Introduction
Heart diseases, including coronary heart disease,
heart failure and cardiomyopathy, have become leading causes of
death in the developed world (1–3).
Important damage of the myocardium is considered irreversible,
since mature cardiomyocytes are highly differentiated and have
limited regenerative capacity (4).
In order to address this issue, cell regeneration therapy has
emerged as a promising new approach for myocardial repair (5,6). In
this direction, a large body of studies have focused on
understanding the mechanisms of cardiomyocyte differentiation using
somatic and embryonic stem (ES) cells, as well as embryonic
carcinoma cells (7–9). The P19 embryonal carcinoma cell line
was derived from a teratocarcinoma induced in C3H/HC mice, and has
been widely used as a model system to study the molecular
mechanisms underlying cellular differentiation (8,10).
Although the P19 cell line has been extensively used to elucidate
the mechanisms governing differentiation of stem cells into
cardiomyocytes (7,8,11),
the role of mitochondria during this process has not been
studied.
Mitochondria are membrane-bound organelles that
possess their own genome and provide the majority of adenosine
triphosphate (ATP) within eukaryotic cells (12,13).
The maternally-inherited, 16.6-kb mitochondrial genome encodes 13
subunits of the electron-transfer chain, 22 transfer (t)RNAs and
two ribosomal (r)RNAs (13,14).
Mitochondria are involved in numerous processes central to cellular
functions, including calcium signaling, growth and differentiation,
cell-cycle control and cell death (15). Previous reports have suggested a
role of mitochondria in ES cell viability and differentiation, a
finding that warrants further investigation (16–18).
This study used P19 embryonal carcinoma cells based
on their ability to differentiate into cardiomyocytes in suspension
culture when exposed to 1% dimethyl sulfoxide (DMSO) (10,19).
We compared the mitochondrial morphology and localization between
undifferentiated and differentiated P19 cells, and quantified the
mitochondrial DNA (mtDNA) copy number, ATP content and reactive
oxygen species (ROS) production during cardiac differentiation. Our
data provide evidence that mitochondria play an important role in
the differentiation of cardiomyocytes.
Materials and methods
Cell cultures and differentiation
The P19 cells used in this study were obtained from
the American Type Culture Collection (ATCC; Manassas, VA, USA), and
were cultured in Gibco® α-modified Eagle’s medium
(α-MEM), supplemented with 10% Gibco® fetal bovine serum
(FBS) (both from Thermo Fisher Scientific Inc., Grand Island, NY,
USA) and 1% penicillin/streptomycin, at 37°C in a 5% CO2
atmosphere. In order to induce cardiac differentiation, P19 cells
that had reached the exponential growth phase were trypsinized and
transferred to 10-cm bacterial dishes containing α-MEM medium
supplemented with 10% FBS and 1% DMSO (Sigma-Aldrich, St. Louis,
MO, USA) at a density of 105 cells/m. The cells were
left to aggregate for 4 days. The cell aggregates were then
transferred into cell culture flasks containing complete medium
containing 90% α-MEM, 10% FBS and 1% penicillin/streptomycin
without DMSO. for an additional 7 days. The morphological changes
in the P19 cells were examined and photographed under an inverted
microscope (Nikon, Tokyo, Japan).
Western blotting
Total protein extracts were isolated from cultured
cells, separated on a 10% sodium dodecyl sulfate gel by
polyacrylamide gel electrophoresis, transferred onto a
nitrocellulose membrane, and blocked in 5% non-fat dried milk in a
phosphate-buffered saline (PBS)/Tween-20 buffer. The membrane was
then incubated with a polyclonal rabbit anti-cTnT antibody (1:500;
Abcam, Cambridge, UK), and a polyclonal rabbit anti-tubulin
antibody (1:10,000; ABR-Affinity BioReagents cat no. AF0524;
Affinity, Shanghai, China), followed by incubation with the
secondary goat anti-rabbit IgG conjugated with horseradish
peroxidase (1:10,000; ZSGB-Bio, Beijing, China).
Electron microscopy
Cultured cells were collected after trypsin
(Gibco-BRL, Carlsbad, CA, USA) digestion, washed in fresh PBS
(Wisent Inc., QC, Canada; pH 7.4), and fixed in a 2.5%
glutaraldehyde-4% paraformaldehyde solution. Cells were then washed
in 0.1 M cacodylate buffer, and post-fixed with 1% osmium tetroxide
and 1.5% potassium ferrocyanide for 1 h. Next, they were washed in
water, stained with 1% aqueous uranyl acetate for 30 min, and
dehydrated through a graded series of ethanol (5 min in 70%, 5 min
in 90%, and 5 min in 100%). Finally, the samples were infiltrated
and embedded in TAAB epon (Marivac Canada Inc., St. Laurent,
Quebec, Canada). Ultrathin sections (60 nm) were obtained on a
Reichert Ultracut S microtome (Leica, Wetzlar, Germany), placed
onto copper grids, stained with uranyl acetate and lead citrate,
and examined under a transmission electron microscope (JEM-1010;
JEOL, Tokyo, Japan), at an accelerating voltage of 80 kV.
