Introduction
microRNAs (miRNAs) are a class of small non-coding
RNAs that are 19–24 nucleotides in length, and crucial in various
biological and pathological processes, including cell
proliferation, differentiation, apoptosis, metabolism, organ
morphology and angiogenesis (1,2).
miRNAs primarily regulate gene expression through the inhibition of
RNA translation by base pairing of their seed region (nucleotides
2–8) to the 30 untranslated regions of the target RNA (3). miRNA can also facilitate the
targeting of specific mRNAs for cleavage, resulting in the
downregulation of target mRNAs (4).
Intratumoral hypoxia is a hallmark of most solid
tumors and results from increased oxygen consumption and/or
insufficient blood supply. A number of the hypoxia-induced cellular
responses are mediated through hypoxia-inducible factors (HIFs),
which regulate genes involved in numerous functions, including
angiogenesis, survival, cell metabolism and invasion. In a previous
study, it was demonstrated that HIF-lα repressed esophageal
squamous cell carcinoma (ESCC) growth in a murine xenograft mouse
model (5). A number of miRNAs that
are induced during hypoxia have been identified; one of these
miRNAs (miR-210) is strongly induced by HIF-lα and has pleiotropic
effects (6,7). It is reported that miR-210 inhibits
proliferation in renal carcinoma, nasopharyngeal, pancreatic and
head and neck cancer (8–10). Therefore, the present study
hypothesized that miR-210, as a downstream target of HIF-1α, may be
involved in cell survival in ESCC.
In the present study, the expression levels of
circulating miR-210 were examined in patients with ESCC, and the
induction of miR-210 in hypoxic ESCC cells was confirmed. The
functional role of miR-210 in the growth of carcinomas and the
mechanism by which it acts were also investigated.
Materials and methods
Patients and plasma samples
Approval from the Medical Ethics Committee of the
Fourth Affiliated Hospital of Hebei Medical University
(Shijiazhuang, China), and written informed consent from all
participants were obtained prior to the start of the study.
Subjects included 22 patients with newly diagnosed ESCC (no prior
treatment) from the Fourth Affiliated Hospital of Hebei Medical
University from September 2012 to May 2013, and 15 healthy
volunteers. All patients with ESCC were treated with 58~64 Gy by
3D-conformal radiation therapy or intensity-modulated radiation
therapy. Up to 8 ml of whole blood was obtained from each patient
prior to radiotherapy, and from the healthy controls. Immediately
subsequent to collection, the blood samples were subjected to
isolation of cell-free nucleic acids using a 3-spin protocol (1,200
g for 30 min, 12,000 g for 5 min and 12,000 g for 5 min) to prevent
contamination by cellular nucleic acids. Plasma samples were then
stored at −80°C until further processing.
Cell culture and transfection
Eca-109 and HEK 293t/17 cells were obtained from Dr.
BE Shan and YM Zhao (Department of Scientific Research Center, the
Fourth Hospital of Hebei Medical University, Shijiazhuang, China).
The cells were cultured in RPMI-1640/Dulbecco’s modified Eagle’s
medium supplemented with 10% fetal bovine serum, 100 U/ml
penicillin and 100 μg/ml streptomycin, were confirmed to be
negative for Mycoplasma and maintained in a humidified 5%
carbon dioxide (CO2) atmosphere at 37°C (all reagents
were purchased from Gibco Life Technologies, Grand Island, NY,
USA). Cells were maintained under hypoxic conditions in the
presence of 1% oxygen using a hypoxia incubator (MiniGalaxy A, RS
Biotech, Scotland, UK), using a continuous flow of a humidified
mixture of 95% N2 and 5% CO2. The cells were
transfected with oligoribonucleotides for miR-210 or negative
control RNA (ncRNA; Ambion Life Technologies, Carlsbad, CA, USA)
using HiPerFect Transfection Reagent (Qiagen, Valencia, CA, USA)
according to the manufacturer’s instructions for
overexpression.
RNA extraction and quantitative
polymerase chain reaction (qPCR)
RNA was isolated from 400 μl serum sample and cells
using the mirVana PARIS kit (Ambion). RNA quality and abundance
were determined following extraction using a 2100 Bioanalyzer
(Agilent Technologies, Santa Clara, CA, USA) and a NanoDrop ND-1000
Spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA),
respectively. The expression of miR-210 was determined by qPCR
using TaqMan MicroRNA Assay kits (Applied Biosystems Life
Technologies, Carlsbad, CA, USA). For synthesis of cDNA, 10 ng of
total RNA for each serum sample was used for the individual assays
in a 15 μl reaction mixture containing 5 μl RNA extract, 0.15 μl
deoxyribonucleotide triphosphate (100 mM), 1 μl MultiScribe reverse
transcriptase (50 U/ml), 1.5 μl 10× reverse transcription buffer,
0.19 μl RNase inhibitor (20 U/ml), 1 μl gene-specific TaqMan primer
and 4.16 μl nucleotide-free water. The reaction mixture was
incubated at 16°C for 30 min, 42°C for 30 min and 85°C for 5 min.
