Introduction
Renal cell carcinoma (RCC) is one of the primary
causes of cancer-associated mortalities, and its incidence is
increasing (1,2). Although there are numerous methods of
detecting localized RCC, in the majority of cases the disease is
difficult to diagnose (3).
Furthermore, it is still controversial whether immunotherapy and
antiangiogenic therapy are effective treatments for RCC (4,5).
Hence, it is necessary to find novel biomarkers for the diagnosis
and treatment of this disease. Additionally, the study of the
pathways involved in the pathogenesis of RCC may offer further
options for the treatment of RCC (6). Smad4 has been identified as a tumor
suppressor gene in various cancer types (7–9). A
higher frequency of Smad4 inactivation was observed in liver
metastases than in extrahepatic metastases (10) and colorectal cancer patients
expressing high Smad4 levels have been shown to have a longer
survival time than the patients with low levels (11). Smad4 increases the levels of
signaling in renal tubulointerstitial cells in a mouse model of
renal disease (12). However, the
potential role and molecular events of Smad4 signaling in RCC have
remained elusive.
Smads are the central components of the
intracellular signaling pathway of transforming growth factor-β
(TGF-β) ligands (13). The
signaling molecules for activins include the two receptor-regulated
Smads (R-Smads) Smad2 and Smad3, which are phosphorylated by ActRIB
and form a heteromeric complex with Smad4 (13). The R-Smad/Smad4 complex
translocates to the nucleus, where forkhead box protein H1 (FOXH1)
interacts with the R-Smad-Smad4 complex to regulate transcription
(14). The function of FOXH1 was
initially identified as binding the Mix.2 gene in Xenopus.
FOXH1 interacts with Smad4 and either phosphorylated Smad2 or Smad3
to form a complex (15). FOXH1
requires Smad to regulate transcription, as it does not contain a
domain for the activation of transcription (16). FOXH1 inhibits the transcription of
estrogen receptors (ERs) and androgen receptors (17). The FOXH1-R-Smad-Smad4 complex
(CFSS) represses the transcriptional activities of ERs. Estrogen
has been reported to induce renal carcinogenesis in Syrian hamsters
(18). Thus, the CFSS may repress
the development of RCC via the inhibition of the transcriptional
activity of estrogen receptors. The aim of the present study was to
investigate whether Smad4 is involved in the progression of
RCC.
Materials and methods
Antibodies and reagents
Rabbit anti-Myc monoclonal antibody (1 mg/ml; 1:500;
used for the co-immunoprecipitation assay) was purchased from
Clontech Laboratories (Mountain View, CA, USA). Mouse anti-Flag M2
monoclonal antibody (1 mg/ml; 1:2,000; used for fluorescence
staining) was from Sigma-Aldrich (St. Louis, MO, USA). Mouse
anti-green fluorescence protein (GFP) monoclonal antibody (1 mg/ml;
1:1,000; used for fluorescence staining) was from Cell Signaling
Technology Inc. (Danvers, MA, USA). Mouse anti-glutathione
S-transferase monoclonal antibody (GST; 0.5 mg/ml; 1:1,000; used
for the assay of the results of GST-pulldown analysis). Mouse
anti-GAPDH monoclonal antibody (1 mg/ml; 1:1,000) and complementary
horseradish peroxidase-labeled goat anti-mouse secondary antibodies
(1 mg/ml; 1:5,000) were purchased from Santa Cruz Biotechnology
(Dallas, Texas, USA). Mouse anti-Smad4 (1:1,000), mouse anti-FOXH1
(1:1,000) and mouse anti-estrogen (1:1,000) monoclonal receptor
antibodies (1 mg/ml each, used for western blot analysis) were
purchased from Shengshi Zhongfang BioSci & Tech (Beijing,
China). Mouse anti-β-actin monoclonal antibody (1:1,000; loading
control for western blot analysis) was purchased from Abcam
(Shanghai, China). Phospho-Smad2 (Ser465/467) (138D4) rabbit
monoclonal antibody (1 mg/ml; 1:1,000; #3108; used for western blot
analysis) was purchased from Cell Signaling Technology, Inc.
