Introduction
Catalase (EC 1.11.1.6), the most abundant protein in
peroxisomes, is ubiquitously present in mammalian and non-mammalian
aerobic cells, which contain a cytochrome system. This enzyme is
important in the removal of toxic hydrogen peroxide from the cell.
Catalase exists as a dumbbell-shaped tetramer of four identical
subunits, which contains a heme at the catalytic center (1). The molecular weights of the catalases
range between 230 and 250 kDa (2).
Aydemir and Kuru demonstrated that catalase activity is stable at a
broad pH range between 5.0 and 10.5, and at temperatures between 10
and 30°C (3). Catalase is widely
used as a typical example of a peroxisomal enzyme during
biochemistry and molecular biology teaching due to its specific
characteristics, including high efficiency, high specificity and
stability.
The crystallization of beef liver catalase was first
reported by Sumner and Dounce in 1937 (4). A number of studies, notably those of
Price et al on rat liver catalase, of Maimoni Gonçalves
et al on human placenta catalase and of Chatterjee and
Sanwal on goat lung catalase, have demonstrated various methods of
catalase purification in mammals (5–7).
Despite developments in molecular cloning and the industrial
production of proteins using ‘artificial’ microorganisms, catalase
purification remains an obligatory step due to the cost and
complexity of these methods in the majority of circumstances
(8).
Although there is now substantial information on
catalase purification, the most common limitation in the
junior-grade undergraduate experimental course is the expense of
the materials and equipment required to perform modern experimental
methods. In Chinese medical universities, junior-grade
undergraduates are required to undertake two or three school years
of study, according to teaching programs, together with their
counterparts in the same class. Several basic courses, including
biochemistry, molecular biology, physiology and immunology are
required during their junior year, from which students are expected
to build on a wide knowledge (9).
As the junior-grade undergraduates of medical schools are
relatively concentrated and are required to learn additional types
of courses, the use of new and advanced technologies in
undergraduate classes is beyond the budget of the majority of
universities and reduce satisfaction in the fundamental courses
(10,11). The present study aimed to describe
a simple method for purifying catalase from mouse liver that relies
solely on ethanol-chloroform treatment, sodium sulfate
fractionation, dialysis and Sephadex G-200 gel filtration
chromatography.
Materials and methods
Animals and reagents
In total, eight 4-month-old Kunming mice of cleaning
grade II, weighing between 20 and 25 g, can be used for each pair
of students. The mice were supplied from the Experimental Animal
Center at Hebei Medical University (Shijiazhuang, China). The
absorbance was obtained using a 752 N UV-Vis spectrophotometer
(Nanjing Everich Medicare Import and Export Co., Ltd, Nanjing,
China) and the relative intensities of the blots were analyzed
using a JD801 imaging analysis system (Jiangsu JEDA
Science-Technology Development Co., Ltd, Nanjing, China). The
rabbit anti-mouse catalase polyclonal antibody and the horseradish
peroxidase labeled goat anti-rabbit immunoglobulin G polyclonal
(cat. no. ZB-2301) were purchased from Zhongshan Golden Bridge
Biotechnology Co., Ltd. (Beijing, China). The standard catalase was
supplied from Beijing Tian Qin Yi He Biotech Co., Ltd. (Beijing,
China. The use of animals was reviewed and approved by the Hebei
Medical University Animal Ethics Committee (Shijiazhuang,
China).
Sodium sulfate fractionation
Sodium sulfate (0.5 mol/l) was added to the
supernatant by 0.06 volume of supernatant, which produced a marked
turbidity and was then left to stand for 1 h at 4°C and centrifuged
at 5,100 × g at 4°C for 10 min. The precipitate was dissolved in
the minimum quantity of 0.1 mol/l sodium phosphate buffer (pH 7.8)
and re-centrifuged at 2,300 × g for 10 min to remove the insoluble
precipitate.
Dialysis
The supernatant was dialyzed against dialysis buffer
(0.1 mol/l acetate buffer, 0.1 mol/l NaCl, 20% ethanol) at 4°C for
12 h, and centrifuged at 5,100 × g for 10 min. The precipitate was
collected by centrifugation (5,100 × g for 10 min) and dissolved in
0.1 mol/l sodium phosphate (pH 7.8). The insoluble material was
then removed by centrifugation (2,300 × g for 10 min).