Measurement of mtDNA concentration by
quantitative (q)PCR
Relative levels of mtDNA were determined using qPCR,
performed on an Applied Biosystems 7500 sequence detection system,
and were quantified using the SDS software (both from Applied
Biosystems, Foster City, CA, USA) in accordance with the
manufacturer’s protocol (20).
Briefly, DNA was isolated from cells using a DNA extraction kit
(cat no. A1120, Promega Corp., Madison, WI, USA) and quantified on
a NanoDrop 2.0 spectrophotometer (Thermo Fisher Scientific Inc.). A
110-nucleotide mtDNA fragment within the cytochrome B gene
(CYTB) was amplified with the following forward primer,
reverse primer, and probe, respectively: 5′-TTT TAT CTG CAT CTG AGT
TTA ATC CTG T-3′, 5′-CCA CTT CAT CTT ACC ATT TAT TAT CGC-3′, and
AGC AAT CGT TCA CCT CCT CTT CCT CCA C. The sequences of the
forward, reverse primer and probe for the amplification of a 291-bp
region of the nuclear gene encoding the 28S rRNA, used for
normalization, were respectively: 5′-GGC GGC CAA GCG TTC ATA G-3′,
5′-AGG CGT TCA GTC ATA ATC CCA CAG-3′, and TGG TAG CTT CGC CCC ATT
GGC TCC T. The PCR product of CYTB had been previously
cloned into the plasmid pMD 18-T (Takara Bio, Inc., Dalian, China)
by qPCR and verified by DNA sequencing (Invitrogen, Shanghai,
China). Plasmid standards (ND1-plasmid pMD 18-T; Takara Bio, Inc.)
of known copy number for ND1 were used to generate a log-linear
standard curve, from which the CYTB copy number was
extrapolated. A standard curve from qPCR results on the plasmids
that contained the 28S fragment (28S-pMD18®-T; Takara
Bio, Inc.) was used to determine the copy number of the studied
28S rRNA fragment. The ratio of mtDNA to nuclear DNA copies
was used to define the concentration of mitochondria per cell.
Assessment of cellular ATP
production
ATP was measured by a luciferase-based luminescence
assay kit (Beyotime Institute of Biotechnology, Nantong, China).
Briefly, cultured cells were homogenized in an ice-cold
ATP-releasing buffer (Beyotime Institute of Biotechnology, cat no.
S0026-4) and centrifuged at 12,000 × g for 10 min. Following
centrifugation, the supernatant was transferred to a fresh tube in
order to measure the ATP concentration. A 20-μl sample along with
100 μl of ATP detection buffer (Beyotime Institute of
Biotechnology, cat no. S0026-1) were assayed in a single-tube
luminometer (Turner BioSystems, Sunnyvale, CA, USA). The standard
curve used to determine the ATP concentration (10−3–1
μM) was prepared from a known amount of ATP, purchased from
Beyotime Institute of Biotechnology. The relative ATP level was
then normalized to the protein concentration of the respective
sample, measured by the bicinchoninic acid assay method.
ROS assay
Intracellular ROS generation was measured using a
2′,7′-dichlorodihydrofluorescein diacetate acetyl ester
(H2-DCFDA) probe (Sigma-Aldrich), as previously
described (21). Briefly, cells
were washed twice with PBS buffer and incubated at 37°C in
prewarmed α-MEM medium containing 5 μM H2-DCFDA. After
30 min, the cells were washed three times with PBS. For flow
cytometry, the cells were trypsinized and centrifuged at 500 × g at
4°C for 5 min, and resuspended in 300 μl PBS. The cells were then
analyzed by fluorescence-activated cell sorting (FACS), using a
FACScan flow cytometer and the CellQuest software (both from BD
Biosciences, San Jose, CA, USA).
Statistical analysis
Statistical significance of differences among
experimental samples was assessed by a one-way analysis of variance
(ANOVA), using the SPSS 16.0 statistical software (IBM, Armonk, NY,
USA). All data were expressed as means ± SD from three independent
experiments. P<0.05 was considered to indicate statistically
significant differences.