Renal cell cultures
The OS-RC-2 human RCC cell line was purchased from
Riken Cell Bank (Tsukuba, Japan). The normal human renal cell line
(HRE) was purchased from Promocell Co. Ltd. (Heidelberg, Germany).
Cells were cultured in a humidified atmosphere of 5% CO2
and 95% air at 37°C in RPMI-1640 medium (Gibco, Inc., Billing, MT,
USA) supplemented with 10% heat-inactivated fetal calf serum (FCS;
Shengma Yuanheng, Beijing, China). RCC and HRE proliferation was
determined by direct counting. For direct counting, 105
cells were seeded, harvested following three days of culture, and
counted using a Hausser Scientific hemocytometer (Hausser
Scientific, Horsham, PA, USA).
Plasmid constructs
Full-length Smad4 (forward, 5′-GAC ATCCATATGGACAATATGTCTATTAC-3′ and
reverse, 5′-GAC TGACTCGAGGTCTAAAGGTTGTGGGTC-3′),
FOXH1 (forward, 5′-GACATCCATATGGGGCCCTGCAGCGGCTC-3′ and
reverse, 5′-GACTGACTCGAGCAGGCTGCACCAGGA GAG-3′)
and estrogen receptor molecules (forward, 5′-GACATC CATATGACCATGACCCTCCACAC-3′ and
reverse, 5′-GAC TGACTCGAGGACTGTGGCAGGGAAACCC-3′),
FOXH1 and estrogen receptor molecules were constructed by
polymerase chain reaction (PCR), followed by subcloning into
various vectors at the sites of NdeI and XhoI
(underlined). PCR was performed using EPPENDORF Mastercycler® nexus
(Eppendorf AG, Hamburg Germany). Cycling conditions were as
follows: 95°C for 1 min, 30 cycles of 95°C for 20 sec, 60°C for 30
sec and 68°C for 2 min and one cycle of 68°C for 10 min. The
vectors were amplified in Escherichia (E.)
coli, isolated using a QIAprep Miniprep kit (Qiagen Inc.,
Chatsworth, CA, USA) and verified using an ABI 3730 automatic DNA
sequencer (Auke Biotech Co., Ltd, Beijing, China) with four dye
fluorescence-based DNA sequencing.
Smad4 expression constructs
The Smad4 gene (accession number, AB043547.1) was
amplified using the following primers: Sense,
5′-GTGAGCTAGCATGGACAATATGTCTATTAC-3′, and antisense,
5′-CTGAGAATTCCTTTATATATGCACTTGG-3′, which generated a 1328-bp
product. The PCR product was cloned into the NheI-EcoRI sites of
the pcDNA3.1 vector according to the manufacturer’s instructions
(TOPO TA Expression kit; Invitrogen, Carlsbad, CA, USA), which was
named as pcDNA3.1-Smad4. The pcDNA3.1-Smad4 plasmid was amplified
in E. coli, isolated using a QIAprep Miniprep kit (Qiagen
Inc.) and verified using an ABI 3730 automatic DNA sequencer with
four dye fluorescence-based DNA sequencing.
RNA interference
The siRNA directed against Smad4
(5′-ATGTGCCATAGACAAGGTGGAG-3′) and the non-target control siRNA
(5′-UUCUCCGAACGUGUCACGU-3′) were synthesized by Shanghai GenePharma
(Shanghai, China).
Transfection of the RCC cell line
OS-RC-2 human RCC cells (2×105 per p-96
plate) were transfected with various vectors. Transfection was
performed on cells in plates at 60% confluence using 9 μl
Lipofectamine 2000™ (Applied Biosystems, Life Technologies, Foster
City, CA, USA). Cells were split 48 h post-transfection and
neomycin-resistant clones (G418; Sigma-Aldrich) were selected.
Resistant colonies were either pooled or cloned by ring
isolation.