Subsequently, 5 μl DNA template was amplified using 10 μl 2× TaqMan
PreAmp Master Mix (Applied Biosystems), 3 μl nuclear-free water,
and 2 μl gene-specific TaqMan primers/probe mix in a final volume
of 20 μl. qPCR was run on the 7500 Fast Real-Time PCR system
(Applied Biosystems). The reaction mixture was incubated at 95°C
for 5 min, followed by 40 cycles of 95°C for 10 sec, 60°C for 30
sec and 72°C for 1 sec. TaqMan qPCR was performed in triplicate.
The qPCR data were normalized to endogenous control genes that were
ideally stably expressed across the analyzed samples to reduce
measurement errors.
Assay of cell proliferation and cell
cycle
Cell counting kit (CCK)-8 and flow cytometric assays
were used to measure cell proliferation and analyze the cell cycle
distribution, respectively. These procedures were conducted with
Eca-109 cells at 24 and 48 h subsequent to transfection, as
described above. The bromodeoxyuridine (EdU) incorporation assay,
which measures cell proliferation, was performed using the EdU
in vitro Imaging kit (Guangzhou RiboBio Co., Ltd.,
Guangzhou, China) according to the manufacturer’s instructions.
Calculations for the analysis of the cell cycle were performed with
ModFit software 2.0 (BD Biosciences, Franklin Lakes, NJ, USA).
Dual-luciferase reporter assay
The region of human polo-like kinase PLK1–3′
untranslated region (UTR), generated by PCR amplification, was
cloned into the pmiR-RB-REPORT luciferase reporter gene plasmid
vector (Guangzhou RiboBio). The primer sequences were as follows:
h-PLK1-wild-type (wt), forward 5′-CCGCTCGAGTAGCTGCCCTCCCCTCCGG-3′
and reverse 5′-GAATGCGGCCGCCTGGCACCCCTCAGGAAATACAAG-3′;
h-PLK1-mutated (mut), forward
5′-TTGGCTTGTGCGTGTTTAAACAGATGTGAATATTC-3′ and reverse
5′-TGTTTAAACACGCACAAGCCAAGGAAAGGACAG-3′. These constructs were
named pmiR-PLK1-wt and pmiR-PLK1-mut. For the reporter assay, HEK
293t/17 cells were seeded onto 24-well plates and transfected with
500 ng of pmiR-PLK1-wt or pmiR-PLK1-mut and 100 nM miR-210 mimics
or ncRNA using lipofectamine 2000 (Invitrogen Life Technologies,
Carlsbad, CA, USA). Following transfection for 48 h, cells were
harvested and assayed with the Dual-Luciferase Reporter Assay
system (Promega, Madison, WI, USA) according to the manufacturer’s
instructions. The tests were performed in triplicate.
Western blot analysis
Transfected cells were harvested for immunoblot
analysis after 48-h incubation. Cells were lysed in Lysis Buffer
(Beyotime Institute of Biotechnology, Jiangsu, China), and protein
concentrations were measured using the Bicinchoninic Acid Protein
Assay kit (Beyotime Institute of Biotechnology). Total protein was
separated by SDS-PAGE using a 12% polyacrylamide gel and
electroblotted onto a polyvinylidenefluoride membrane (EMD
Millipore, Billerica, MA, USA). The membrane was immunoblotted
overnight at 4°C with the following primary antibodies: Rabbit
monoclonal antibody against human PLK1 (1:500; Cell Signaling
Technology, Inc., Danvers, MA, USA) and mouse monoclonal antibody
against human β-actin (1:2,000; Beyotime Institute of
Biotechnology). A secondary antibody, horseradish
peroxidase-conjugated goat immunoglobulin G (1:1,000; Beyotime
Institute of Biotechnology), was incubated with the membrane for 1
h following three washes with Tris-buffered saline with Tween 20.
Signals were detected with UltraECL Western Blot Detection reagent
(Beyotime Institute of Biotechnology). The images were obtained on
Kodak film (Rochester, NY, USA) and quantified with Quantity One
software 4.4 (Bio-Rad Laboratories, Hercules, CA, USA). All
experiments were performed in triplicate.
