Introduction
Lung cancer, predominantly non-small-cell lung
cancer (NSCLC, comprising 80% of lung cancers), is the leading
cause of cancer mortality worldwide (1). Despite the advances made in the
diagnosis and treatment of lung cancer in the last few decades, the
prognosis of lung cancer remains very poor. Patients with
early-stage NSCLC who undergo complete tumor resection develop
distant metastases in 50–70% of cases, resulting in 5-year survival
rates of ~40% (2–4). Although the tumor-node-metastasis (TNM)
staging system is the best prognostic index for resectable NSCLC,
patients with the same pathological stage of the disease exhibit a
great variability in recurrence and survival rates (5). Therefore, there is an urgent need to
identify appropriate molecular markers that are associated with the
prognosis of patients with lung cancer.
In a large number of types of human cancer, the
tumor protein 53 (TP53) gene is the most frequently mutated gene
[identified in ~50% of cases of NSCLC (6,7)]. The
TP53 gene contains 11 exons that encode a 53 kDa nuclear
phosphoprotein, termed p53, which exerts an essential role in cell
cycle control and apoptosis. In response to oncogenic cellular
stresses, such as deoxyribonucleic acid (DNA) damage, p53 protein
acts as a transcription factor that induces the expression of
downstream genes, including p21 and BCL2-associated X protein
(BAX), which are involved in cell cycle arrest, DNA repair and
apoptosis. It has previously been reported that p53 protein
overexpression may be an important prognostic marker of decreased
survival rates (8,9). Among these studies, an accumulation of
abnormal p53 protein was detected in the cell nuclei by performing
routine immunohistochemistry (IHC). However, the measurement of p53
expression by IHC has led to inconsistent conclusions, not only due
to variations in the understanding of the term ‘overexpression’,
but also since the accumulation of p53 usually corresponds with the
class of TP53 gene mutation that results in tumors with a
frame-shift or non-sense mutations, and the p53 protein is
therefore not generally detectable by IHC (10,11).
Furthermore, p53 protein concentrations are increased in certain
tumor types that lack any mutations resulting from DNA damage, as
would be caused by, e.g. ionizing radiation or chemotherapeutic
agents, and this may act as a physiological response to allow for
DNA repair (12). Therefore, results
obtained from IHC analysis alone are insufficient to permit an
evaluation of the prognostic importance of TP53 gene mutation.
In recent years, a large number of studies have been
performed to evaluate the impact of TP53 mutations on the prognosis
of patients with NSCLC; however, the results of these studies
remain controversial (13,14). Several studies indicated that
patients with mutations of TP53 survived for a shorter period of
time (15–18), whereas others reported that there was
no significant correlation between TP53 mutation and the survival
rate (19–22). The present study aimed to present a
meta-analysis of the available data on the prognostic significance
of TP53 gene mutations in patients with NSCLC. Due to the
limitations of IHC, this study analyzed data exclusively extracted
from studies employing SSCP (single-stranded conformational
polymorphism) or DNA sequencing to detect mutations of this gene.
The results of the present study may provide a clearer
understanding of the prognostic importance of TP53 mutations in
NSCLC, and its association with clinicopathological features and
clinical outcomes.
Materials and methods
Literature searches
All relevant articles were retrieved by searching
the PubMed, Embase and the Central Registry of Controlled Trials of
the Cochrane Library databases using a combination of the terms
‘TP53’, ‘p53’, ‘p53 protein’, ‘p53 mutation’, ‘lung’,
‘non-small-cell lung cancer’ and ‘NSCLC’. An additional search in
Google Scholar, and a manual search through the reference lists of
pertinent reviews, were additionally performed. Two authors (JC. G
and J. W.) performed the searches independently of each other. No
language or date restrictions were set in the search.
Inclusion and exclusion criteria
Studies considered to be eligible for the present
meta-analysis were required to meet the following criteria: i)
Published trials of any study design were included that examined
the prognostic influence of TP53 mutations in NSCLC; ii) the
subjects had not undergone chemotherapy or radiotherapy prior to
surgery or biopsy, which might have eliminated the effects due to
the TP53 gene; iii) the study had employed DNA techniques for TP53
mutational analysis; iv) the clinical outcomes had been stratified
on the basis of TP53 mutation status; and v) information on the
primary outcome of survival [i.e. overall survival (OS)] was
accessible. Studies failing to meet these inclusion criteria were
excluded.
Outcome measures, data extraction and
quality assessment
The primary outcome for the primary meta-analysis
was OS. Data for OS were extracted as the hazard ratios (HRs) of
patients with TP53 mutations compared with those with wild-type
TP53 in NSCLC and its 95% confidence interval (CI) from the
subgroup analysis. If the HR and its variance were available
directly in an individual trial, these values were subsequently
used. However, since a large number of trials did not report this
information directly, appropriate data, such as P-values of the
log-rank test, were extracted to estimate the log HR and its
variance using the previously reported methods (23,24), and
the time-to-event data were extracted from the survival curves.
Kaplan-Meier curves were read using Engauge Digitizer version 4.1
(free software downloaded from http://sourceforge.net). Data combination was
performed using RevMan version 5.1 (free software downloaded from
http://www.cochrane.org). The log HR and its
variance were pooled using an inverse variance-weighted average,
and the results are presented as HR and 95% CI. The data collection
and assessment of methodological quality were performed according
to the QUORUM and the Cochrane Collaboration guidelines (http://www.cochrane.de). The data on lead author,
patient status, study category, pathological type, TP53 mutation
status, smoking status and OS were extracted by two investigators
(JC. G and J. W.) independently. Three reviewers (JC. G, J. W. and
JW X.) used the Newcastle-Ottawa scale specific to cohort study to
assess all included studies (25).
Discrepancies were discussed with a fourth author (Y.B. Z.) in
order to reach a consensus.
Publication bias
An extensive search strategy was designed in order
to minimize the potential publication bias. Graphical funnel plots
were generated to visually assess a publication bias. The
statistical methods used to detect funnel plot asymmetry were the
rank correlation test of Begg and Mazumdar (26) and the regression asymmetry test of
Egger et al (27).
Statistical analysis
HRs for OS with 95% CIs were pooled. Heterogeneity
across the studies was assessed using a forest plot and the
inconsistency statistic (I2). The random-effects model
was employed in case of potential heterogeneity, and to avoid
underestimation of standard errors of pooled estimates in our
meta-analyses. All calculations were performed using STATA version
11.0 (Stata Corp., College Station, TX, USA). Subgroup analysis was
performed according to the respective study type and treatment
line. An HR value <1 represented a greater benefit for those
without TP53 mutations in terms of the OS value. All CIs had a
two-sided probability coverage of 95%. P<0.05 was considered to
indicate a statistically significant value.