Sephadex G-200 gel filtration
chromatography
The supernatant was applied to a Sephadex G-200
column (1.8×32 cm; Suolaibao Bio-technology Co., Ltd, Shanghai,
China), which was previously equilibrated using 0.1 mol/l sodium
phosphate (pH 7.8). As the liver is rich in catalase, it was
possible to use tiny columns of packed materials. The enzyme was
eluted with the same buffer at a flow rate of 0.3 ml/min. The peak
eluted from the Sephadex G-200 revealed two bands following
absorbance analysis at 280 and 406 nm, which were characteristic
for the protein and the heme group, respectively. The fractions
surrounding the peak, with an OD406/280 ratio exceeding
1.08, were then pooled.
Enzyme assay
Catalase activity was measured using a potassium
permanganate titration method (10). For assay of the activity, the
enzyme preparation of each purification step was diluted with 0.1
mol/l sodium phosphate buffer (pH 7.8). The diluted enzyme
preparation was then added to the buffer containing 40 mmol/l
H2O2 and incubated at 25°C for 10 min. The
reaction was then terminated using 95% sulfuric acid and the
remaining H2O2 was titrated using 2 mmol/l
potassium permanganate (2KMnO4 +
5H2O2 + 3H2SO4 → 2MnSO4
+ K2SO4 + 5O2↑ + 8H2O). One unit
of catalase activity was defined as the quantity of enzyme able to
decompose 1 μmol H2O2/min under standard
assay conditions.
The protein concentration was measured using the
method detailed by Lowry et al, which used bovine serum
albumin (Beinuo Biotech Co., Ltd., Shanghai, China) as the standard
protein (12).
Enzyme kinetics
The catalase activity was measured by the increase
in hydrogen peroxide concentration ranging between 8 mmol/l and 40
mmol/l in 0.1 mol/l sodium phosphate(pH 7.8), according to the
method of titration previously described (13). The apparent Km
H2O2 value was estimated using a
Lineweaver-Burk plot.
Determination of the molecular weights of
the subunits
The molecular weights of the subunits, assuming a
globular structure for the catalase, was determined by SDS-PAGE
electrophoresis. The enzyme preparation was separated by SDS-PAGE
using a 2.5% stacking gel and a 12% running gel. The proteins in
the running gel were then made visible by staining with Coomassie
Brilliant Blue R-250 (Beinuo Biotech Co., Ltd.). The molecular
weight calibration markers were: 94.0, 66.2, 45.0, 35.0, 24.0, 20.0
and 14.4 KDa.
Western blot analysis
The purified enzyme was identified by western blot
analysis, in which 4.0 μg protein was loaded into each well,
separated on SDS-PAGE gels and transferred onto a nitrocellulose
membrane. The rabbit anti-mouse catalase antibody was the added to
bind to the epitope of the target protein and the primary antibody
was recognized by the relative secondary antibody labeled with
horseradish peroxidase. The relative intensities of the blots were
quantified by densitometry using the JD801 imaging analysis
system.
Results
Catalase purification
The purification profile of the catalase from mouse
liver is shown in Table I. The
highest specific activity (1.68×105 U/mg) was obtained
from the Sephadex G-200 column. Catalase was purified 31.8-fold
with an 18.3% yield. Aydemir and Kulin purified catalase from
chicken erythrocyte cytosolic fractions 139.59-fold with a 1.68%
yield (3). The difference in the
purification fold may be due to the present study using only gel
filtration chromatography, which increases the ease of performance
and reduces the cost of use in the student laboratory.
 | Table IPurification of mouse liver
catalase. |
Table I
Purification of mouse liver
catalase.