Quantitative reverse transcription PCR
(RT-qPCR)
Total RNA was isolated from the cells using
QIAshredder and RNeasy Mini kits (Qiagen, Inc.). An initial strand
of cDNA was synthesized from 500 ng of RNA extracts in a volume of
20 μl using avian myeloblastosis virus reverse transcriptase XL
(Takara Biotechnology Co., Ltd., Dalian, China) priming with random
9-mers at 42°C for 10 min. The cDNA strand was stored at 20°C prior
to use. Smad2, Smad4, FOXH1 and estrogen receptor transcriptional
levels were estimated using RT-qPCR and qPCR. qPCR was conducted
using SYBR Green I Master mix in a Light-Cycler 480, both obtained
from Roche (Mannheim, Germany). RNA was isolated from the
non-transfected and transfected OS-RC-2 cells, followed by cDNA
synthesis and data analysis as described previously (7). The primers for qPCR were as follows:
Sense, 5′-TACTATGTCTACTTCCTGAG-3′, and antisense,
5′-CAAGGAAAATAAAACATACC-3′ for Smad2; sense,
5′-ATTGATCTCTCAGGATTAAC-3′, and antisense,
5′-GTGGTAGTGCTGTTATGATG-3′ for Smad4; sense,
5′-ACTGAAGCTGGCCCAGATCA-3′ and antisense,
5′-GGCCCAGGTCCTTGGCGAAG-3′ for FOXH1; sense,
5′-TACCAATGACAAGGGAAGTA-3′ and antisense,
5′-TGTTTCAACATTCTCCCTCC-3′ for estrogen receptor and sense,
5′-CCCTTCATTGACCTCAACTAC-3′, and antisense,
5′-CCACCTTCTTGATGTCATCAT-3′ for GAPDH. GAPDH was used as an
internal control. The AmpliTaq Gold enzyme (Applied Biosystems,
Foster City, CA, USA) was activated by heating for 10 min at 95°C,
and all genes were amplified by 50 cycles of 15 sec at 95 °C,
followed by 1 min at 60°C.
To normalize for differences in the amount of total
RNA added to each reaction mixture, GAPDH was used as an endogenous
control. The data represent the average expression levels of the
target genes, relative to GAPDH, from three independent
cultures.
Renal cancer tissue collection
The study of human subjects and informed consent
documents were approved by The Human Research Ethics Committee of
The Second Affiliated Hospital of Harbin University (Harbin,
China). From May 7th, 2011 to September 8th, 2012, a total of 102
RCC patients and 40 healthy subjects were recruited at The Second
Affiliated Hospital of Harbin University. Potential confounding and
mediating factors were identified in the associations between
sedentary lifestyles and adiposity, which may be closely associated
with the risk of RCC. During the survey, the body mass index (BMI)
of participants was calculated using the formula: BMI = weight
[kg]/(height [m])2.
The diagnostic requirements were included in the RCC
group. The diagnostic criteria for RCC were used as in a previous
study (19). There are four stages
of RCC (stages I–IV). A number of tissue samples were obtained from
The Second Affiliated Hospital of Harbin University with prior
approval from the Institutional Review Board. Renal cancer or
normal tissue biopsies were collected via a cystoscope and
maintained below −80°C. A total of 142 tissue samples (normal
tissue, n=40; RCC tissue, n=32 for stage I, n=28 for stage II, n=24
for stage III and n=18 for stage IV) were distinguished by a
pathologist experienced in renal cancer at the Department of
Urology, Harbin Medical University.
Western blot analysis
RCC and HRE cells were homogenized in
radioimmunoprecipitation assay buffer (Millipore, Billerica, MA,
USA), consisting of 150 mM sodium chloride, 1% NP-40, 0.5% sodium
deoxycholate, 0.1% SDS, 50 mM Tris-HCl (pH 8.0) and cOmplete Mini
Protease Inhibitor Cocktail (Roche). Once the debris was removed,
the resulting supernatants were boiled and mixed with an equal
volume of 20% glycerol containing 0.02% bromophenol blue (Beijing
F&F Chemical Industrial Co., Ltd., Beijing, China). Proteins
were separated by 12% SDS-PAGE (Beijing JingKeHongDa Biotechnology
Co., Ltd) and transferred onto a polyvinylidene difluoride membrane
(Millipore, Billerica, MA, USA). The membranes were blocked with 5%
skim med milk in Tris-buffered saline with Tween 20 [10 mM Tris (pH
7.5), 100 mM NaCl and 0.1% Tween 20; Dako, Carpinteria, CA, USA]
and incubated with primary antibodies for Smad4, FOXH1, estrogen
receptor, β-actin and phospho-Smad2 (Ser465/467) (138D4), in TBST
with 0.5% skimmed milk overnight at 4°C. The membrane was treated
with a peroxidase-conjugated secondary antibody (1:3,000) (GE
Healthcare, Pittsburgh, PA, USA). Immunoreactive bands were
visualized by enhanced chemiluminescence (RPN2132; GE Healthcare)
in the chamber of the Chemiluminescence Analyzer system CLA-FS4
(TohokuTM Electronic lnc., Miyagi, Japan), and
quantified by densitometry with Image J software version 1.45
(National Institutes of Health, Bethesda, MD, USA).