Results
Cardiac differentiation of P19 cells
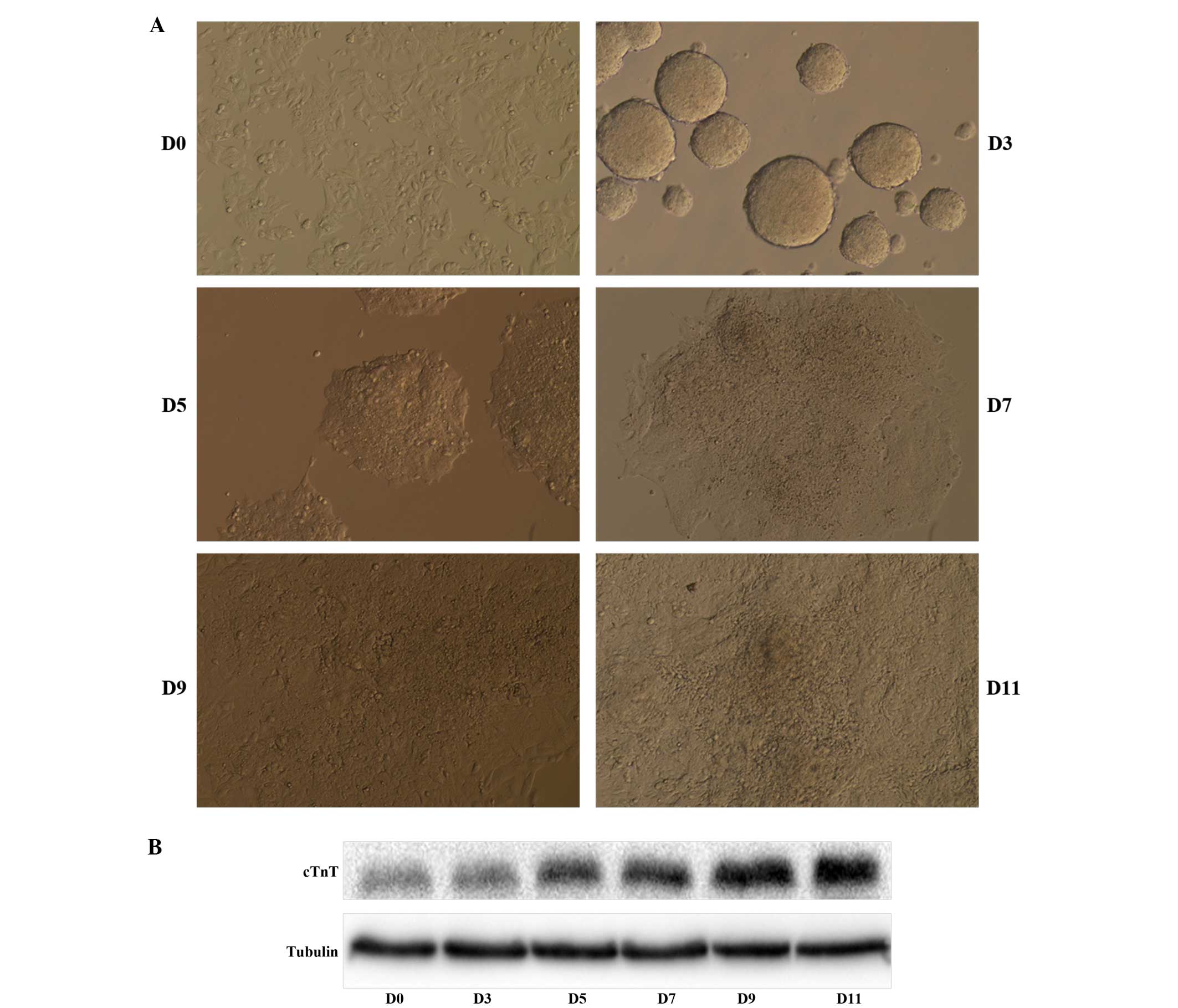
Upon induction with 1% DMSO, cell aggregation, and a
total of 11 days in culture, P19 cells differentiated into beating
cardiomyocytes (Fig. 1A). When the
cells were observed under a phase contrast microscope,
differentiated cells displayed a denser cytoplasm, while the
undifferentiated cells were relatively small. The first beating
cardiomyocytes were observed on day 9 and were abundant by day 11.
In order to confirm the differentiation of P19 cells into mature
cardiomyocytes, we analyzed the expression of the cardiomyocyte
protein marker cTnT, encoding the cardiac-specific isoform of
troponin T. Western blot analysis showed that the protein level of
cTnT increased throughout cell differentiation (Fig. 1B).