Statistical analysis
All data were calculated as the mean ± standard
deviation. SPSS version 18.0 (SPSS, Inc., Chicago, IL, USA) was
used for statistical analysis. Comparisons between treatment groups
and controls were conducted with Student’s t-test. P<0.05 was
considered to indicate a statistically significant difference.
Results
Circulating miR-210 overexpression in
esophageal cancer
A total of 37 participants were involved in the
present study, including 22 patients with ESCC and 15 healthy
individuals. The clinicopathological characteristics of the study
population are summarized in Table
I. The ranges of the Ct values of miR-210 in the ESCC and
control groups were 22.8–34.7 and 23.6–37.5, respectively. Fig. 1 presents the ΔCt values obtained
from the two groups. The miR-210 serum concentration was elevated
in the ESCC group compared with the control group. No significant
association was found between miR-210 levels and gender, age, tumor
location and differentiation levels (Table I).
 | Table ISerum miR-210 expression and
clinicopathological factors. |
Table I
Serum miR-210 expression and
clinicopathological factors.
| Clinical factors | Patients (n) | Serum miR-210
expression median (25th,75th percentile) | P-value |
|---|
| Gender | | | 0.195 |
| Male | 18 | 0.00040 (0.00018,
0.00057) | |
| Female | 4 | 0.00066 (0.00030,
0.00088) | |
| Age | | | 0.493 |
| ≥55 | 17 | 0.00040 (0.00021,
0.00060) | |
| <55 | 5 | 0.00054 (0.00023,
0.00084) | |
| Tumor location | | | 0.898 |
| Upper | 3 | 0.00039 (0.00008,
0.00092) | |
| Middle | 12 | 0.00045 (0.00013,
0.00062) | |
| Lower | 7 | 0.00041 (0.00027,
0.00076) | |
| Differentiation | | | 0.861 |
| Good | 1 | 0.00054 | |
| Moderate | 14 | 0.00040 (0.00018,
0.00061) | |
| Poor | 7 | 0.00041 (0.00020,
0.00090) | |
miR-210 is induced by hypoxia
To assess whether hypoxia regulates miR-210
expression in ESCC, Eca-109 cells were exposed to 1% oxygen for