GST pull-down assay
Bacteria-expressed GST or GST-Smad4 proteins were
immobilized on glutathione-Sepharose 4B beads (GE Healthcare),
washed, and the beads were incubated with FOXH1. The beads were
washed with GST binding buffer (100 mM NaCl, 50 mM NaF, 2 mM EDTA
and 1% Nonidet P40; Beijing JingKeHongDA Biotechnology Co., Ltd,
Beijing, China) and proteins were eluted, followed by western
blotting.
Immunoprecipitation
Cells were harvested and lysed in HEPES lysis buffer
[20 mM HEPES (pH 7.2), 50 mM NaCl, 0.5% Triton X-100, 1 mM NaF, 1
mM dithiothreitol; Beijing JingKeHongDA Biotechnology Co., Ltd].
The lysate was incubated with the antibodies for Myc and GST for 3
h at 4°C, and then Protein A/G-plus Agarose was added. The
immunoprecipitates were washed three times with lysis buffer and
analyzed by western blotting.
Fluorescence microscopy
At 24 h post-transfection, the cells were fixed with
2% paraformaldehyde for 10 min, rinsed with phosphate-buffered
saline (PBS), and permeabilized with 1% Triton X-100 for 10 min.
The cells were then rinsed with PBS and incubated with monoclonal
antibody for 1 h, followed by incubation with secondary antibody
for 1 h. The nuclei of the cells were stained with 0.1 g/ml DAPI
and the cells were observed under a fluorescence microscope
(Eclipse E600; Nikon Corp. Toyko, Japan).
Statistical analysis
Values are presented as the mean ± standard
deviation. Statistical analyses were performed using SPSS 20.0
software (International Business Machines, Armonk, NY, USA) and
variables were compared using the Students t-test. P<0.05
was considered to indicate a statistically significant difference
between values.
Results
Smad4 decreases the growth rate of RCC
cells
The growth rate of the OS-RC-2 RCC cells transfected
with Smad4 was reduced by 20% compared with that of the
non-transfected cell lines (P<0.01) (Fig. 1A). Conversely, in the RCC cells
transfected with siRNA targeting Smad4, the growth rate was
increased by 15% compared with that of the corresponding
non-transfected cell lines (P<0.05) (Fig. 1A). These results indicated that
Smad4 inhibits the proliferation of RCC.
Smad4 expression increases FOXH1
depresses estrogen receptor mRNA levels in RCC cells
Compared with HRE cells, in OS-RC-2 human RCC cells,
the mRNA expression levels of Smad4 were reduced, while the
expression levels of estrogen receptor were enhanced (P<0.01)
(Fig. 1B). Compared with those of
the controls, mRNA levels of Smad4 and FOXH1 were also reduced in
RCC cells; however, the difference was not statistically
significant. When Smad4 was silenced in RCC cells, the mRNA
expression levels of of FOXH1 were slightly reduced and the
expression levels of estrogen receptor were markedly increased
(P<0.01). Conversely, when Smad4 was highly expressed in RCC,
the mRNA expression levels of FOXH1 were markedly increased and the
expression levels of estrogen receptor were markedly reduced
(P<0.01) (Fig. 1B). Smad2 mRNA
levels were the same in RCC and HRE cells and were unaffected by
Smad4 levels (P>0.05).