| Purification
step | Total activity
(U) | Total protein
(mg) | Specific activity
(U/mg protein)a | Purification
fold | Yield (%) |
|---|
| Homogenate
liquid |
1.16×106 | 219.7 |
5.29×103 | 1.0 | 100.0 |
| Salting-out
liquid |
9.70×105 | 24.1 |
4.05×104 | 7.7 | 83.6 |
| Dialysate |
6.72×105 | 4.4 |
1.52×105 | 28.7 | 57.9 |
| Sephadex G-200 gel
filtration chromatography |
2.12×105 | 1.3 |
1.68×105 | 31.8 | 18.3 |
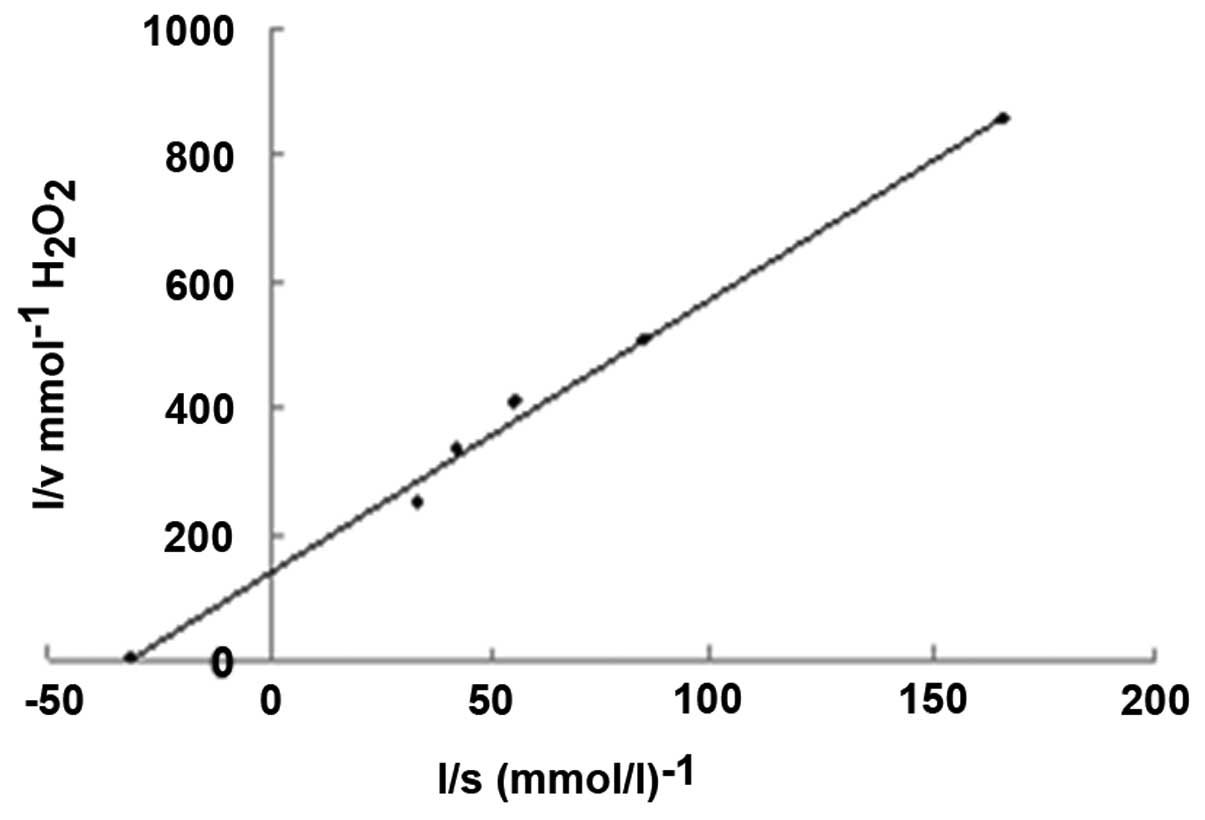
The Km value of the enzyme was 32.3 mmol/l, measured
using a Lineweaver-Burk plot at pH 7.8 and 25°C (Fig. 1). The lower Km value of 32.3 mmol/l
for mouse liver catalase compared with 93 mmol/l for bovine liver
catalase demonstrated a greater affinity of catalase towards
H2O2 (14).