different time periods. Among the different settings, the
hypoxia-induced miR-210 expression was discernible following 24 h
(Fig. 2).
miR-210 inhibits cancer cell
proliferation by inducing cell cycle arrest in G2/M
phase
The functional role of miR-210 in ESCC was assessed
by adding synthetic miR-210 to the Eca-109 cell line, in which
miR-210 expression is low (Fig.
2). The effect of miR-210 on the proliferation of ESCC cells
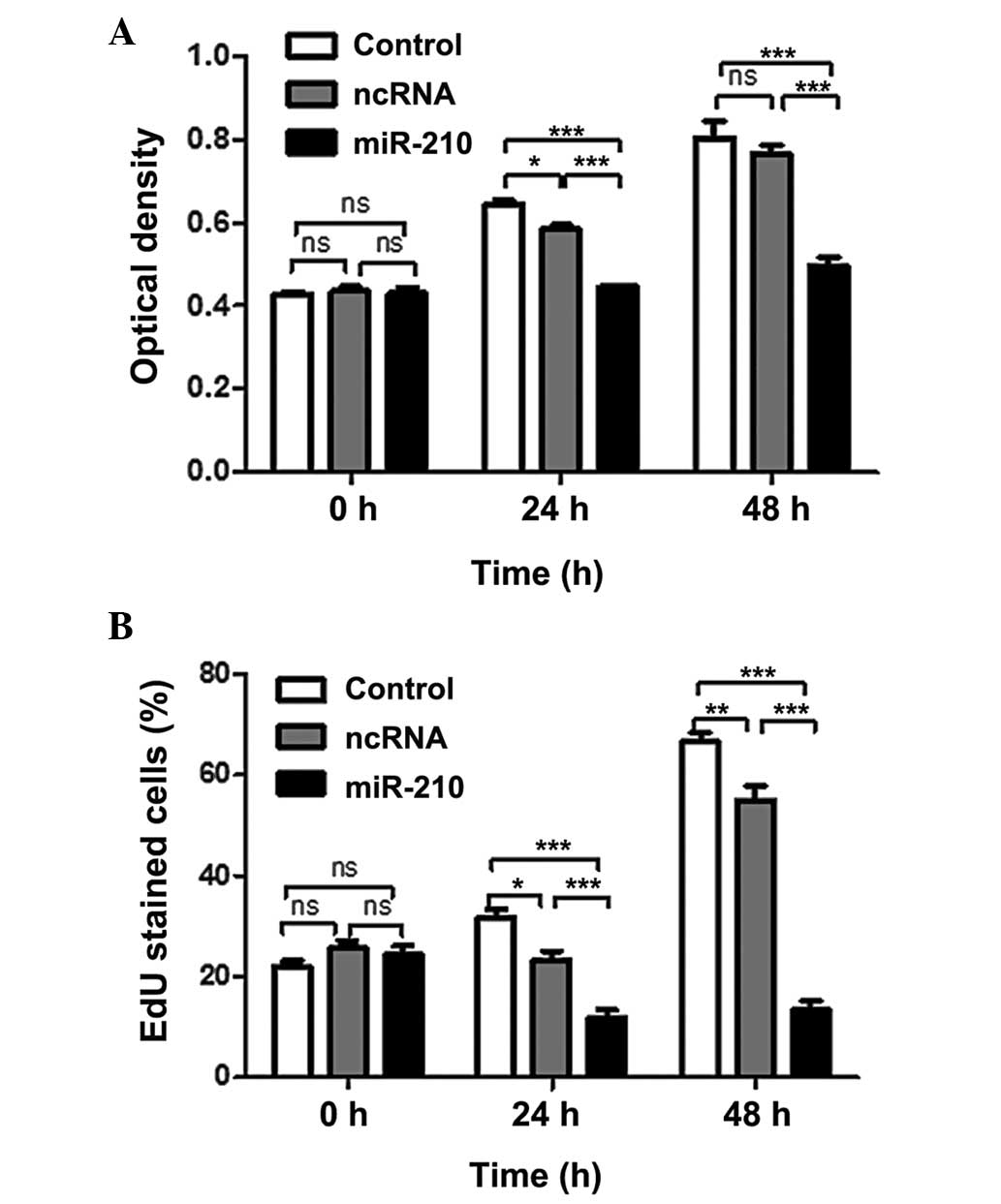
was examined by CCK-8 assay. Transfection of miR-210 significantly
reduced the proliferation rate of cancer cells (Fig. 3A). In addition, an EdU
incorporation assay was performed, and the results indicated that
miR-210 significantly reduced the uptake of EdU (Fig. 3B). These results suggested that
miR-210 negatively regulated cancer cell proliferation. Following
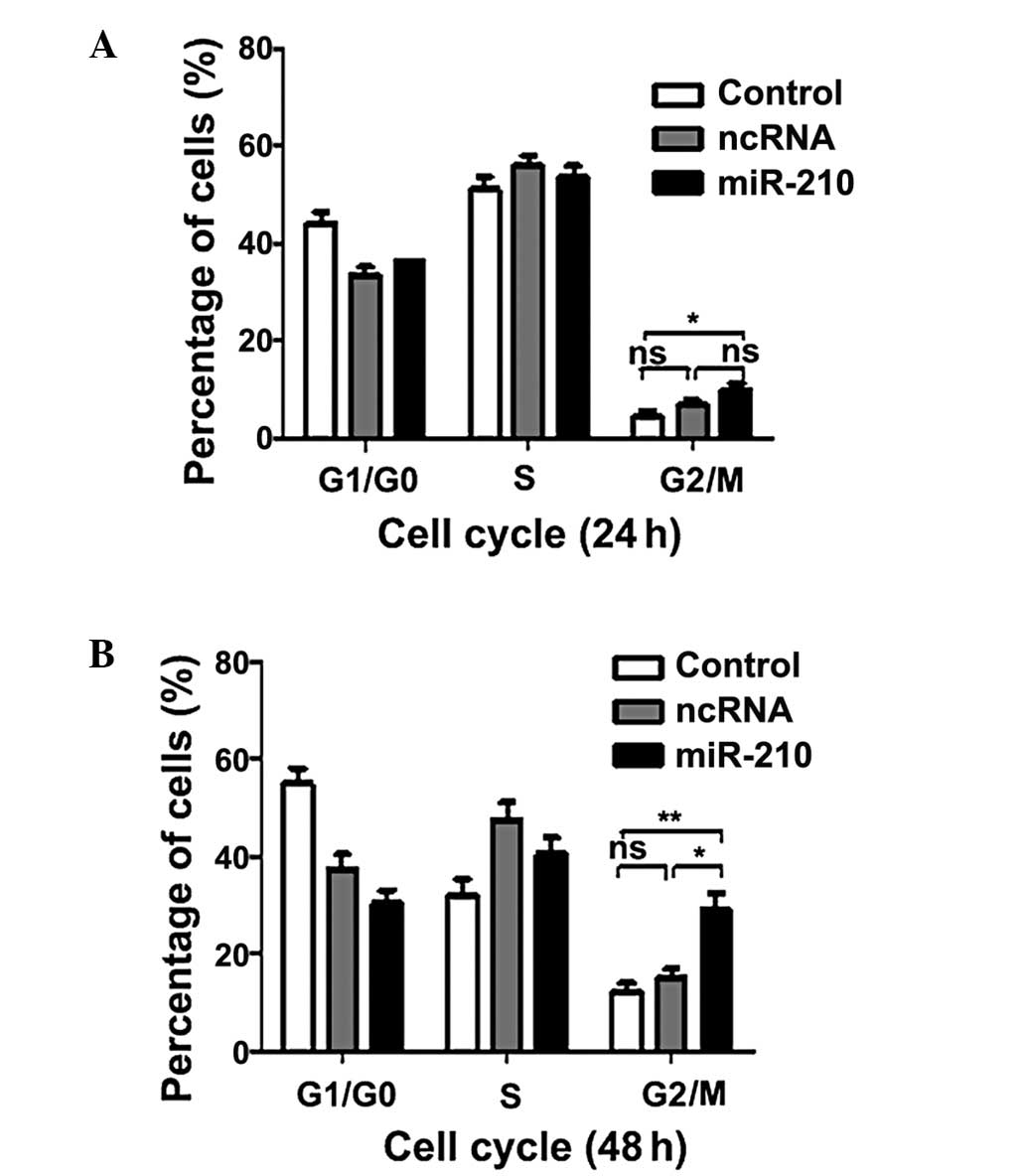
this, the effect of miR-210 on the cell cycle was investigated. As
shown in Fig. 4, transfection of
miR-210 resulted in a significant increase the G2/M
phase population. The results suggested that miR-210 may inhibit
proliferation of ESCC cells inducing G2/M phase
arrest.
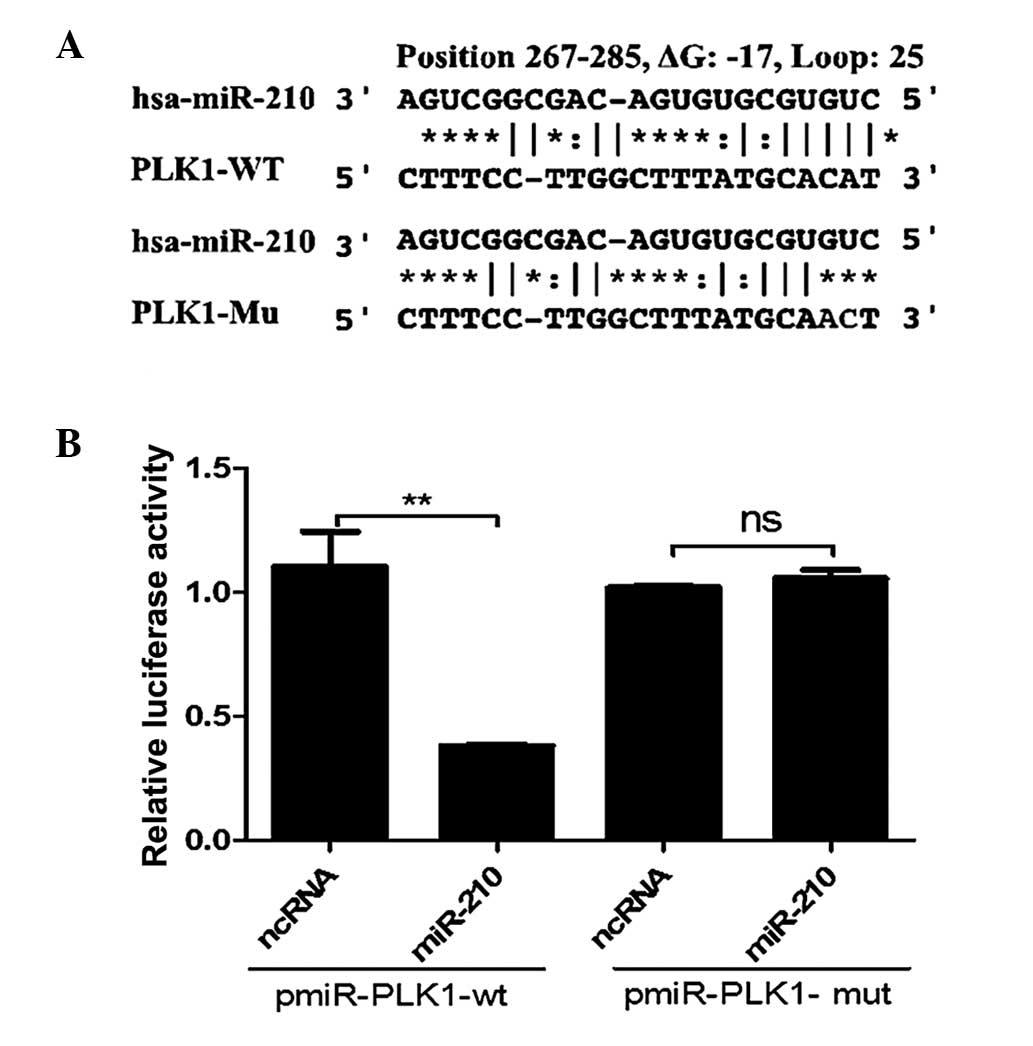
miR-210 targets the PLK1 3′-UTR
directly
To explore the underlying mechanism of the
association between miR-210 expression and cell cycle arrest, the
FindTar3 algorithm (http://bio.sz.tsinghua.edu.cn/) was used to identify
targets of miR-210, taking into consideration whether the predicted
genes are likely to be potential candidates linked to the cell
cycle. One of the common targets identified by the program was
PLK1, which is critical during G2/M transitions of the
normal cell cycle. To validate the target prediction, the direct
interaction between miR-210 and the 3′UTR of PLK1 mRNA was
assessed. As presented in Fig. 5,
when miR-210 oligos were transfected into 293T/17 cells with the
reporter construct pGL3-Luc-PLK1, luciferase activity was reduced