Smad4 expression increases FOXH1
depresses estrogen receptor protein levels in RCC cells
Compared with HRE cells, in OS-RC-2 human RCC cells,
the protein levels of Smad4 and FOXH1 were reduced, while the
levels of estrogen receptor were significantly enhanced (Fig. 2). The protein levels of
phospho-Smad2 were stable, indicating that the levels of Smad4 did
not affect the degree of phosphorylation of Smad2. When Smad4 was
silenced, the protein levels of Smad2 did not change and the
protein levels of FOXH1 were significantly reduced, while estrogen
receptor levels were markedly increased compared with those of the
HRE cells (P<0.01). Conversely, when Smad4 was highly expressed
in RCC, the protein levels of FOXH1 were significantly increased
and levels of estrogen receptor were markedly reduced compared with
those in the HRE cells (P<0.01). These results suggested that
Smad4 is necessary for the repression of the protein expression of
estrogen receptor.
Risk of RCC is associated with gender,
age, BMI and sedentary lifestyle
A total of 142 subjects (n=40 for normal renal
tissues, 18 females/22 males; n=32 for stage I RCC, 14 females/18
males; n=28 for stage II RCC, 12 females/16 males; n=24 for stage
III RCC, 10 females/14 males; and n=18 for stage IV RCC, 7
females/11 males) (Table I) were
recruited. When considering the potential confounding factors for
increasing the risk of RCC, gender and age were the notable
contributing factors (Table I)
(P<0.05). Older participants and male participants were more
likey to have a higher risk of RCC. The present study found
associations between the BMI and sedentary lifestyles or daily food
intake. The BMI increased with a more sedentary lifestyle
(P<0.01) and was additionally associated with the total daily
calorie intake. A sedentary lifestyle and high daily food intake
were incrementally detrimental to the BMI, which in turn was
associated with the development of RCC.
 | Table ICharacteristics of the patients with
renal cell carcinoma and the healthy controls. |
Table I
Characteristics of the patients with
renal cell carcinoma and the healthy controls.
| | Healthy | Stage I | Stage II | Stage III | Stage IV |
|---|
| Male | Cases (n) | 48 | 29 | 27 | 20 | 13 |
| Age (years) | 47.5±6.8 | 57.3±5.5 | 59.4±9.3 | 57.1±8.0 | 62.3±9.3 |
| BMI
(kg/m2) | 24.7±5.1 | 25.5±8.1 | 26.3±6.6 | 26.9±6.1 | 27.7±7.7 |
| Daily calorie intake
(kcal) | 2248±366 | 2298±388 | 2314±319 | 2345±221 | 2495±332 |
| Sedentary time
(h/day) | 6.5±4.7 | 7.6±5.2 | 8.1±5.7 | 9.0±6.1 | 9.3±6.6 |
| Female | Cases (n) | 32 | 25 | 18 | 15 | 12 |
| Age (years) | 57.9±7.5 | 59.8±6.3 | 60.7±7.6 | 65.1±8.3 | 70.2±8.0 |
| BMI
(kg/m2) | 24.9±5.8 | 26.9±7.8 | 27.8±9.8 | 28.8±9.9 | 29.9±10.1 |
| Daily calorie intake
(kcal) | 1975±283 | 1992±297 | 2012±321 | 2032±351 | 2149±397 |
| Sedentary time
(h/day) | 6.9±5.3 | 8.1±5.8 | 8.7±6.1 | 9.5±5.1 | 10.3±5.6 |
The relative protein levels of
phospho-Smad2, Smad4, FOXH1 and estrogen receptor in RCC and HRE
tissues
Following the investigation of RCC patient
characteristics, the relative protein levels of phospho-Smad2,
Smad4, FOXH1 and estrogen receptor were assessed in RCC and HRE
tissues. Compared with HRE tissues, in RCC tissues, the protein
levels of Smad4 and FOXH1 were reduced, while the levels of
estrogen receptor were enhanced (Fig.
2B). The protein expression levels of FOXH1 were significantly
reduced, while the expression levels of estrogen receptor were
markedly increased (P<0.01) with increasing RCC stage.
