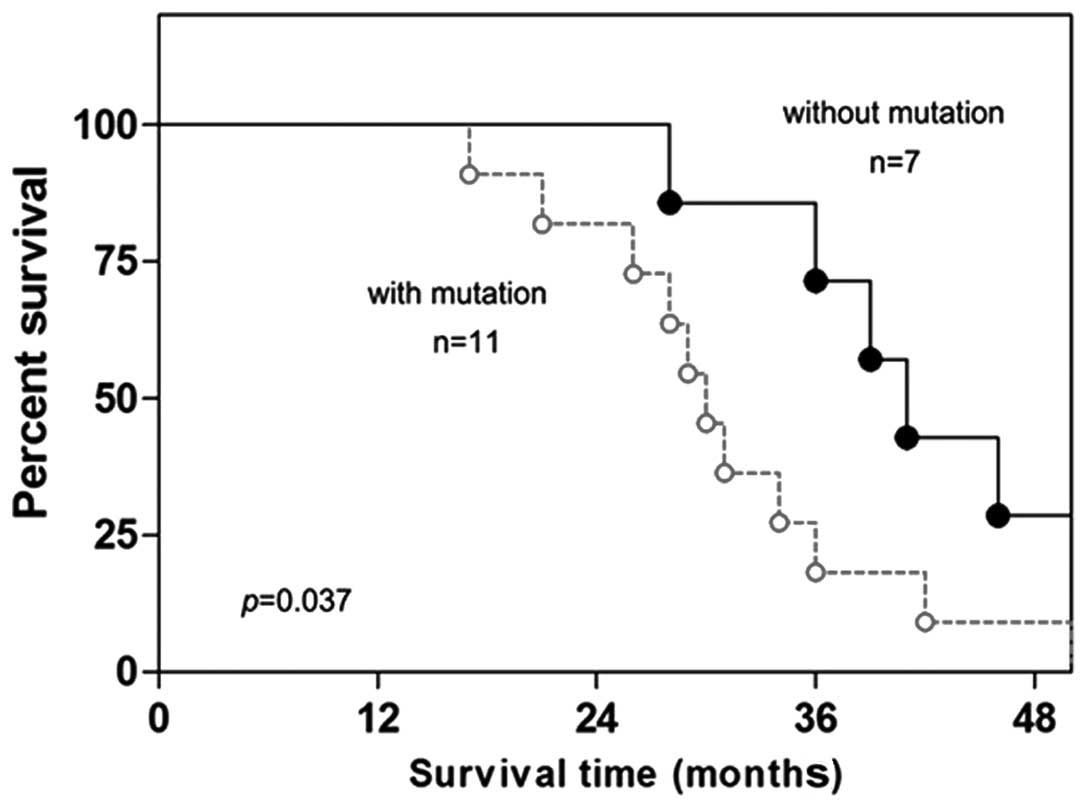
Promoter mutation of miR-7 is
associated with poor survival of lung cancer patients
Finally, the possible value of miR-7 promoter
mutations in the prognosis of lung cancer patients was analyzed.
There was no association between mutation and age at diagnosis,
gender, or smoking status (Table
II). miR-7 promoter mutations were more common in
adenocarcinoma (ADC) (13/17, 76.5%) compared with squamous cell
carcinoma (7/13, 53.8%) and large-cell carcinoma (5/9, 55.6%),
although the difference was not statistically significant
(P=0.357). However, a statistically significant association was
observed between the mutations and stage II–IV disease according to
the American Joint Committee on Cancer (AJCC), with miR-7 promoter
mutations being present at a higher frequency in stage II–IV
(25/32, 78.1%) compared with stage I disease (2/7, 40.0%)
(P=0.020). Finally, the Kaplan-Meier long-rank analysis revealed
that the presence of promoter mutation of miR-7 was associated with
poorer overall survival (Fig. 5,
P=0.037).
 | Table II.Association of miR-7 promoter
mutations in cancer tissue with the clinicopathological
characteristics of lung cancer patients (n=39). |
Table II.