by >50% compared with the activity following transfection with
scramble oligos.
miR-210 represses expression of PLK1
The present study demonstrated that miR-210 targets
the PLK1 3′UTR directly. In order to further explore the
association between miR-210 and PLK1, protein expression levels of
PLK1 in human esophageal squamous cells were assessed by western
blotting. As presented in Fig. 6,
the levels of PLK1 protein expression in transfected cells with
miR-210 oligos were significantly reduced compared with the
expression in cells with stable integration of the scramble
sequence after 48 h.
Discussion
miR-210 is reportedly induced under hypoxic
conditions and regulated by HIFs as a transcriptional target
(11). The present study provided
support for this in ESCC. In previous studies, altered expression
of miR-210 was reported in other malignant tissues, including head
and neck, pancreas, breast and ovarian cancer (10,12–15);
however, the expression of circulating miR-210 has rarely been
discussed. In the present study, it was demonstrated for the first
time, to the best of our knowledge, that the expression of
circulating miR-210 is upregulated in patients with ESCC. Another
study on the role of circulating miRNAs in cancer and their
potential utility as prognostic markers has emerged. The
correlation of circulating miR-210 levels with breast cancer
mortality rates is evident, according to a study by Jung et
al (16), who reported that
plasma miR-210 levels correlated with sensitivity to trastuzumab,
tumor presence and lymph node metastases in patients with breast
cancer. Another previous study indicated a statistically
significant four-fold increase in circulating levels of miR-210 in
patients with pancreatic cancer, compared with levels in normal
controls (17). It must be taken
into account that the above three studies were conducted on small
cohorts, but the presence of elevated miR-210 in the plasma of