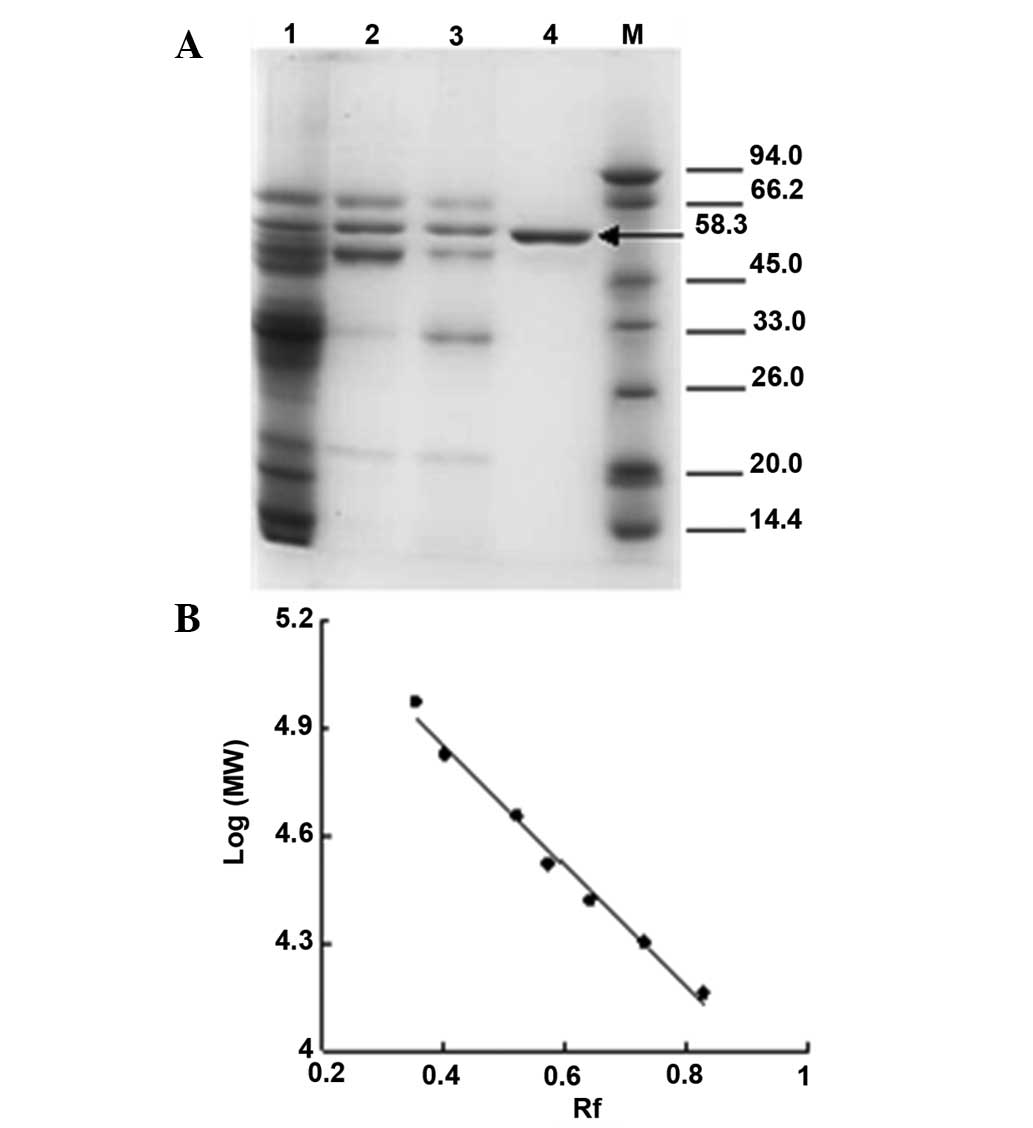
The molecular weights of the subunits of mouse liver
catalase were determined using SDS-PAGE electrophoresis (Fig. 2). The SDS-PAGE revealed one
polypeptide band at the expected position at a mass of 58.3 KDa.
Furuta et al observed subunits with a similar molecular mass
of 59.758 KDa (15). The value
corresponds to the major monomeric form of the enzyme.