Results
Study identification and
selection
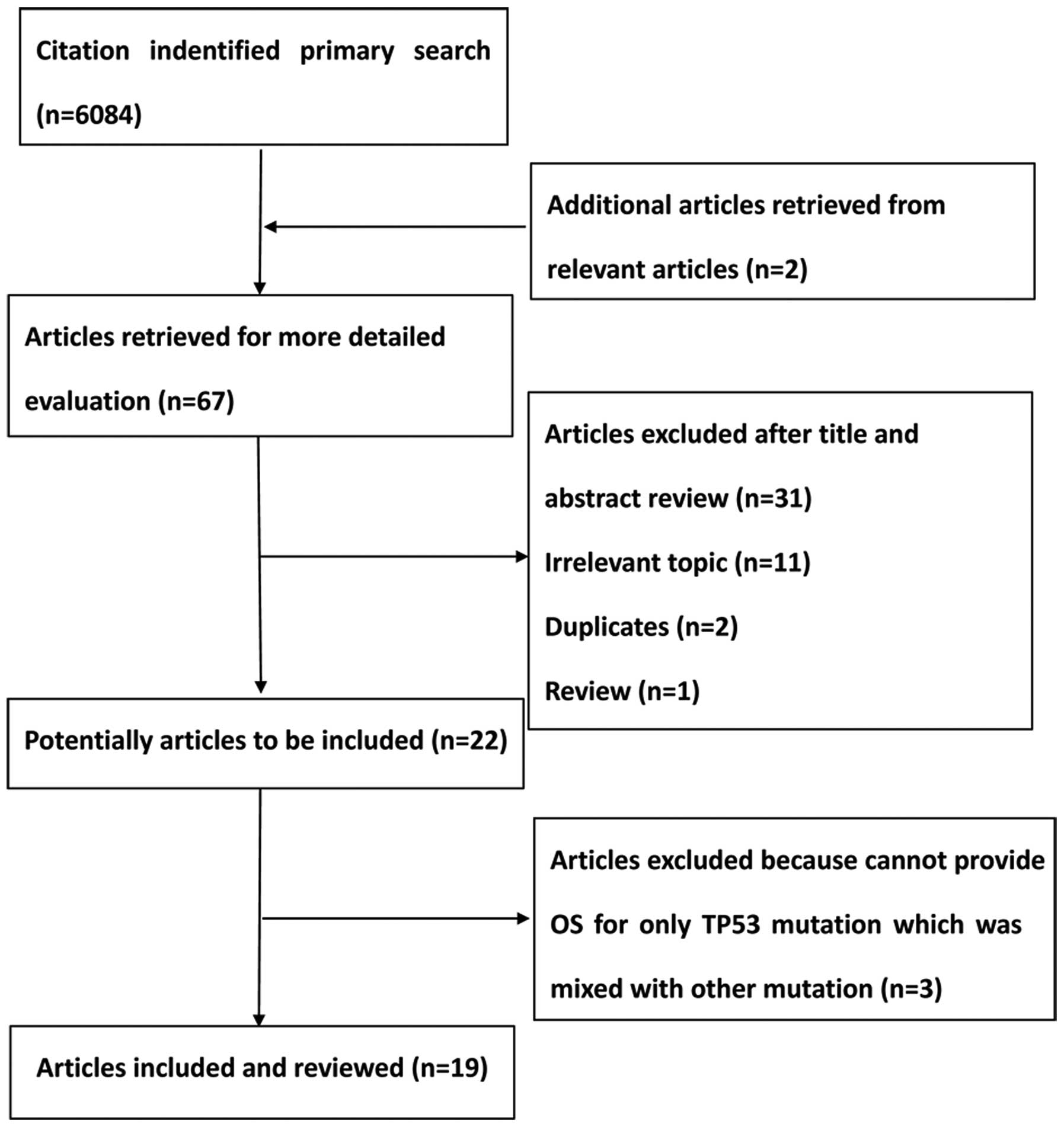
A total of 6,084 citations were identified from the
PubMed, Embase and the Central Registry of Controlled Trials of the
Cochrane Library databases. Following a review by all the authors,
19 studies (9,15–22,28–39) were
identified that fulfilled the inclusion criteria and were eligible
with complete and validated data for meta-analysis. Fig. 1 shows a summary of the various stages
of the performed literature searches in a flow chart.
Characteristics of the studies and
quality assessment
The main characteristics of the 19 studies between
1994 and 2015 that were eligible for the meta-analysis are shown in
Table I. Among these studies, 3,371
patients with NSCLC without therapy prior to surgery or biopsy were
involved, and these were stratified according to TP53 mutation
status. Patients possessing TP53 mutations were categorized as a
TP53 mutation cohort (n=1406), whereas the remaining patients had
the wild-type TP53 gene (n=1965). The Newcastle-Ottawa scale scores
of the included studies were >5, and the methodological quality
of the 19 eligible studies is shown in Table II.
 | Table I.Characteristics of the included
studies for the meta-analyses. |
Table I.
Characteristics of the included
studies for the meta-analyses.
|
| Pathological
type |
|
|---|
|
|
|
|
|---|
| First
author/year | Study type | Methods of
detection | Sequence | Patient status | Gender ratio
(M/F) | Clinical stage | AC | SCC | Others | TP53 mutation
status (sample size) | HR estimation | HR for OS (95%
CI)a | Refs. |
|---|
| Lee et al,
2015 | Pro | PCR+direct
sequencing | Exons | Surgery | 1:1 | I: 67; II/IIIA:
40 | 85 | 22 | 0 | Wild-type
(n=107) | Surv.