patients with cancer suggests that this may be a vital factor to
consider, and may further advance the understanding of the
underlying pathogenesis of multiple types of cancer.
In order to enhance the understanding of the
function of miR-210 in esophageal squamous cells, the consequences
of miR-210 overexpression in Eca-109 squamous cells cultured under
normoxic conditions were examined. The inhibition of proliferation
implied by the CCK8 and EdU incorporation assay is consistent with
previous similar studies in renal carcinoma and nasopharyngeal
cancer cells (8,9). Additionally, overexpression of
miR-210 in pancreatic and head and neck cancer cell lines has been
demonstrated to delay tumor cell growth in a murine xenograft mouse
model (10). Consistent with this
observation, transfection of miR-210 was indicated to significantly
induce G2/M phase cell cycle arrest. Several other
studies have also demonstrated that miR-210 inhibited tumor cell
proliferation by inducing G0/G1 and/or
G2/M phase cell cycle arrest. In contrast with other
solid tumors, miR-210 is frequently underexpressed in cases of
ovarian cancer. This potentially leads to increased expression of
the transcription factor E2F3, which participates in regulation of
the cell cycle (14). Likewise,
miR-210 is downregulated in ESCC. miR-210 inhibits cancer cell
proliferation and induces G0/G1 phase cell
cycle arrest by uninhibiting fibroblast growth factor receptor-like
1 (FGFRL1), which in turn accelerates cell cycle progression
(15). However, it cannot
generally be stated that miR-210 induction in hypoxia negatively
regulates cell cycle progression and proliferation. In hepatic
cancer cells, miR-210 activates the myc pathway via downregulation
of the c-Myc antagonist MNT, and loss of myc has been demonstrated
to abolish miR-210-mediated override of hypoxia-induced cell cycle
arrest (18). Therefore, the net
impact of miR-210 on cell cycle regulation may be
context-dependent.