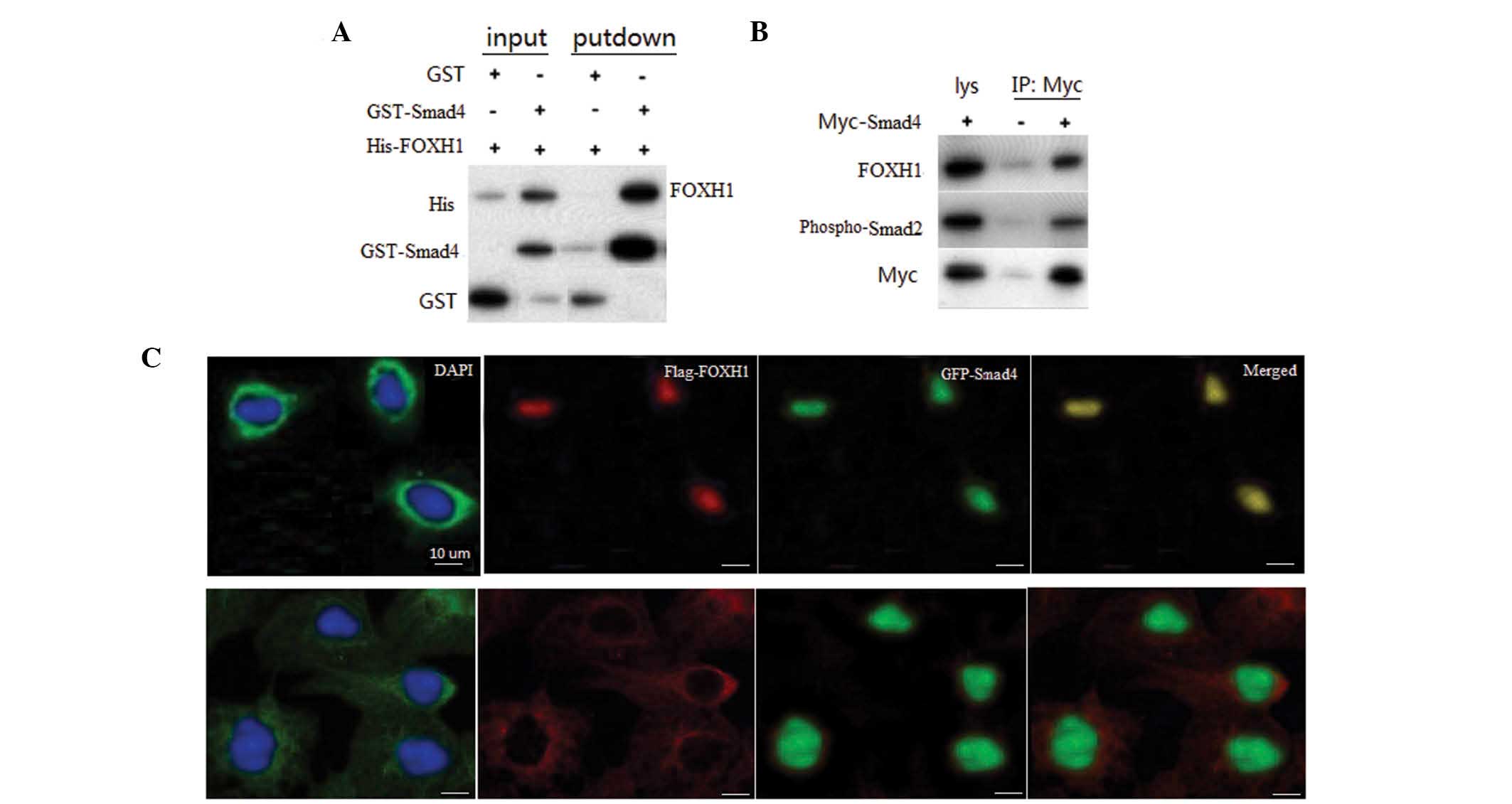
Smad4 interacts with FOXH1
To confirm the interaction between Smad4 and FOXH1,
in vitro GST pull-down assays were performed with
recombinant Smad4 and FOXH1. A specific interaction of Smad4 with
FOXH1 was observed, but not with GST alone (Fig. 3A). To assess whether Smad4
interacts with FOXH1, a co-immunoprecipitation (Co-IP) assay was
performed in primary neuronal cells, and the results revealed an
association between Smad4 and Myc-FOXH1 (Fig. 3B). The interaction between Smad4
and FOXH1 in cultured cells indicated that these two types of
protein may localize in the same subcellular compartment. To assess
the subcellular localization of Smad4 and FOXH1, RCC cells were
cotransfected with GFP-Smad4 and Flag-FOXH1. When coexpressed,
Smad4 and FOXH1 were colocalized in the nuclei of RCC cells
(Fig. 2C, top panel). In normal
renal cells, Smad4 was primarily located in cytoplasm, while FOXH1
was localized in the nuclei.