curvesb | 1.38
(0.73–2.61) |
(19) |
|
|
|
| 2–11 |
| 6:3 | I: 40; II/IIIA:
22 | 32 | 34 | 0 | TP53 mutation
(n=66) |
|
|
|
| Molina-Vila et
al, 2014 | Retro | PCR+Sanger
sequencing | NA | Che | NA | NA | NA | NA | NA | Wild-type
(n=225) | HR | 1.45
(0.95–2.22) |
(32) |
|
|
|
|
|
| NA | NA |
|
|
| TP53 mutation
(n=93) |
|
|
|
| Ma et al,
2013 | Pro | PCR+direct
sequencing | Exons | Surgery or
surgery-Che | 3:7 | I: 115; II: 79;
III: 109 | 117 | 156 | 30 | Wild-type
(n=303) | HR | 1.08
(0.86–1.37) |
(21) |
|
|
|
| 4–8 |
| 6:6 | I: 58; II: 51; III:
112 | 58 | 136 | 27 | TP53 mutation
(n=221) |
|
|
|
| Scoccianti et
al, 2012 | Pro | DHPLC+2th
PCR+bi-directional sequencing | Exons | Surgery | 4:9 | I: 90; II: 28; III:
9 | 85 | 41 | 3 | Wild-type
(n=129) | HR | 0.95
(0.64–1.40) |
(20) |
|
|
|
| 4–10 |
| 5:7 | I: 82; II: 32; III:
8 | 48 | 69 | 4 | TP53 mutation
(n=121) |
|
|
|
| Chien et al,
2010 | Retro | PCR+direct
sequencing | Exons | Surgery | NA | NA | NA | NA | NA | Wild-type
(n=216) | HR | 1.16
(0.87–1.55) |
(22) |
|
|
|
| 5–8 |
|
|
|
|
|
| TP53 mutation
(n=90) |
|
|
|
| Regina et
al, 2009 | Pro | PCR+direct
sequencing | Exons | Surgery | 5:6 | I/II: 18; III: 11;
IV: 4 | 22 | 7 | 4 | Wild-type
(n=33) | HR | 0.67
(0.44–1.00) |
(30) |
|
|
|
| 5–8 |
| 9 | I/II: 10; III: 5;
IV: 5 | 10 | 6 | 4 | TP53 mutation
(n=20) |
|
|
|
| Kosaka et
al, 2009 | Pro | PCR+direct
sequencing | Exons | Surgery | 1:4 | I: 158; II–V:
76 | 234 | 0 | 0 | Wild-type
(n=234) | Surv. curves | 1.50
(1.02–2.50) |
(15) |
|
|
|
| 4–10 |
| 2:3 | I: 77; II–V:
65 | 142 | 0 | 0 | TP53 mutation
(n=142) |
|
|
|
| Ludovini et
al, 2008 | Pro | PCR | Exons 5–8 | Surgery | 0:6 | I/II: 31; III:
4 | 18 | 12 | 5 | Wild-type
(n=76) | HR | 2.3
(0.80–6.60) |
(37) |
|
|
|
|
|
| 7:2 | I/II: 29; III:
12 | 10 | 25 | 6 | TP53 mutation
(n=41) |
|
|
|
| Tsao et al,
2007 | Pro | PCR+direct
sequencing | Exons |
Surgery-Observation | NA | NA | NA | NA | NA | Wild-type
(n=40) | HR | 1.15
(0.75–1.77) |
(9) |
|
|
|
| 5–9 |
|
|
|
|
|
| TP53 mutation
(n=200) |
|
|
|
| Ahrendt et
al, 2003 | Pro | PCR+direct
sequencing | Exons 5–9 | Surgery | 1 | I: 48; II: 19; III:
17 | 34 | 25 | 25 | Wild-type
(n=84) | HR | 1.56 (1.0–2.4) |
(34) |
|
|
|
|
|
| 1;8 | I: 58; II: 28; III:
18 | 39 | 52 | 13 | TP53 mutation
(n=104) |
|
|
|
| Bria et al,
2015 | Retro | Multiple PCR+direct
sequencing | Exons | Gefitini
b-Surgery/surgery | NA | III/IV: 7 | NA | NA | NA | Wild-type
(n=8) | HR | 1.36
(0.24–7.26) |
(49) |
|
|
|
| 5–8 |
|
| III/IV: 11 |
|
|
| TP53 mutation
(n=11) |
|
|
|
| Tomizawa et
al, 1999 | Pro | PCR-SSCP
sequencing | Exons | Surgery | NA | I: 61 | NA | NA | NA | Wild-type
(n=61) | Surv. curves | 2.21
(0.78–6.23) |
(18) |
|
|
|
| 5–8 |
|
| I: 39 |
|
|
| TP53 mutation
(n=39) |
|
|
|
| Vega et al,
1997 | Pro | PCR-SSCP
sequencing | Exons | Surgery | NA | I: 30; II: 4; III:
21 | 40 | 17 | 7 | Wild-type
(n=64) | Surv. curves | 1.46
(0.60–3.55) |
(33) |
|
|
|
| 5–9 |
|
| I: 7; II: 1; III:
9 | 3 | 12 | 2 | TP53 mutation
(n=17) |
|
|
|
| Huang et al,
1997 | Pro | PCR-SSCP
sequencing | Exons | Surgery | 2:3 | I: 46; II: 10; III:
37 | 67 | 22 | 1 | Wild-type
(n=93) | Surv. curves | 1.34
(0.76–2.37) |
(31) |
|
|
|
| 5–8 |
| 4:1 | I: 24; II: 7; III:
20 | 21 | 27 | 6 | TP53 mutation
(n=51) |
|
|
|
| Ohno et al,
1997 | Pro | PCR-SSCP
sequencing | Exons | Surgery | 1:2 | I: 29; II: 11; III:
13 | 12 | 39 | 2 | Wild-type
(n=53) | Surv. curves | 2.02
(0.75–5.44) |
(36) |
|
|
|
| 5–9 |
| 1.6 | I: 8; II: 5; III:
8 | 8 | 11 | 2 | TP53 mutation
(n=21) |
|
|
|
| Fukuyama et
al, 1996 | NA | PCR-SSCP
sequencing | Exons | Surgery | 1.2 | I/II: 69; III/IV:
33 | 69 | 21 | 2 | Wild-type
(n=102) | HR | NA |
(16) |
|
|
|
| 5–8 |
| 5.3 | I/II: 38; III/IV:
19 | 25 | 26 | 6 | TP53 mutation
(n=57) |
|
|
|
| Top et al,
1995 | Pro | PCR-SSCP
sequencing | Exons | NA | 1.8 | I: 10; II: 6; III:
1 | 14 | 2 | 1 | Wild-type
(n=17) | Surv. curves | 2.35
(0.65–8.51) |
(28) |
|
|
|
| 5–8 |
| 3.6 | I: 22; II: 7; III:
8 | 18 | 7 | 12 | TP53 mutation
(n=37) |
|
|
|
| Mitsudomi et
al, 1995 | Pro | PCR-SSCP
sequencing | Exons |
| NA | NA | 27 | 10 | NA | Wild-type
(n=82) | HR | 1.18
(0.60–2.30) |
(35) |
|
|
|
| 5–8 |
|
|
| 13 | 7 | NA | TP53 mutation
(n=44) |
|
|
|
| Kashii et
al, 1994 | NA | PCR-SSCP
sequencing | Exons | Surgery | 1.1 | I: 25; II: 5; III:
7; IV:1 | 27 | 4 | 7 | Wild-type
(n=38) | HR | 2.0
(0.88–4.55) |
(29) |
|
|
|
| 5–9 |
| 1.6 | I: 9; II: 4; III:
16; IV:2 | 20 | 5 | 6 | TP53 mutation
(n=31) |
|
|
|
 | Table II.Quality assessment of eligible
studies using the Newcastle-Ottawa quality assessment scale. |
Table II.