Clinical data also present this discrepancy. Marked
reduction in the levels of miR-210 have been observed, particularly
in poorly differentiated ESCCs (15). By contrast, overexpression of
miR-210 in tumor tissues has been correlated with poor prognosis in
breast, head and neck and pancreatic cancer (10,12,13,19).
A likely explanation is that miR-210 is the most robustly induced
miRNA under hypoxic conditions. It is possible that upregulated
miR-210 expression levels only reflect the in vivo status of
tumor hypoxia. However, given the well-established roles of tumor
hypoxia in predicting a poor prognosis in patients with cancer, it
is likely that the increased expression of miR-210 also correlates
with the poor prognoses. Based on the evidence from the present and
previous studies, it was hypothesized that miR-210 acts in the
regulation of cell survival through multiple mechanisms, and
further careful study of the function of miR-210 in different
genetic backgrounds and human tumor types is required.
It is well-established that miR-210 induces
G0/G1 phase cell cycle arrest by targeting
E2F3 and FGFRL1. However, the underlying mechanism remains elusive.
In the present study, PLK1, which is a critical regulator of
mitosis at several levels, was investigated as a candidate target
of miR-210. In the present study, it was demonstrated that miR-210
targeted the PLK1 3′UTR directly and suppressed its expression in
Eca-109 squamous cells. Previous studies have demonstrated that
PLK1 facilitates the activation of cyclin D kinase 1 (Cdk1)/cyclin
B by activating cell division cycle 25C (Cdc25C) (20). The Cdc25 phosphatase family are
essential in G2/M transitions of the normal cell cycle.
Among them, Cdc25B and C are required for entry into mitosis
(21,22). Activation of Cdk1/cyclin B is
initiated by Cdc25B during the G2/M transition, while
the full activation of Cdk1/cyclin B is governed by Cdc25C at the
onset of mitosis. The active Cdk1/cyclin B then phosphorylates
Cdc25B and C, resulting in an irreversible autoamplification loop
that drives cells into mitosis (23). PLK1 also induces a further increase
in Cdk1 activity by inhibiting Myt1 and inducing the degradation of
Wee1 through phosphorylation (24,25).
Aside from mitotic entry, PLK1 controls multiple processes during
mitotic progression. PLK1 phosphorylates ninein-like protein and
kizuna to promote the recruitment of g-tubulin ring complexes and
keep the integrity of the centrosomes (25,26).
PLK1 regulates sister chromatid resolution through promoting the
removal of cohesins in prophase and promoting their cleavage by
separase. PLK1 is also essential for cytokinesis as it activates
the Rho guanosin triphosphatase, an activator of the actomyosin
ring that encourages the contraction of the contractile ring and
promotes cytokinesis (24).
Together, these findings indicated that miR-210 may exert its
tumor-suppressive effect in ESCC mainly by targeting PLK1. This
conclusion was supported by a recent study demonstrating that
miR-210 disturbs mitotic progression of nasopharyngeal cancer
through regulating a group of mitosis-associated genes, including
PLK1 (9).
In conclusion, the results of the present study was
the first to reveal that circulating miR-210 levels were elevated
in patients with ESCC and that it may potentially serve as a useful
biomarker for ESCC diagnosis. miR-210 inhibits the proliferation of
ESCC cells by inducing G2/M phase cell cycle arrest, and
the effects of miR-210 are mediated mainly by the targeting of
PLK1. The data suggested that miR-210 may be critical in the
proliferation of ESCC and that miR-210 and its targets may serve as
therapeutic targets of ESCC.
References
|
1
|
Krol J, Loedige I and Filipowicz W: The
widespread regulation of microRNA biogenesis, function and decay.