Association of miR-7 promoter
mutations in cancer tissue with the clinicopathological
characteristics of lung cancer patients (n=39).
|
| Promoter mutation
status, patient no. (%) |
|
|---|
|
|
|
|
|---|
| Clinicopathological
characteristics | Positive | Negative | P-value |
|---|
| Age, years |
|
| 0.396 |
| <60
(n=15) | 10 (66.7) | 5
(33.3) |
|
| >60
(n=24) | 15 (62.5) | 9
(37.5) |
|
| Gender |
|
| 0.635 |
| Male
(n=33) | 21 (63.6) | 12 (36.4) |
|
| Female
(n=6) | 4
(66.7) | 2
(33.3) |
|
| Smoking status |
|
| 0.493 |
| Ever
(n=32) | 21 (65.6) | 11 (34.4) |
|
| Never
(n=7) | 4
(57.1) | 3
(42.9) |
|
| Histologic
types |
|
| 0.357 |
| SCC
(n=13) | 7
(53.8) | 6
(46.2) |
|
| ADC
(n=17) | 13 (76.5) | 4
(23.3) |
|
| LCC
(n=9) | 5
(55.6) | 4
(44.4) |
|
| Pathological
stage |
|
| 0.020 |
| I
(n=7) | 2
(28.6) | 5
(71.4) |
|
| II–IV
(n=32) | 25 (78.1) | 7
(21.9) |
|
Discussion
miR-7, a unique member of the miRNA family, has been
proposed to be a new type of tumor suppressor gene, or oncomiRNA,
in several tumor types (13,18–20). In
lung cancer, recent evidence demonstrated that the expression of
miR-7 is decreased in cancer tissues (14). Moreover, miR-7 was found to inhibit
the growth and metastasis of lung cancer cells and induce their
apoptosis (21). Mechanistic evidence
demonstrated that miR-7 was able to regulate the transduction of
the Akt pathway, which is crucial for the growth and metastasis of
tumor cells (22,23). In line with these findings, our
previous study also demonstrated that the overexpression of miR-7
inhibited the growth and metastasis of lung cancer cells (17). Consistently, more recent evidence
further demonstrated that restoration of miR-7 expression
suppressed the tumorigenicity of lung cancer cells in vivo
(24) and overexpression of miR-7
improved the sensitivity of lung cancer cells to paclitaxel
(25). These data suggested that
miR-7 may be an important regulator and have multiple functions in
the development of lung cancer. Of note, a recent study reported
that stable overexpression of miR-7 promoted the growth and
migration of a lung cancer cell clone by regulating the expression
of Kruppel-like factor 4 (26).
Combing these data indicated that the precise role of miR-7 in the
development of lung cancer is complex, and closely associated with
expression level, cellular context and growth conditions, as well
as the specific targeted genes. Therefore, investigating the
regulation of expression of miR-7 in cancers may be of significant
value. However, to date, the mechanism underlying the altered
expression of miR-7 remains largely unknown.
In the present study, we identified two mutation
sites in the miR-7 promoter in lung cancer. Moreover, there was a
positive association between the −617 and −604 sites. Importantly,
the expression level of miR-7 in lung cancer with mutations of the
promoter sites was clearly decreased. Moreover, the transcriptional
activity driven by the mutated promoter from lung cancer tissues
was significantly reduced. These results suggested that the reduced
miR-7 expression in lung cancer is, at least in part, due to a
defect in the promoter of the gene. In addition, mutations were not
detected in all cancer tissues; in fact, no mutation was found in
~35.9% of lung cancer patients. This finding suggests that other
contributing factors are also involved in the altered expression of
the miR-7 gene in lung cancer. Possible non-mutational causes for
the reduced expression of miR-7 in lung cancer include the presence
of transcription activators or repressors, as well as
post-transcriptional factors. In fact, it was previously reported
that c-Myc may bind to the promoter region of miR-7 and enhance
miR-7 expression in lung cancer cells (13). Our recent findings further
demonstrated that the human R antigen post-transcriptionally
regulates the expression of miR-7 (17). In addition, recent evidence indicated
that histone methylation or CpG methylation may affect the
expression of distinct miRNA molecules in certain types of cancers
(27,28). Therefore, the predominant mechanism
involved in the repressed expression of the miR-7 gene remains to
be fully elucidated in future studies.