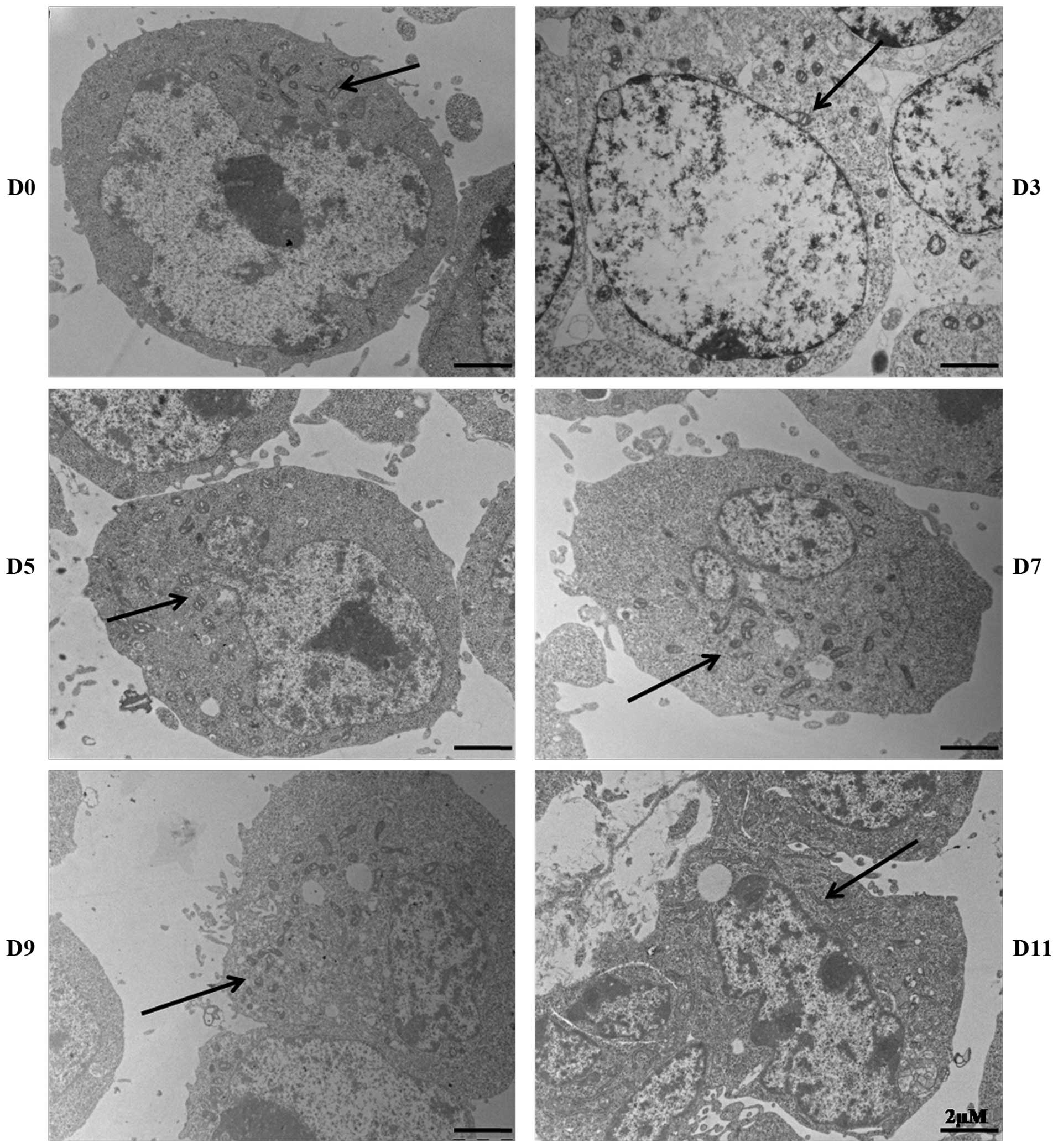
Mitochondrial morphology and
localization
The morphology and cellular localization of
mitochondria during P19 cell differentiation were investigated by
electron microscopy (Fig. 2).
Clustering of mitochondria was observed in undifferentiated P19
cells, with individual mitochondria displaying granular and
thread-like patterns. During the aggregation stage of
differentiation, we observed that round mitochondria dispersed
around the nucleus; these mitochondria appeared smaller compared to
those of undifferentiated cells. Following this stage, the number
of mitochondria increased, and they were found dispersed throughout
the cytoplasm, with the majority appearing rod-shaped, with
extended cristae.
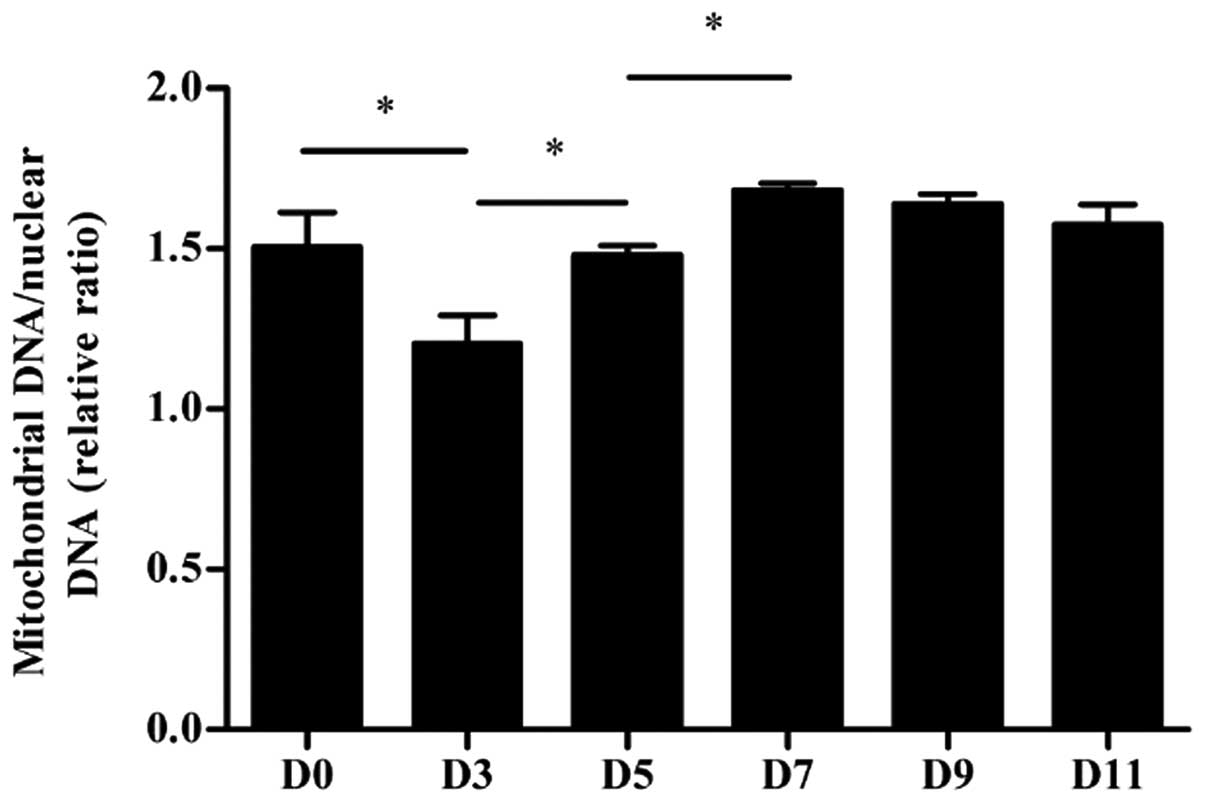
Mitochondrial DNA copy number
The mtDNA copy number per mitochondrion is