Quality assessment of eligible
studies using the Newcastle-Ottawa quality assessment scale.
| First
author/year |
Selectiona |
Comparabilityb |
Outcomec | Total (quality)
scored | Refs. |
|---|
| Lee et al,
2015 | 4 | 2 | 1 | 7 | (19) |
| Molina-Vila et
al, 2014 | 3 | 1 | 2 | 6 | (31) |
| Ma et al,
2013 | 4 | 2 | 1 | 7 | (21) |
| Scoccianti et
al, 2012 | 4 | 2 | 2 | 8 | (20) |
| Chien et al,
2010 | 3 | 1 | 2 | 6 | (22) |
| Regina et
al, 2009 | 4 | 2 | 3 | 9 | (29) |
| Kosaka et
al, 2009 | 4 | 2 | 2 | 8 | (15) |
| Ludovini et
al, 2008 | 4 | 2 | 2 | 8 | (36) |
| Tsao et al,
2007 | 3 | 1 | 3 | 7 |
(9) |
| Ahrendt et
al, 2003 | 4 | 2 | 2 | 8 | (33) |
| Bria et al,
2015 | 4 | 2 | 2 | 8 | (49) |
| Tomizawa et
al, 1999 | 4 | 2 | 3 | 9 | (18) |
| Vega et al,
1997 | 4 | 2 | 3 | 9 | (32) |
| Huang et al,
1997 | 4 | 2 | 3 | 9 | (30) |
| Ohno et al,
1997 | 4 | 2 | 3 | 9 | (35) |
| Fukuyama et
al, 1996 | 4 | 2 | 3 | 9 | (16) |
| Top et al,
1995 | 4 | 2 | 3 | 9 | (27) |
| Mitsudomi et
al, 1995 | 3 | 1 | 2 | 6 | (34) |
| Kashii et
al, 1994 | 4 | 2 | 3 | 9 | (28) |
Meta-analyses of the wild-type and
TP53 mutation groups in terms of OS
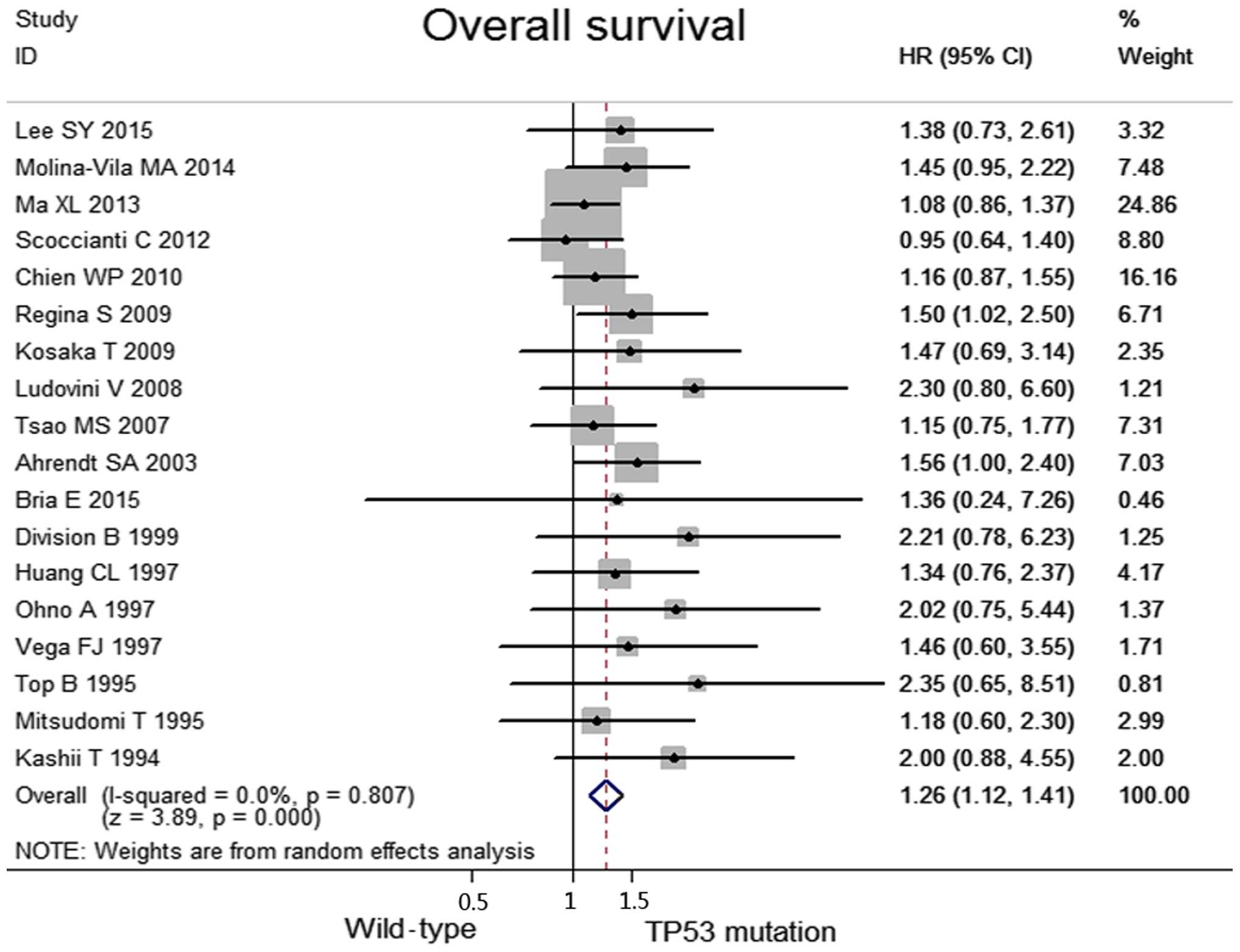
No heterogeneity was observed among the included
studies regarding the OS (I2=0.0%, P=0.81). Taken
together, when compared with the TP53 mutation group, the wild-type
group was associated with significantly higher OS values (HR, 1.26;
95% CI, 1.12–1.41, P<0.0001; Fig.