Smad4 forms a complex with phospho-Smad2 and enters
the nucleus. This complex interacts with FOXH1 and downregulates
the transcriptional activity of estrogen receptor. Therefore, the
downregulation of estrogen receptor is likely to inhibit the
progression of RCC, as estrogen has been reported to induce renal
carcinogenesis in Syrian hamsters (18).
Discussion
Members of the Smad family are able to activate the
transcription of downstream genes by transducing extracellular
signals from TGF-β to the nucleus. There are eight vertebrate
Smads, Smad1–8 (13), all of which
are classified into three major categories: Receptor-regulated
Smads (R-Smads), which include Smad1-3, −5 and −8/9 (20); Smad4, the only common-mediator Smad
(co-Smad), which interacts with R-Smads to affect signaling
(21); and Smad6 and −7, the
inhibitory Smads (I-Smad), which repress the activities of R-Smads
and co-Smads (22). Thus, as Smad4
is a separate type of Smad, its function is different from that of
the other Smads. Smad4 interacts with phospho-Smad2 or −3 to form
an R-Smad/Smad4 complex (23).
Although Smad4 has been reported to interact with phospho-Smad2
(24), the present study found
that the phosphorylation of Smad2 was not affected by the protein
levels of Smad4.
FOXH1 is a transcription factor that mediates
signaling by TGF-β, activin and nodal (24). The biological roles of FOXH1 are
diverse. The nodal-FOXH1 signaling pathway has a central role in
the anterior-posterior patterning and node formation in mice
(25). In addition, FOXH1 has been
identified as an androgen receptor repressor (26). FOXH1 does not contain a
transcriptional activation domain and requires Smad interaction for
transcriptional regulation (16).
The formation of the CFSS is necessary for the repression of the
transcriptional activities of estrogen receptors.
A number of potential treatments have been explored
for inhibiting the progression of RCC. For example, sorafenib has
been shown to reduce the risk of RCC-associated mortality; however,
the benefit was not statistically significant according to the
O’Brien-Fleming threshold (27).
Furthermore, sorafenib has a number of adverse side effects, which
commonly include diarrhea, rashes, fatigue and hand-foot skin
reactions. Hypertension and cardiac ischemia are rare serious
side-effects that were more common in patients receiving sorafenib
(27). Hence, it is necessary to
search for novel therapies with few adverse side-effects. Smad4 is
present in the human body and has few side-effects. Thus, Smad4 may
be developed as a potential drug for the treatment of RCC. In
addition, the present study found that a high daily food intake and
sedentary lifestyles were factors that contributed to an increased
risk of RCC. Thus, daily physical exercise and a low calorie intake
are also important to reduce the risk of RCC.