considered to be constant in all mammalian cell types (22). We examined the mtDNA copy number of
the undifferentiated and differentiated P19 cells harvested at
different time points using qPCR (Fig.
3). Our results show that the mtDNA copy number was
significantly higher on days 5 and 7 (both P<0.05) compared to
earlier stages of differentiation, i.e. days 3 and 5, respectively,
and there was no significant difference in mtDNA copy number during
the remaining stages of cardiomyocyte differentiation
(P>0.05).
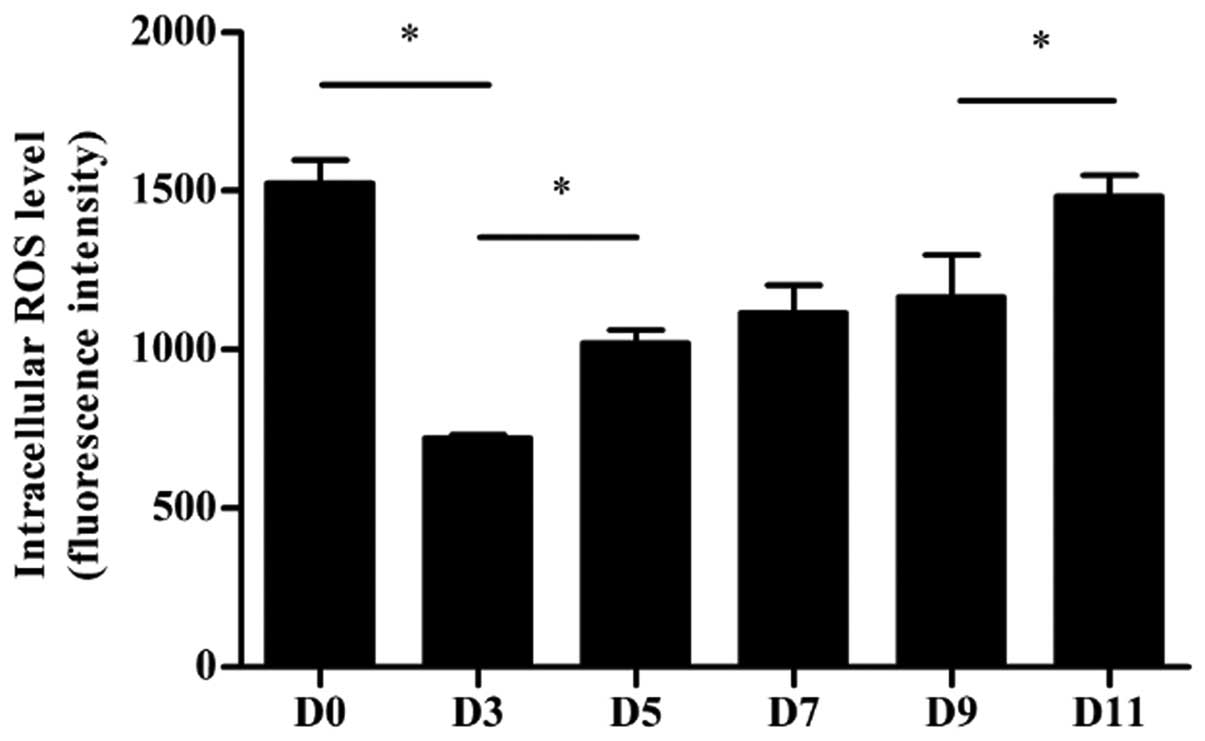
Intracellular ROS levels in
differentiated P19 cells
The intracellular levels of ROS were measured in
differentiating P19 cells, using the fluorescent probe DCFDA
(Fig. 4). Undifferentiated P19
cells had a relatively high intracellular ROS level, which was
significantly reduced in aggregating cells on day 3 (P<0.05).