2). Data concerning the response rates were unavailable in the
majority of the studies; consequently, they were not referred to as
outcome endpoints.
Subgroup analyses and sensitivity
analyses
When stratifying patients according to clinical
stage (early stage, including the I/II stages, vs. advanced stage,
including the II–IV stages), pathological type (adenocarcinoma vs.
non-adenocarcinoma) and methods of detection (PCR-SSCP vs. others),
the observed results indicated that significant benefits of OS in
the wild-type group were identified in the subgroup involving
patients with NSCLC in the early stage, including the I/II phase
(HR, 1.93; 95% CI, 1.17–3.19; P=0.01; heterogeneity:
I2=0.0%, P=0.976) and patients with adenocarcinoma (HR,
3.06; 95% CI, 1.66–5.62, P=0.00; heterogeneity: I2=0.0%,
P=0.976). No significant differences were identified with the
methods of detection. All the results from the above subgroups are
shown in Table III.
 | Table III.Summary of the results of the
subgroup analyses results. |
Table III.
Summary of the results of the
subgroup analyses results.
|
|
|
|
| Effect size | Heterogeneity |
|---|
|
|
|
|
|
|
|
|---|
| Outcome | Subgroup | No. of studies | HR (95%
CI)a | Z | P-value | I2 | P-value |
|---|
| Overall
survival | PCR-SSCP and other
methods | 11 | 1.21
(1.07–1.38) | 3.03 | 0.002 | 0.0% | 0.702 |
|
| PCR-SSCP | 7 | 1.56
(1.15–2.12) | 2.86 | 0.004 | 0.0% | 0.880 |
|
| Adenocarcinoma | 4 | 3.06
(1.66–5.62) | 3.60 | 0.000 | 0.0% | 0.976 |
|
|
Non-adenocarcinoma | 5 | 1.25
(0.57–2.74) | 0.56 | 0.574 | 0.0% | 0.990 |
|
| Early stage
(I/II) | 4 | 1.93
(1.17–3.19) | 2.56 | 0.011 | 0.0% | 0.976 |
|
| Advanced stage
(II/III/IV) | 4 | 0.76
(0.55–1.05) | 1.09 | 0.095 | 0.0% | 0.781 |
Publication bias
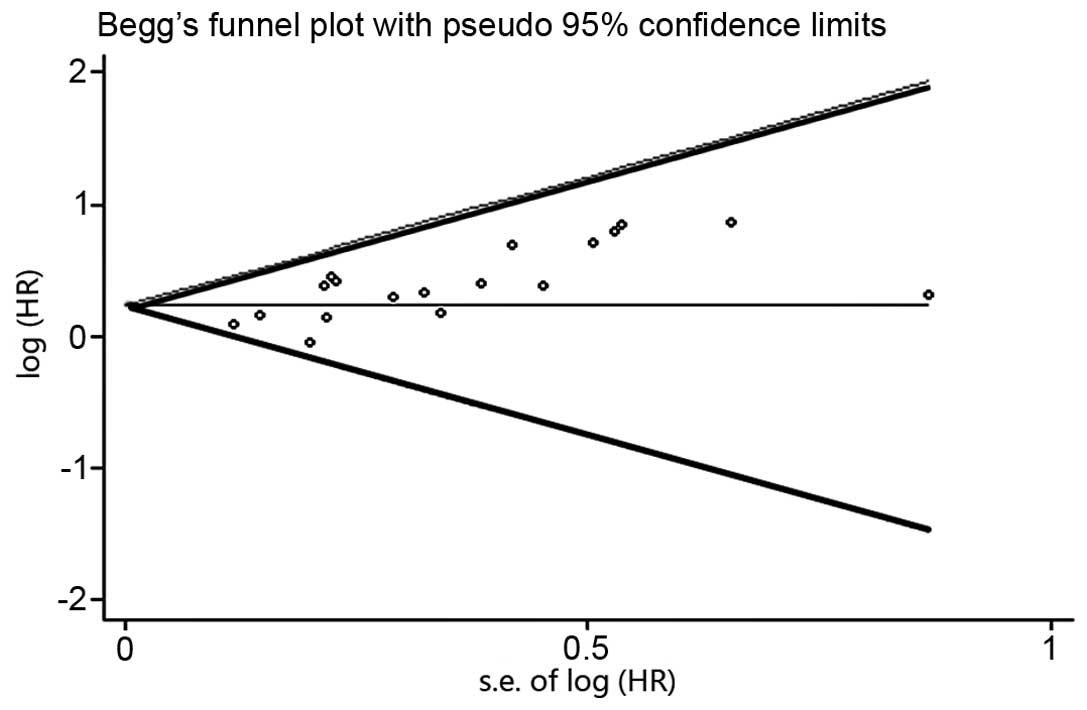
With regard to the publication bias, the funnel plot
revealed an almost symmetrical distribution, as shown in Fig. 3. This therefore suggested that no
clear publication bias was present in this meta-analysis.
Discussion
For patients with NSCLC, the association of TP53
mutations with prognostic significance has yet to be fully
elucidated. A meta-analysis incorporating all the available data
from correlative studies provides a useful method for addressing
this question. We performed the present study, and identified that
the patients with TP53 mutations indeed have markedly worse
survival rates compared with those without the mutations,
especially for patients with NSCLC in the early stages, or with
adenocarcinoma.
Theoretically, SSCP is less sensitive as a technique
compared with direct dideoxynucleotide sequencing, as it failed to
identify mutations in 14–38% of the tumors in which TP53 mutations
were detected by the latter technique (40,41).