Accumulating data suggests that the mutation or
methylation of the promoter region of certain molecules, which was
found to be associated with major prognostic factors, has emerged
as a useful novel biological marker for the prognosis of cancer
patients. Similarly, miR-7 was reported to be downregulated in
various cancers, including pancreatic and breast cancer, and was
closely associated with the metastatic status of the patients
(12,29). In the present study, we observed that
the frequency of miR-7 promoter mutation was higher in the ADC type
of lung cancer, although the difference was not statistically
significant. Moreover, miR-7 promoter mutations were also
associated with stage II–IV of lung cancer. Finally, we further
demonstrated a significant correlation between miR-7 promoter
mutation and poor prognosis of lung cancer patients, indicating
that miR-7 promoter mutation is of significant prognostic value.
Similarly, it has been reported that miR-7 exhibits high diagnostic
accuracy and may be a helpful adjunct to thyroid fine-needle
aspiration biopsy (30). In addition,
we also noted that there was no significant association between
miR-7 promoter mutation and other factors, such as smoking status.
Therefore, a Cox regression analysis including other prognostic
factors, such as tumor size, lymph node status and adjuvant
therapy, which were not investigated in the present study, may be
of value for validation of the significance of promoter mutation of
miR-7 in the prognosis of lung cancer patients.
In conclusion, the present study demonstrated that
there are mutation sites in the miR-7 promoter region in lung
cancer tissues, which alter the expression of miR-7. Moreover, the
mutation sites of the miR-7 promoter are closely associated with
the poor prognosis of lung cancer patients. These data may provide
a novel insight in the mechanism underlying the altered expression
of distinct miRNA molecules in lung cancer, and may be helpful in
the development of novel prognostic methods and therapeutic targets
against lung cancer.
Acknowledgements
This study was supported by the Program for New
Century Excellent Talents in University, Ministry of Education of
China (grant no. NCET-12-0661), the National Natural Science
Foundation of China (grant nos. 31370918 and 81372347) and the
International Cooperation Foundation of Guizhou Province (grant no.
2010-7031).
Glossary
Abbreviations
Abbreviations:
|
miRNA
|
microRNA
|
|
miR-7
|
miRNA-7
|
|
HuR
|
human R antigen
|
|
Mu
|
mutation
|
|
WT
|
wild-type
|
References
|
1
|
Osada H and Takahashi T: Genetic
alterations of multiple tumor suppressors and oncogenes in the
carcinogenesis and progression of lung cancer. Oncogene.
21:7421–7434. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Carneiro JG, Couto PG, Bastos-Rodrigues L,
Bicalho MA, Vidigal PV, Vilhena A, Amaral NF, Bale AE, Friedman E
and De Marco L: Spectrum of somatic EGFR, KRAS, BRAF, PTEN
mutations and TTF-1 expression in Brazilian lung cancer patients.
Genet Res (Camb). 96:e0022014.PubMed/NCBI
|
|
3
|
Zöchbauer-Müller S, Fong KM, Virmani AK,
Geradts J, Gazdar AF and Minna JD: Aberrant promoter methylation of
multiple genes in non-small cell lung cancers. Cancer Res.
61:249–255. 2001.PubMed/NCBI
|
|
4
|
Esteller M, Sanchez-Cespedes M, Rosell R,
Sidransky D, Baylin SB and Herman JG: Detection of aberrant
promoter hypermethylation of tumor suppressor genes in serum DNA
from non-small cell lung cancer patients. Cancer Res. 59:67–70.