From that day onwards, the ROS level gradually increased, and
returned to baseline levels on day 11.
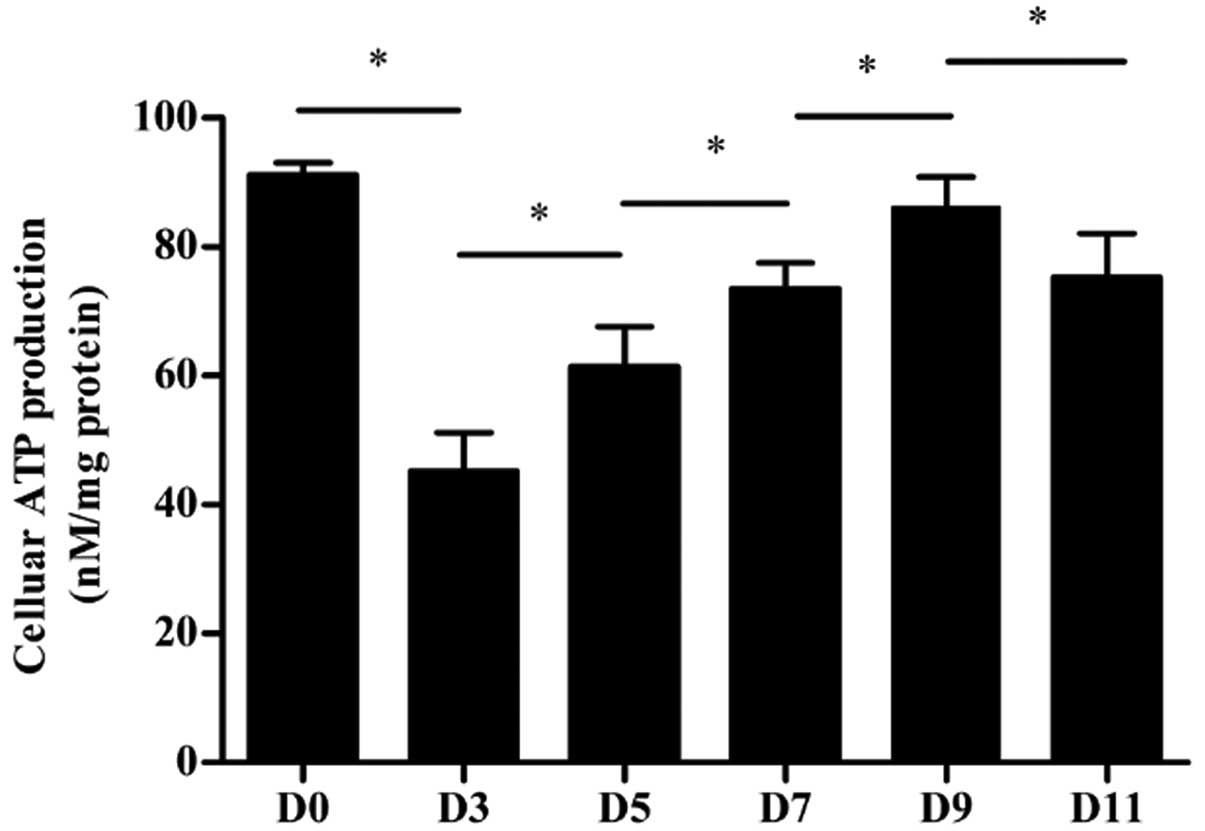
Cellular ATP production
The concentration of cellular ATP was measured in
differentiating P19 cells (Fig.
5). The intracellular ATP content of differentiated cells on
day 3 was decreased compared to that of undifferentiated P19 cells
on day 0 (P<0.05). On the following days, the intracellular ATP
production gradually increased, although the total cellular ATP
level was slightly decreased on day 11 compared to day 9
(P<0.05).
Discussion
Since adult human heart cells have very limited
regenerative capacity, the application of stem cell-based cardiac
regeneration to repair the failing heart has attracted a great deal
of attention (23). ES cells,
induced pluripotent stem cells, and embryonic carcinoma cells have
become major models for the elucidation of the mechanisms
underlying the process of differentiation into cardiomyocytes
(8,9). Due to the fact that the adult heart
has a very high energy demand, as it continuously contracts to
supply the body with blood (24),
the role of mitochondria during cardiac differentiation has emerged
as an important study area.
Advances in electron microscopy have allowed to
reveal the considerable structural diversity of the mitochondria,
which change shapes under different physiological conditions
(25). These different
configurations are believed to relate to mitochondrial function
(26). In our study, mitochondria
were clustered and appeared as granular and thread-like organelles
in undifferentiated P19 cells, which is different from the
mitochondrial morphology and distribution in ES cells (25). This difference in morphology may be
explained by the origin of the P19 embryonal carcinoma cells. Given
that they are derived from a teratocarcinoma induced in C3H/HC mice
(10), these cells are expected to
possess features of both tumor and stem cells. Accordingly, P19
have energy requirements consistent with the maintenance of the
pluripotent state, as well as the need for continuous growth.