However, when the subgroup analysis in methods of detection was
performed in the present study, the two methods revealed a very
similar, significant predictive value of TP53 mutations, which
indicated that these methods were not the key factor affecting the
association between TP53 mutations and the survival rate. Based on
data compiled in the International Agency for Research on Cancer
(IARC) TP53 database (http://p53.iarc.fr/), mutations were significantly
(P<0.01) associated with histology (21). In the study of Ludovini et al
(37), 55% of the patients with
NSCLC possessed TP53 mutations, and the incidence of the mutations
was higher in squamous-cell carcinomas and in smokers compared with
those in adenocarcinomas and non-smokers, as previously reported by
Fong et al (42). In the
studies of Fukuyama et al (16) and Kashii et al (29), it was stated that TP53 mutations were
an unfavorable prognostic factor in patients with adenocarcinoma,
although not in patients with squamous cell carcinoma (SCC), in
spite of its higher frequency (16,29), a
conclusion which has been borne out by the results in the present
study. On further analysis, the tumors with wild-type TP53 more
often had a K-ras mutation (P=0.036), which is known to constitute
an unfavorable prognostic factor in lung adenocarcinoma. Huang
et al (31) reported that it
is important to evaluate mutations of TP53 and K-ras
simultaneously, for the purpose of predicting the prognosis of
patients and determining appropriate treatments, particularly in
patients with adenocarcinoma. Furthermore, TP53 gene mutations are
considered to occur relatively early in the dysplastic epithelium
in the histogenesis of SCC, whereas they may occur relatively late
in adenocarcinoma, as suggested by the above results, hence
providing a different impact on the prognosis of patients (43). In addition, for SCC, Vega et
al (33) identified a markedly
poor clinical evolution when the TP53 mutation was located in exon
5 (an independent parameter of borderline importance), with the
group of patients with SCC having this alteration exhibiting the
worst prognosis. These facts may suggest that TP53 mutations exert
a different role in adenocarcinoma compared with SCC.
Mitsudomi et al (17) identified a much greater prognostic
effect of TP53 mutations in patients with more advanced disease
(stages IIIB and IV). There is a tendency that the prognostic value
of TP53 is more significant for patients with early-stage disease
compared with those in the advanced stage, as included in the
present study. Furthermore, Tomizawa et al (18) reported that, although TP53 expression
has no correlation with the survival rate, the presence of TP53
mutations in tumors was significantly associated with decreased
survival rates. A prospective study also suggested that TP53
mutation predicts poor survival in patients with stage I NSCLC,
although not in patients with advanced NSCLC (44). Similarly, stage I patients with
wild-type TP53 in the study by Chien et al (22) had better overall survival rates for
lung cancer compared with those who bore TP53 mutations, although
such a result was not identified in patients with advanced NSCLC
(22). The present study suggests
that TP53 mutations are associated with a higher risk of eventual
patient mortality in patients with stage I NSCLC. From a biological
viewpoint, TP53 and K-ras mutations may represent very early events
in lung carcinogenesis (20), which
consequently have an important role for prediction at early stage.
When tumors progress and become increasingly complex, it is
difficult for tumor behavior to be defined by a single genetic
abnormality. At the present time, the obtained results do not
readily provide the explanation for these discrepancies.
TP53 mutations were also classified into two groups:
Disruptive and non-disruptive (45),
on the basis of the degree of disturbance of the protein structure
predicted from the crystal structure of the TP53-DNA complex
(46). Poeta et al (45) reported that a disruptive TP53
alteration, as compared with the wild-type, had an independent,
significant association with decreased survival. In the study of
Lee et al (19), neither
disruptive nor non-disruptive mutations were significantly
associated with the survival rate of the patients. However, these
various TP53 genotypes were not mentioned in other studies that
were included in this meta-analysis. Therefore, in future studies,
it will be important to take into consideration TP53 mutations and
the TP53 genotype in assessing the prognosis and predictive
importance of the gene status of TP53 in NSCLC.
Notably, to the best of our knowledge, this is the
first study to comprehensively answer the prognostic value of TP53
mutations detected by molecular techniques in patients with NSCLC.
Nevertheless, there exist several limitations. First, data for the
objective response rate (ORR) and the disease control rate (DCR)
were not available in all the included studies, and an absence of
the short-term prognosis value does not preclude that mutations
have significance as predictors of the response to specific forms
of therapies. Secondly, after searching in the PubMed, Embase and
the Central Registry of Controlled Trials of the Cochrane Library
databases, publication bias remains, since positive results tend to
be accepted by journals, whereas negative results are often
rejected, or not even submitted. In addition, since p53 mutations
occur frequently in the so-called ‘hot-spot’ region of exons 5–8,
only the hot-spot will have been examined to evaluate the frequency
of TP53 mutations in the majority of studies, whereas meta-analyses
have determined that 13.6% of the mutations occur outside exons 5–8
(47–49). Therefore, further studies are
warranted to ensure the robustness of the conclusions of the
present study.
In conclusion, TP53 mutations may be an indicator
for poor prognosis in only a subset of patients. The present study
also suggested that the role of TP53 alterations may therefore
differ between that observed in adenocarcinomas and SCC. The
presence of these mutations may define a subset of patients with
NSCLC appropriate for investigational therapeutic strategies. In
the future, it may be possible to apply our expanding knowledge of
the molecular genetics of these lesions in order to improve the
survival rates and quality of life of patients suffering from this
disease.
Acknowledgments
The present study was supported by grants from the
National Natural Science foundation of China (no. 81570008, to
Yanbin Zhou), the Natural Science Foundation of Guangdong Province
of China (no. 2014A030313052; to Yanbin Zhou), and Science and
Technology Program of Guangzhou, China (no. 2014J4100132; to Yanbin
Zhou).
Glossary
Abbreviations
Abbreviations:
|
NSCLC
|
non-small cell lung cancer
|
|
TNM
|
tumor-node-metastasis
|
|
DNA
|
deoxyribonucleic acid
|
|
IHC
|
immunohistochemistry
|
|
SSCP
|
single-stranded conformational
polymorphism
|
|
OS
|
overall survival
|
|
HR
|
hazard ratio
|
|
CI
|
confidence interval
|
|
IARC
|
international agency for research on
cancer
|
|
SCC
|
squamous cell carcinoma
|
|
BAX
|
Bcl2-associated X protein
|
|
ORR
|
objective response rate
|
|
DCR
|
disease control rate
|
References
|
1
|
Jemal A, Siegel R, Ward E, Hao Y, Xu J,
Murray T and Thun MJ: Cancer statistics, 2008. CA Cancer J Clin.
58:71–96. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ihde DC and Minna JD: Non-small cell lung
cancer. Part II: Treatment. Curr Probl Cancer. 15:105–154. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Farjah F, Flum DR, Ramsey SD, Heagerty PJ,
Symons RG and Wood DE: Multi-modality mediastinal staging for lung
cancer among medicare beneficiaries. J Thorac Oncol. 4:355–363.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Goldstein I, Marcel V, Olivier M, Oren M,
Rotter V and Hainaut P: Understanding wild-type and mutant p53
activities in human cancer: New landmarks on the way to targeted
therapies. Cancer Gene Ther. 18:2–11. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Singhal S, Vachani A, Antin-Ozerkis D,
Kaiser LR and Albelda SM: Prognostic implications of cell cycle,
apoptosis, and angiogenesis biomarkers in non-small cell lung
cancer: A review. Clin Cancer Res. 11:3974–3986. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Toyooka S, Tsuda T and Gazdar AF: The TP53
gene, tobacco exposure, and lung cancer. Hum Mutat. 21:229–239.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Greenblatt MS, Bennett WP, Hollstein M and
Harris CC: Mutations in the p53 tumor suppressor gene: Clues to
cancer etiology and molecular pathogenesis. Cancer Res.