1999.PubMed/NCBI
|
|
5
|
Kocaturk CI, Gunluoglu MZ, Cansever L,
Demir A, Cinar U, Dincer SI and Bedirhan MA: Survival and
prognostic factors in surgically resected synchronous multiple
primary lung cancers. Eur J Cardiothorac Surg. 39:160–166. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yanaihara N, Caplen N, Bowman E, Seike M,
Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et
al: Unique microRNA molecular profiles in lung cancer diagnosis and
prognosis. Cancer Cell. 9:189–198. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Boeri M, Verri C, Conte D, Roz L, Modena
P, Facchinetti F, Calabrò E, Croce CM, Pastorino U and Sozzi G:
MicroRNA signatures in tissues and plasma predict development and
prognosis of computed tomography detected lung cancer. Proc Natl
Acad Sci USA. 108:3713–3718. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Huang CJ, Nguyen PN, Choo KB, Sugii S, Wee
K, Cheong SK and Kamarul T: Frequent co-expression of miRNA-5p and
−3p species and cross-targeting in induced pluripotent stem cells.
Int J Med Sci. 11:824–833. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hu Z, Chen X, Zhao Y, Tian T, Jin G, Shu
Y, Chen Y, Xu L, Zen K, Zhang C, et al: Serum microRNA signatures
identified in a genome-wide serum microRNA expression profiling
predict survival of non-small-cell lung cancer. J Clin Oncol.
28:1721–1726. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xu K, Chen Z, Qin C and Song X: miR-7
inhibits colorectal cancer cell proliferation and induces apoptosis
by targeting XRCC2. Onco Targets Ther. 7:325–332. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li Y, Li Y, Liu Y, Xie P, Li F and Li G:
PAX6, a novel target of microRNA-7, promotes cellular proliferation
and invasion in human colorectal cancer cells. Dig Dis Sci.
59:598–606. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kong X, Li G, Yuan Y, He Y, Wu X, Zhang W,
Wu Z, Chen T, Wu W, Lobie PE, et al: MicroRNA-7 inhibits
epithelial-to-mesenchymal transition and metastasis of breast
cancer cells via targeting FAK expression. PLoS One. 7:e415232012.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chou YT, Lin HH, Lien YC, Wang YH, Hong
CF, Kao YR, Lin SC, Chang YC, Lin SY, Chen SJ, et al: EGFR promotes
lung tumorigenesis by activating miR-7 through a Ras/ERK/Myc
pathway that targets the Ets2 transcriptional repressor ERF. Cancer
Res. 70:8822–8831. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xiong S, Zheng Y, Jiang P, Liu R, Liu X,
Qian J, Gu J, Chang L, Ge D and Chu Y: PA28gamma emerges as a novel
functional target of tumour suppressor microRNA-7 in non-small-cell
lung cancer. Br J Cancer. 110:353–362. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rai K, Takigawa N, Ito S, Kashihara H,
Ichihara E, Yasuda T, Shimizu K, Tanimoto M and Kiura K: Liposomal
delivery of MicroRNA-7-expressing plasmid overcomes epidermal
growth factor receptor tyrosine kinase inhibitor-resistance in lung
cancer cells. Mol Cancer Ther. 10:1720–1727. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xu L, Wen Z, Zhou Y, Liu Z, Li Q, Fei G,
Luo J and Ren T: microRNA-7-regulated TLR9 signaling-enhanced
growth and metastatic potential of human lung cancer cells by
altering the phosphoinositide-3-kinase, regulatory subunit 3/Akt
pathway. Mol Biol Cell. 24:42–55. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li YJ, Wang CH, Zhou Y, Liao ZY, Zhu SF,
Hu Y, Chen C, Luo JM, Wen ZK and Xu L: TLR9 signaling repressed
tumor suppressor miR-7 expression through up-regulation of HuR in
human lung cancer cells. Cancer Cell Int. 13:902013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yu Z, Ni L, Chen D, Zhang Q, Su Z, Wang Y,
Yu W, Wu X, Ye J, Yang S, et al: Identification of miR-7 as an
oncogene in renal cell carcinoma. J Mol Histol. 44:669–677. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang X, Hu S, Zhang X, Wang L, Zhang X,
Yan B, Zhao J, Yang A and Zhang R: MicroRNA-7 arrests cell cycle in
G1 phase by directly targeting CCNE1 in human hepatocellular
carcinoma cells. Biochem Biophys Res Commun. 443:1078–1084. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Okuda H, Xing F, Pandey PR, Sharma S,
Watabe M, Pai SK, Mo YY, Iiizumi-Gairani M, Hirota S, Liu Y, et al:
miR-7 suppresses brain metastasis of breast cancer stem-like cells
by modulating KLF4. Cancer Res. 73:1434–1444. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xiong S, Zheng Y, Jiang P, Liu R, Liu X
and Chu Y: MicroRNA-7 inhibits the growth of human non-small cell
lung cancer A549 cells through targeting BCL-2. Int J Biol Sci.