Following the aggregation stage of differentiation, the dynamic
distribution and morphology of the mitochondria were similar to
that of mitochondria of human embryonic stem cells (hESCs)
(25), and consistent with the
energy demands of cells differentiating into cardiomyocytes.
Two key features of the mitochondria are the facts
that they are the only organelles in animal cells containing their
own genome, and that the multicopy mtDNA is maternally inherited
(27). In this study, we observed
an initial decline in the mtDNA copy number in aggregating cells
relative to undifferentiated P19 cells, followed by a gradual
increase back to baseline levels during differentiation. It is
possible that the low mtDNA copy number in aggregating cells may be
required to maintain their proliferative capacity in suspension
culture, while the subsequent increase between days 3 and 7 is in
accordance with the observed changes in mitochondrial distribution
and morphology. We interpret the stable mtDNA copy number from day
7 onwards as an indication that the majority of P19 embryonal
carcinoma cells had successfully committed to the cardiomyocyte
lineage.
Mitochondria are considered to be the major cellular
sources of ROS and ATP, and thus, the ROS and ATP cellular levels
were here used as markers of mitochondrial function in the P19
cells differentiating into cardiomyocytes. Furthermore, the
production of ROS by mitochondria (28) or the nonphagocytic NADPH oxidases
(29), are known to play a key
role in the differentiation of ES cells into the cardiomyocyte
lineage (30–32). Consistent with the dynamic changes
in mitochondrial number and mtDNA copy number, we observed that
intracellular ROS dropped to its lowest level on day 3 of
differentiation, and then gradually returned to baseline levels. As
a metabolic by-product and a common signal in cellular processes,
fluctuations in the intracellular ROS level may reflect changes in
ROS signaling involved in P19 cell differentiation, or may be a
consequence of the differentiation into high-energy demand
cardiomyocytes. Similarly, intracellular ATP also declined to its
lowest level on day 3, and then steadily increased as the process
of differentiation into cardiomyocytes progressed and the energy
demands became higher. Interestingly, on day 11 of differentiation,
intracellular ATP production was again slightly, but significantly,
reduced. We hypothesize that this late decrease in the ATP level
reflects higher rates of ATP consumption relative to the rates of
ATP synthesis, reflecting the increased energy demand in the
beating cardiomyocyte.
In summary, our results demonstrate that during the
differentiation of P19 embryonal carcinoma cells into
cardiomyocytes, mitochondria undergo dynamic changes in their
morphology and function. Specifically, we suggest that mitochondria
of undifferentiated cells transform from a primitive status with
immature morphology and low function into aggregating cells, and
then cells with gradually enhanced function along the
differentiation process. Our study is the first to indicate, to the
best of our knowledge, that dynamic mitochondrial changes occur
throughout the differentiation of P19 cell-derived cardiomyocytes.
This study thus offers a glimpse into the mechanisms underlying P19
embryonal carcinoma cell differentiation into cardiomyocytes, and
may be useful in the optimization of protocols for the in
vitro differentiation of cardiomyocytes.
Acknowledgements
This study was supported by grants from the National
Natural Science Foundation of China (no. 81070138), the Natural
Science Foundation of Jiangsu Province, China (no. BK2010582), and
the Talent Foundation of Jiangsu Province, China (no. WSN8020).
References
|
1
|
Gavin JB, Maxwell L and Edgar SG:
Microvascular involvement in cardiac pathology. J Mol Cell Cardiol.
30:2531–2540. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Klingel K, Sauter M, Bock CT, Szalay G,
Schnorr JJ and Kandolf R: Molecular pathology of inflammatory
cardiomyopathy. Med Microbiol Immunol. 193:101–107. 2004.
View Article : Google Scholar
|
|
3
|
Sabharwal NK and Lahiri A: Role of
myocardial perfusion imaging for risk stratification in suspected
or known coronary artery disease. Heart. 89:1291–1297. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Koudstaal S, Jansen Of Lorkeers SJ,
Gaetani R, et al: Concise review: heart regeneration and the role
of cardiac stem cells. Stem Cells Transl Med. 2:434–443. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Itescu S, Schuster MD and Kocher AA: New
directions in strategies using cell therapy for heart disease. J
Mol Med (Berl). 81:288–296. 2003.PubMed/NCBI
|
|
6
|
Kehat I and Gepstein L: Human embryonic
stem cells for myocardial regeneration. Heart Fail Rev. 8:229–236.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lev S, Kehat I and Gepstein L:
Differentiation pathways in human embryonic stem cell-derived
cardiomyocytes. Ann NY Acad Sci. 1047:50–65. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
van der Heyden MA and Defize LH: Twenty
one years of P19 cells: what an embryonal carcinoma cell line
taught us about cardiomyocyte differentiation. Cardiovasc Res.