Enzyme identification
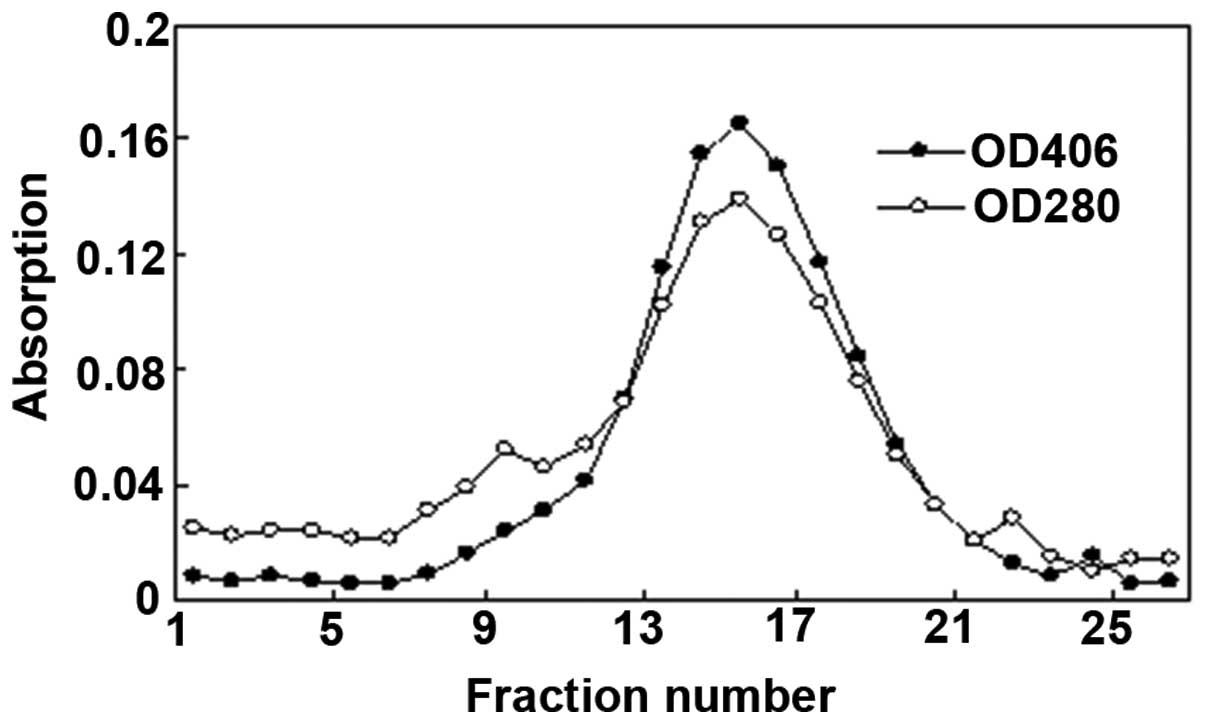
This catalase enzyme was confirmed using two
techniques. Fig. 3 shows the
results of the separation of catalase and other proteins by
Sephadex G-200 gel filtration chromatography. The absorption
spectrum of the purified enzyme revealed two major peaks at 280 nm
(due to the protein) and at 406 nm (due to the heme group). The
overlapping peak of 280 and 406 nm was collected and the optical
density 406/280 ratio exceeded 1.08. In addition, western blot
analysis was used to identify the enzyme on the basis of the
affinity between a certain antigen and its relative antibody. As
shown in Fig. 4, one band was
observed in the enzyme preparation at the same position as the
standard catalase protein control.
Discussion
The present study demonstrated a straightforward
method for purifying catalase from mouse liver that relies solely
on ethanol-chloroform treatment, sodium sulfate fractionation,
dialysis and Sephadex G-200 gel filtration chromatography. Al-Bar
and Omar described a method to purify camel liver catalase by the
preparation of crude extract, ammonium sulfate precipitation and
chromatography on diethylaminoethyl cellulose-sepharose (16). Although camel liver is rich in
catalase, it is difficult to obtain camel liver, which limits its
use for junior-grade undergraduates. Yang and DePierre presented a
method of purification from mouse liver catalase by immobilized
metal ion affinity chromatography (17), however, this was expensive and
complex to perform due to the requirement of prior treatment to
materials. By comparison, the purification procedure described in
the present study did not require specific equipment and was
performed using the material and apparatus present in the majority
of student laboratories, making it adaptable to the junior-grade
undergraduate experimental course.
It is possible that the method described in the
present study, can be performed, with data analyzed and discussed,
in 2–3 days as the liver is rich in catalase, the method is
sensitive and few materials are used. In addition, due to the high
stability of catalase, the flexibility of the procedure enables it
to be finished continuously or discontinuously in stages. In the
majority of China’s medical schools, the experimental course is not
a separate course, but is included in the fundamental courses
taught during theoretical knowledge teaching (18). The majority of junior-grade
undergraduates only have a 4-hour biochemistry and molecular
biology experiment course every week and are unable to complete a
large purification experiment in one occasion; therefore, the
method presented in the present study many be more popular as
students are able to complete the purification procedure in phases
according to their experiment course arrangements. An example of a
schedule for the overall operation procedure is shown in Fig. 5.
In addition, the method introduces several
fundamental laboratory theories to students in terms of
biochemistry and molecular biology. These theories include that of
dialysis, the process of separating molecules in solution by
differences in their rate of diffusion through a semipermeable
membrane and chromatography, the technique of separating components
of a mixture according to the different affinities for a fixed or
stationary phase and their differential solubility in a moving or
mobile phase. Students are able to acquire two separation
techniques through the entire procedure. In addition to the
theoretical knowledge inherent to the purification procedure, the
purified enzyme can be used to learn certain basic enzyme
characterization techniques, including western blotting,
determination of protein activity and concentration and protein
molecular weight determination.