7:805–814. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kefas B, Godlewski J, Comeau L, Li Y,
Abounader R, Hawkinson M, Lee J, Fine H, Chiocca EA, Lawler S, et
al: microRNA-7 inhibits the epidermal growth factor receptor and
the Akt pathway and is down-regulated in glioblastoma. Cancer Res.
68:3566–3572. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Webster RJ, Giles KM, Price KJ, Zhang PM,
Mattick JS and Leedman PJ: Regulation of epidermal growth factor
receptor signaling in human cancer cells by microRNA-7. J Biol
Chem. 284:5731–5741. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li J, Zheng Y, Sun G and Xiong S:
Restoration of miR-7 expression suppresses the growth of Lewis lung
cancer cells by modulating epidermal growth factor receptor
signaling. Oncol Rep. 32:2511–2516. 2014.PubMed/NCBI
|
|
25
|
Liu R, Liu X, Zheng Y, Gu J, Xiong S,
Jiang P, Jiang X, Huang E, Yang Y, Ge D, et al: MicroRNA-7
sensitizes non-small cell lung cancer cells to paclitaxel. Oncol
Lett. 8:2193–2200. 2014.PubMed/NCBI
|
|
26
|
Meza-Sosa KF, Pérez-García EI,
Camacho-Concha N, López-Gutiérrez O, Pedraza-Alva G and
Pérez-Martínez L: miR-7 promotes epithelial cell transformation by
targeting the tumor suppressor KLF4. PLoS One. 9:e1039872014.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ben Gacem R, Ben Abdelkrim O, Ziadi S, Ben
Dhiab M and Trimeche M: Methylation of miR-124a-1, miR-124a-2 and
miR-124a-3 genes correlates with aggressive and advanced breast
cancer disease. Tumour Biol. 35:4047–4056. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fiaschetti G, Abela L, Nonoguchi N, Dubuc
AM, Remke M, Boro A, Grunder E, Siler U, Ohgaki H, Taylor MD, et
al: Epigenetic silencing of miRNA-9 is associated with HES1
oncogenic activity and poor prognosis of medulloblastoma. Br J
Cancer. 110:636–647. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Singh S, Chitkara D, Kumar V, Behrman SW
and Mahato RI: miRNA profiling in pancreatic cancer and restoration
of chemosensitivity. Cancer Lett. 334:211–220. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kitano M, Rahbari R, Patterson EE, Xiong
Y, Prasad NB, Wang Y, Zeiger MA and Kebebew E: Expression profiling
of difficult-to-diagnose thyroid histologic subtypes shows distinct
expression profiles and identify candidate diagnostic microRNAs.
Ann Surg Oncol. 18:3443–3452. 2011. View Article : Google Scholar : PubMed/NCBI
|