Nat Rev Genet. 11:597–610. 2010.PubMed/NCBI
|
|
2
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nilsen TW: Mechanisms of microRNA-mediated
gene regulation in animal cells. Trends Genet. 23:243–249. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lim LP, Lau NC, Garrett-Engele P, et al:
Microarray analysis shows that some microRNAs downregulate large
numbers of target mRNAs. Nature. 433:769–773. 2005. View Article : Google Scholar
|
|
5
|
Jing SW, Wang YD, Kuroda M, et al: HIF-1α
contributes to hypoxia-induced invasion and metastasis of
esophageal carcinoma via inhibiting E-cadherin and promoting MMP-2
expression. Acta Med Okayama. 66:399–407. 2012.
|
|
6
|
Hua Z, Lv Q, Ye W, et al: MiRNA-directed
regulation of VEGF and other angiogenic factors under hypoxia. PLoS
One. 1:e1162006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kulshreshtha R, Ferracin M, Wojcik SE, et
al: A microRNA signature of hypoxia. Mol Cell Biol. 27:1859–1867.
2007. View Article : Google Scholar
|
|
8
|
Nakada C, Tsukamoto Y, Matsuura K, et al:
Overexpression of miR-210, a downstream target of HIF1α, causes
centrosome amplification in renal carcinoma cells. J Pathol.
224:280–288. 2011.PubMed/NCBI
|
|
9
|
He J, Wu J, Xu N, et al: MiR-210 disturbs
mitotic progression through regulating a group of mitosis-related
genes. Nucleic Acids Res. 41:498–508. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Huang X, Ding L, Bennewith KL, et al:
Hypoxia-inducible mir-210 regulates normoxic gene expression
involved in tumor initiation. Mol Cell. 35:856–867. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Huang X, Le QT and Giaccia AJ: MiR-210 -
micromanager of the hypoxia pathway. Trends Mol Med. 16:230–237.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gee HE, Camps C, Buffa FM, et al:
hsa-mir-210 is a marker of tumor hypoxia and a prognostic factor in
head and neck cancer. Cancer. 116:2148–2158. 2010.PubMed/NCBI
|
|
13
|
Camps C, Buffa FM, Colella S, et al:
hsa-mir-210 is induced by hypoxia and is an independent prognostic
factor in breast cancer. Clin Cancer Res. 14:1340–1348. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Giannakakis A, Sandaltzopoulos R, Greshock
J, et al: miR-210 links hypoxia with cell cycle regulation and is
deleted in human epithelial ovarian cancer. Cancer Biol Ther.
7:255–264. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tsuchiya S, Fujiwara T, Sato F, et al:
MicroRNA-210 regulates cancer cell proliferation through targeting
fibroblast growth factor receptor-like 1 (FGFRL1). J Biol Chem.
286:420–428. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jung EJ, Santarpia L, Kim J, et al: Plasma
microRNA 210 levels correlate with sensitivity to trastuzumab and
tumor presence in breast cancer patients. Cancer. 118:2603–2614.
2012. View Article : Google Scholar
|
|
17
|
Ho AS, Huang X, Cao H, et al: Circulating
miR-210 as a novel hypoxia marker in pancreatic cancer. Transl
Oncol. 3:109–113. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang Z, Sun H, Dai H, et al: MicroRNA
miR-210 modulates cellular response to hypoxia through the MYC
antagonist MNT. Cell Cycle. 8:2756–2768. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Toyama T, Kondo N, Endo Y, et al: High
expression of microRNA-210 is an independent factor indicating a
poor prognosis in Japanese triple-negative breast cancer patients.
Jpn J Clin Oncol. 42:256–263. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Donaldson MM, Tavares AA, Hagan IM, Nigg
EA and Glover DM: The mitotic roles of Polo-like kinase. J Cell
Sci. 114:2357–2358. 2001.PubMed/NCBI
|
|
21
|
Boutros R, Lobjois V and Ducommun B: CDC25
phosphatases in cancer cells: key players? Good targets? Nat Rev
Cancer. 7:495–507. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gabrielli BG, De Souza CP, Tonks ID, et
al: Cytoplasmic accumulation of cdc25B phosphatase in mitosis
triggers centrosomal microtubule nucleation in HeLa cells. J Cell
Sci. 109:1081–1093. 1996.
|
|
23
|
Boutros R, Dozier C and Ducommun B: The
when and wheres of CDC25 phosphatases. Curr Opin Cell Biol.
18:185–191. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Archambault V and Glover DM: Polo-like
kinases: conservation and divergence in their functions and
regulation. Nat Rev Mol Cell Biol. 10:265–275. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lens SM, Voest EE and Medema RH: Shared
and separate functions of polo-like kinases and aurora kinases in
cancer. Nat Rev Cancer. 10:825–841. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Casenghi M, Meraldi P, Weinhart U, et al:
Polo-like kinase 1 regulates Nlp, a centrosome protein involved in
microtubule nucleation. Dev Cell. 5:113–125. 2003. View Article : Google Scholar : PubMed/NCBI
|