54:4855–4878. 1994.PubMed/NCBI
|
|
8
|
Steels E, Paesmans M, Berghmans T, Branle
F, Lemaitre F, Mascaux C, Meert AP, Vallot F, Lafitte JJ and
Sculier JP: Role of p53 as a prognostic factor for survival in lung
cancer: A systematic review of the literature with a meta-analysis.
Eur Respir J. 18:705–719. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tsao MS, Aviel-Ronen S, Ding K, Lau D, Liu
N, Sakurada A, Whitehead M, Zhu CQ, Livingston R, Johnson DH, et
al: Prognostic and predictive importance of p53 and RAS for
adjuvant chemotherapy in non small-cell lung cancer. J Clin Oncol.
25:5240–5247. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bodner SM, Minna JD, Jensen SM, D'Amico D,
Carbone D, Mitsudomi T, Fedorko J, Buchhagen DL, Nau MM, Gazdar AF,
et al: Expression of mutant p53 proteins in lung cancer correlates
with the class of p53 gene mutation. Oncogene. 7:743–749.
1992.PubMed/NCBI
|
|
11
|
Lohmann D, Ruhri C, Schmitt M, Graeff H
and Höfler H: Accumulation of p53 protein as an indicator for p53
gene mutation in breast cancer. Occurrence of false-positives and
false-negatives. Diagn Mol Pathol. 2:36–41. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Harris CC and Hollstein M: Clinical
implications of the p53 tumor-suppressor gene. N Engl J Med.
329:1318–1327. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mitsudomi T, Hamajima N, Ogawa M and
Takahashi T: Prognostic significance of p53 alterations in patients
with non-small cell lung cancer: A meta-analysis. Clin Cancer Res.
6:4055–4063. 2000.PubMed/NCBI
|
|
14
|
Huncharek M, Kupelnick B, Geschwind JF and
Caubet JF: Prognostic significance of p53 mutations in non-small
cell lung cancer: A meta-analysis of 829 cases from eight published
studies. Cancer Lett. 153:219–226. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kosaka T, Yatabe Y, Onozato R, Kuwano H
and Mitsudomi T: Prognostic implication of EGFR, KRAS, and TP53
gene mutations in a large cohort of Japanese patients with
surgically treated lung adenocarcinoma. J Thorac Oncol. 4:22–29.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fukuyama Y, Mitsudomi T, Sugio K, Ishida
T, Akazawa K and Sugimachi K: K-ras and p53 mutations are an
independent unfavourable prognostic indicator in patients with
non-small-cell lung cancer. Br J Cancer. 75:1125–1130. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mitsudomi T, Oyama T, Kusano T, Osaki T,
Nakanishi R and Shirakusa T: Mutations of the p53 gene as a
predictor of poor prognosis in patients with non-small-cell lung
cancer. J Natl Cancer Inst. 85:2018–2023. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tomizawa Y, Kohno T, Fujita T, Kiyama M,
Saito R, Noguchi M, Matsuno Y, Hirohashi S, Yamaguchi N, Nakajima T
and Yokota J: Correlation between the status of the p53 gene and
survival in patients with stage I non-small cell lung carcinoma.
Oncogene. 18:1007–1014. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lee SY, Jeon HS, Hwangbo Y, Jeong JY, Park
JY, Lee EJ, Jin G, Shin KM, Yoo SS, Lee J, et al: The influence of
TP53 mutations on the prognosis of patients with early stage
non-small cell lung cancer may depend on the intratumor
heterogeneity of the mutations. Mol Carcinog. 54:93–101. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Scoccianti C, Vesin A, Martel G, Olivier
M, Brambilla E, Timsit JF, Tavecchio L, Brambilla C, Field JK and
Hainaut P: European Early Lung Cancer Consortium: Prognostic value
of TP53, KRAS and EGFR mutations in nonsmall cell lung cancer: The
EUELC cohort. Eur Respir J. 40:177–184. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ma X, Rousseau V, Sun H, Lantuejoul S,
Filipits M, Pirker R, Popper H, Mendiboure J, Vataire AL, Le
Chevalier T, et al: Significance of TP53 mutations as predictive
markers of adjuvant cisplatin-based chemotherapy in completely
resected non-small-cell lung cancer. Mol Oncol. 8:555–564. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chien WP, Wong RH, Cheng YW, Chen CY and
Lee H: Associations of MDM2 SNP309, transcriptional activity, mRNA
expression, and survival in stage I non-small-cell lung cancer
patients with wild-type p53 tumors. Ann Surg Oncol. 17:1194–1202.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Parmar MK, Torri V and Stewart L:
Extracting summary statistics to perform meta-analyses of the
published literature for survival endpoints. Stat Med.
17:2815–2834. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Williamson PR, Smith CT, Hutton JL and
Marson AG: Aggregate data meta-analysis with time-to-event
outcomes. Stat Med. 21:3337–3351. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Stang A: Critical evaluation of the
Newcastle-Ottawa scale for the assessment of the quality of
nonrandomized studies in meta-analyses. Eur J Epidemiol.
25:603–605. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Begg CB and Mazumdar M: Operating
characteristics of a rank correlation test for publication bias.
Biometrics. 50:1088–1101. 1994. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Egger M, Smith G Davey, Schneider M and
Minder C: Bias in meta-analysis detected by a simple, graphical
test. BMJ. 315:629–634. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Top B, Mooi WJ, Klaver SG, Boerrigter L,
Wisman P, Elbers HR, Visser S and Rodenhuis S: Comparative analysis
of p53 gene mutations and protein accumulation in human
non-small-cell lung cancer. Int J Cancer. 64:83–91. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kashii T, Mizushima Y, Lima C, Noto H,
Sato H, Saito H, Kusajima Y, Kitagawa M, Sugiyama S and Kobayashi
M: Evaluation of prognostic-significance of p53 gene alterations in
patients with surgically resected lung-cancer. Int J Oncol.