In conclusion, the present study demonstrated the
purification of mouse liver catalase using basic equipment, common
to the majority of junior-grade undergraduate laboratories.
Throughout this purification process, students are able to gain
experience of laboratory techniques, acquire valuable data
analyzing skills and understand research ideas, which are highly
relevant to biochemistry and molecular biology. In addition, the
procedure described has the potential to improve the comprehensive
and manipulative ability of students, and develop their sense of
innovation in their junior years.
Acknowledgements
This study was supported by the Higher Education
Reform Project of Hebei Province of China. The authors would like
to thank Dr Lingling Jiang of the Institute of Basic Medicine for
their critical evaluation.
References
|
1
|
Kirkman HN and Gaetani GF: Mammalian
catalase: a venerable enzyme with new mysteries. Trends Biochem
Sci. 32:44–50. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Baird MB, Massie HR and Birnbaum LS:
Presence of a high-molecular-weight form of catalase in enzyme
purified from mouse liver. Biochem J. 163:449–453. 1977.PubMed/NCBI
|
|
3
|
Aydemir T and Kuru K: Purification and
partial characterization of catalase from chicken erythrocytes and
the effect of various inhibitors on enzyme activity. Turk J Chem.
27:85–97. 2003.
|
|
4
|
Sumner JB and Dounce AL: Crystalline
catalase. Science. 85:366–367. 1937. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Price VE, Sterling WR, Tarantola VA,
Hartley RW Jr and Rechcigl M: The kinetics of catalase synthesis
and destruction in vivo. J Biol Chem. 237:3468–3475.
1962.PubMed/NCBI
|
|
6
|
Maimoni Gonçalves V, Cezar de Cerqueira,
Leite L, Raw I and Cabrera-Crespo J: Purification of catalase from
human placenta. Biotechnol Appl Biochem. 29:73–77. 1999.PubMed/NCBI
|
|
7
|
Chatterjee U and Sanwal GG: Purification
and characterization of catalase from goat (Capra capra) lung. Mol
Cell Biol. 126:125–133. 1993.
|
|
8
|
Alenkin D, Yermekbayeva L, Mujib S,
Vesterberg A, Newman E, Yamazaki K, Cossar D and Dhe-Paganon S: A
centrifugation-free high-throughput protein purification system
using in-line microfluidization. Protein Expr Purif. 79:204–209.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lam TP, Wan XH and Ip MS: Current
perspectives on medical education in China. Med Educ. 40:940–949.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang HF and Zhan HJ: The research progress
in determination of catalase activity. Science and Technology
Innovation Herald. 19:7–8. 2009.
|
|
11
|
Xiao WW, Ma WL and Zhang XM: Present
situation of biochemical experiment teaching and its reform
strategies. Xi Bei Yi Xue Jiao Yu. 15:270–271. 2007.(In
Chinese).
|
|
12
|
Lowry OH, Rosebrough NJ, Farr AL and
Randall RJ: Protein measurement with the Folin phenol reagent. J
Biol Chem. 193:265–275. 1951.PubMed/NCBI
|
|
13
|
Dixon M: The graphical determination of Km
and Ki. Biochem J. 129:197–202. 1972.PubMed/NCBI
|
|
14
|
Switala J and Loewen PC: Diversity of
properties among catalases. Arch Biochem Biophys. 401:145–154.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Furuta S, Hayashi H, Hijikata M, Miyazawa
S, Osumi T and Hashimoto T: Complete nucleotide sequence of cDNA
and deduced amino acid sequence of rat liver catalase. Proc Natl
Acad Sci USA. 83:313–317. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Al-Bar and Omar AM: Characterization of
partially purified catalase from camel (Camelus dromedarius) liver.
Afr J Biotechnol. 11:9633–9640. 2012.
|
|
17
|
Yang Q and DePierre JW: Rapid one-step
isolation of mouse liver catalase by immobilized metal ion affinity
chromatography. Protein Expr Purif. 12:277–283. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Schwarz MR, Wojtczak A and Zhou T: Medical
education in China’s leading medical schools. Med Teach.
26:215–222. 2004. View Article : Google Scholar : PubMed/NCBI
|