58:292–302. 2003.PubMed/NCBI
|
|
9
|
Wei H, Juhasz O, Li J, Tarasova YS and
Boheler KR: Embryonic stem cells and cardiomyocyte differentiation:
phenotypic and molecular analyses. J Cell Mol Med. 9:804–817. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
McBurney MW, Jones-Villeneuve EM, Edwards
MK and Anderson PJ: Control of muscle and neuronal differentiation
in a cultured embryonal carcinoma cell line. Nature. 299:165–167.
1982. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Skerjanc IS: Cardiac and skeletal muscle
development in P19 embryonal carcinoma cells. Trends Cardiovasc
Med. 9:139–143. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Näveri L, Näveri H and Härkönen M:
Myocardial energy metabolism. Ann Chir Gynaecol. 76:3–11. 1987.
|
|
13
|
Birky CW Jr: Uniparental inheritance of
mitochondrial and chloroplast genes: mechanisms and evolution. Proc
Natl Acad Sci USA. 92:11331–11338. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Anderson S, Bankier AT, Barrell BG, et al:
Sequence and organization of the human mitochondrial genome.
Nature. 290:457–465. 1981. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Osellame LD, Blacker TS and Duchen MR:
Cellular and molecular mechanisms of mitochondrial function. Best
Pract Res Clin Endocrinol Metab. 26:711–723. 2012. View Article : Google Scholar
|
|
16
|
Bavister BD: The mitochondrial
contribution to stem cell biology. Reprod Fertil Dev. 18:829–838.
2006. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Oh SK, Kim HS, Ahn HJ, et al: Derivation
and characterization of new human embryonic stem cell lines:
SNUhES1, SNUhES2, and SNUhES3. Stem Cells. 23:211–219. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
St John JC, Ramalho-Santos J, Gray HL, et
al: The expression of mitochondrial DNA transcription factors
during early cardiomyocyte in vitro differentiation from human
embryonic stem cells. Cloning Stem Cells. 7:141–153.
2005.PubMed/NCBI
|
|
19
|
Edwards MK, Harris JF and McBurney MW:
Induced muscle differentiation in an embryonal carcinoma cell line.
Mol Cell Biol. 3:2280–2286. 1983.PubMed/NCBI
|
|
20
|
Kaaman M, Sparks LM, van Harmelen V, Smith
SR, Sjolin E, Dahlman I and Arner P: Strong association between
mitochondrial DNA copy number and lipogenesis in human white
adipose tissue. Diabetologia. 50:2526–2533. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Maxwell DP, Wang Y and McIntosh L: The
alternative oxidase lowers mitochondrial reactive oxygen production
in plant cells. Proc Natl Acad Sci USA. 96:8271–8276. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Robin ED and Wong R: Mitochondrial DNA
molecules and virtual number of mitochondria per cell in mammalian
cells. J Cell Physiol. 136:507–513. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Christoffels V: Regenerative medicine:
muscle for a damaged heart. Nature. 474:585–586. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Poirier VL: Can we develop a permanent
pulsatile rotary blood pump? Yes, we can. Artif Organs. 20:475–480.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cho YM, Kwon S, Pak YK, et al: Dynamic
changes in mitochondrial biogenesis and antioxidant enzymes during
the spontaneous differentiation of human embryonic stem cells.
Biochem Biophys Res Commun. 348:1472–1478. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mannella CA: Structural diversity of
mitochondria: functional implications. Ann NY Acad Sci.
1147:171–179. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Giles RE, Blanc H, Cann HM and Wallace DC:
Maternal inheritance of human mitochondrial DNA. Proc Natl Acad Sci
USA. 77:6715–6719. 1980. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Crespo FL, Sobrado VR, Gomez L, Cervera AM
and McCreath KJ: Mitochondrial reactive oxygen species mediate
cardiomyocyte formation from embryonic stem cells in high glucose.
Stem Cells. 28:1132–1142. 2010.PubMed/NCBI
|
|
29
|
Buggisch M, Ateghang B, Ruhe C, Strobel C,
Lange S, Wartenberg M and Sauer H: Stimulation of ES-cell-derived
cardiomyogenesis and neonatal cardiac cell proliferation by
reactive oxygen species and NADPH oxidase. J Cell Sci. 120:885–894.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sauer H, Rahimi G, Hescheler J and
Wartenberg M: Role of reactive oxygen species and
phosphatidylinositol 3-kinase in cardiomyocyte differentiation of
embryonic stem cells. FEBS Lett. 476:218–223. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Schmelter M, Ateghang B, Helmig S,
Wartenberg M and Sauer H: Embryonic stem cells utilize reactive
oxygen species as transducers of mechanical strain-induced
cardiovascular differentiation. FASEB J. 20:1182–1184. 2006.
View Article : Google Scholar
|
|
32
|
Sharifpanah F, Wartenberg M, Hannig M,
Piper HM and Sauer H: Peroxisome proliferator-activated receptor
alpha agonists enhance cardiomyogenesis of mouse ES cells by
utilization of a reactive oxygen species-dependent mechanism. Stem
Cells. 26:64–71. 2008. View Article : Google Scholar
|