6:123–128. 1995.PubMed/NCBI
|
|
30
|
Regina S, Valentin JB, Lachot S, Lemarié
E, Rollin J and Gruel Y: Increased tissue factor expression is
associated with reduced survival in non-small cell lung cancer and
with mutations of TP53 and PTEN. Clin Chem. 55:1834–1842. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Huang CL, Taki T, Adachi M, Konishi T,
Higashiyama M, Kinoshita M, Hadama T and Miyake M: Mutations of p53
and K-ras genes as prognostic factors for non-small cell lung
cancer. Int J Oncol. 12:553–563. 1998.PubMed/NCBI
|
|
32
|
Molina-Vila MA, Bertran-Alamillo J, Gascó
A, Mayo-de-las-Casas C, Sánchez-Ronco M, Pujantell-Pastor L,
Bonanno L, Favaretto AG, Cardona AF, Vergnenègre A, et al:
Nondisruptive p53 mutations are associated with shorter survival in
patients with advanced non-small cell lung cancer. Clin Cancer Res.
20:4647–4659. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Vega FJ, Iniesta P, Caldés T, Sanchez A,
López JA, de Juan C, Diaz-Rubio E, Torres A, Balibrea JL and Benito
M: p53 Exon 5 mutations as a prognostic indicator of shortened
survival in non-small-cell lung cancer. Br J Cancer. 76:44–51.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ahrendt SA, Hu Y, Buta M, McDermott MP,
Benoit N, Yang SC, Wu L and Sidransky D: p53 mutations and survival
in stage I non-small-cell lung cancer: Results of a prospective
study. J Natl Cancer Inst. 95:961–970. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mitsudomi T, Oyama T, Nishida K, Ogami A,
Osaki T, Nakanishi R, Sugio K, Yasumoto K and Sugimachi K: p53
nuclear immunostaining and gene mutations in non-small-cell lung
cancer and their effects on patient survival. Ann Oncol. 6:(Suppl
3). S9–S13. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ohno A, Hirashima T, Kubo A, Masuda N,
Takada M, Fujiwara H, Yasumitsu T, Kikui M, Fukuoka M and Nakagawa
K: p53 status and prognosis in stage I–IIIa non-small cell lung
cancer. Int J Oncol. 10:521–528. 1997.PubMed/NCBI
|
|
37
|
Ludovini V, Pistola L, Gregorc V, Floriani
I, Rulli E, Piattoni S, Di Carlo L, Semeraro A, Darwish S,
Tofanetti FR, et al: Plasma DNA, microsatellite alterations, and
p53 tumor mutations are associated with disease-free survival in
radically resected non-small cell lung cancer patients: A study of
the perugia multidisciplinary team for thoracic oncology. J Thorac
Oncol. 3:365–373. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Horio Y, Takahashi T, Kuroishi T, Hibi K,
Suyama M, Niimi T, Shimokata K, Yamakawa K, Nakamura Y, Ueda R, et
al: Prognostic significance of p53 mutations and 3p deletions in
primary resected non-small cell lung cancer. Cancer Res. 53:1–4.
1993.PubMed/NCBI
|
|
39
|
Schiller JH, Adak S, Feins RH, Keller SM,
Fry WA, Livingston RB, Hammond ME, Wolf B, Sabatini L, Jett J, et
al: Lack of prognostic significance of p53 and K-ras mutations in
primary resected non-small-cell lung cancer on E4592: A laboratory
ancillary study on an eastern cooperative oncology group
prospective randomized trial of postoperative adjuvant therapy. J
Clin Oncol. 19:448–457. 2001.PubMed/NCBI
|
|
40
|
Tolbert DM, Noffsinger AE, Miller MA,
DeVoe GW, Stemmermann GN, Macdonald JS and Fenoglio-Preiser CM: p53
immunoreactivity and single-strand conformational polymorphism
analysis often fail to predict p53 mutational status. Mod Pathol.
12:54–60. 1999.PubMed/NCBI
|
|
41
|
Meinhold-Heerlein I, Ninci E, Ikenberg H,
Brandstetter T, Ihling C, Schwenk I, Straub A, Schmitt B,
Bettendorf H, Iggo R and Bauknecht T: Evaluation of methods to
detect p53 mutations in ovarian cancer. Oncology. 60:176–188. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Fong KM, Kida Y, Zimmerman PV, Ikenaga M
and Smith PJ: Loss of heterozygosity frequently affects chromosome
17q in non-small cell lung cancer. Cancer Res. 55:4268–4272.
1995.PubMed/NCBI
|
|
43
|
Sozzi G, Miozzo M, Donghi R, Pilotti S,
Cariani CT, Pastorino U, Porta G Della and Pierotti MA: Deletions
of 17p and p53 mutations in preneoplastic lesions of the lung.
Cancer Res. 52:6079–6082. 1992.PubMed/NCBI
|
|
44
|
Ahrendt SA, Hu Y, Buta M, McDermott MP,
Benoit N, Yang SC, Wu L and Sidransky D: p53 mutations and survival
in stage I non-small-cell lung cancer: Results of a prospective
study. J Natl Cancer Inst. 95:961–970. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Poeta ML, Manola J, Goldwasser MA,
Forastiere A, Benoit N, Califano JA, Ridge JA, Goodwin J, Kenady D,
Saunders J, et al: TP53 mutations and survival in squamous-cell
carcinoma of the head and neck. N Engl J Med. 357:2552–2561. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Cho Y, Gorina S, Jeffrey PD and Pavletich
NP: Crystal structure of a p53 tumor suppressor-DNA complex:
Understanding tumorigenic mutations. Science. 265:346–355. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Chiba I, Takahashi T, Nau MM, D'Amico D,
Curiel DT, Mitsudomi T, Buchhagen DL, Carbone D, Piantadosi S, Koga
H, et al: Mutations in the p53 gene are frequent in primary,
resected non-small cell lung cancer. Lung Cancer Study Group.
Oncogene. 5:1603–1610. 1990.PubMed/NCBI
|
|
48
|
Soussi T and Béroud C: Assessing TP53
status in human tumours to evaluate clinical outcome. Nat Rev
Cancer. 1:233–240. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Bria E, Pilotto S, Amato E, Fassan M,
Novello S, Peretti U, Vavalà T, Kinspergher S, Righi L, Santo A, et
al: Molecular heterogeneity assessment by next-generation
sequencing and response to gefitinib of EGFR mutant advanced lung
adenocarcinoma. Oncotarget. 6:12783–12795. 2015. View Article : Google Scholar : PubMed/NCBI
|