References
|
1
|
Sheehan JP, Sun MH, Kondziolka D,
Flickinger J and Lunsford LD: Radiosurgery in patients with renal
cell carcinoma metastasis to the brain: long-term outcomes and
prognostic factors influencing survival and local tumor control. J
Neurosurg. 98:342–349. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ljungberg B, Hanbury DC, Kuczyk MA, et al;
European Association of Urology Guideline Group for renal cell
carcinoma. Renal cell carcinoma guideline. Eur Urol. 51:1502–1510.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Linehan WM: The genetic basis of kidney
cancer: implications for management and use of targeted therapeutic
approaches. Eur Urol. 61:896–898. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jacobs JF, Nierkens S, Figdor CG, de Vries
IJM and Adema GJ: Regulatory T cells in melanoma: the final hurdle
towards effective immunotherapy? Lancet Oncol. 13:e32–e42. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
de Jesus-Gonzalez N, Robinson E, Moslehi J
and Humphreys BD: Management of antiangiogenic therapy-induced
hypertension. Hypertension. 60:607–615. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhu Y, Xu L, Zhang J, et al: Klotho
suppresses tumor progression via inhibiting PI3K/Akt/GSK3β/Snail
signaling in renal cell carcinoma. Cancer Sci. 104:663–671. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kim BG, Li C, Qiao W, et al: Smad4
signalling in T cells is required for suppression of
gastrointestinal cancer. Nature. 441:1015–1019. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang LH, Kim SH, Lee JH, et al:
Inactivation of SMAD4 tumor suppressor gene during gastric
carcinoma progression. Clin Cancer Res. 13:102–110. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Müller N, Reinacher-Schick A, Baldus S, et
al: Smad4 induces the tumor suppressor E-cadherin and P-cadherin in
colon carcinoma cells. Oncogene. 21:6049–6058. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Losi L, Bouzourene H and Benhattar J: Loss
of Smad4 expression predicts liver metastasis in human colorectal
cancer. Oncol Rep. 17:1095–1099. 2007.PubMed/NCBI
|
|
11
|
Alazzouzi H, Alhopuro P, Salovaara R, et
al: SMAD4 as a prognostic marker in colorectal cancer. Clin Cancer
Res. 11:2606–2611. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Goto Y, Manabe N, Uchio-Yamada K, et al:
Augmented cytoplasmic Smad4 induces acceleration of TGF-beta1
signaling in renal tubulointerstitial cells of hereditary nephrotic
ICGN mice with chronic renal fibrosis; possible role for
myofibroblastic differentiation. Cell Tissue Res. 315:209–221.
2004. View Article : Google Scholar
|
|
13
|
Derynck R and Zhang YE: Smad-dependent and
Smad-independent pathways in TGF-beta family signalling. Nature.
425:577–584. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
ten Dijke P and Hill CS: New insights into
TGF-β-Smad signalling. Trends Biochem Sci. 29:265–273. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hoodless PA, Tsukazaki T, Nishimatsu S,
Attisano L, Wrana JL and Thomsen GH: Dominant-negative Smad2
mutants inhibit activin/Vg1 signaling and disrupt axis formation in
Xenopus. Dev Biol. 207:364–379. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Attisano L and Wrana JL: Smads as
transcriptional co-modulators. Curr Opin Cell Biol. 12:235–243.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yum J, Jeong HM, Kim S, et al:
PKA-mediated stabilization of FoxH1 negatively regulates ERalpha
activity. Mol Cells. 28:67–71. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bhat HK, Hacker HJ, Bannasch P, Thompson
EA and Liehr JG: Localization of estrogen receptors in interstitial
cells of hamster kidney and in estradiol-induced renal tumors as
evidence of the mesenchymal origin of this neoplasm. Cancer Res.
53:5447–5451. 1993.PubMed/NCBI
|
|
19
|
Tsui K-H, Shvarts O, Smith Rb, Figlin Ra,
de Kernion J and Belldegrun A: Prognostic indicators for renal cell
carcinoma: a multivariate analysis of 643 patients using the
revised 1997 TNM staging criteria. J Urol. 163:1090–1095. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wu J-W, Hu M, Chai J, et al: Crystal
structure of a phosphorylated Smad2: Recognition of phosphoserine
by the MH2 domain and insights on Smad function in TGF-beta
signaling. Mol Cell. 8:1277–1289. 2001. View Article : Google Scholar
|
|
21
|
Shi Y, Hata A, Lo RS, Massagué J and
Pavletich NP: A structural basis for mutational inactivation of the
tumour suppressor Smad4. Nature. 388:87–93. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Itoh F, Asao H, Sugamura K, Heldin C-H,
ten Dijke P and Itoh S: Promoting bone morphogenetic protein
signaling through negative regulation of inhibitory Smads. EMBO J.
20:4132–4142. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ju W, Ogawa A, Heyer J, et al: Deletion of
Smad2 in mouse liver reveals novel functions in hepatocyte growth
and differentiation. Mol Cell Biol. 26:654–667. 2006. View Article : Google Scholar :
|
|
24
|
Yao Z, Fenoglio S, Gao DC, et al: TGF-beta
IL-6 axis mediates selective and adaptive mechanisms of resistance
to molecular targeted therapy in lung cancer. Proc Natl Acad Sci
USA. 107:15535–15540. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yamamoto M, Meno C, Sakai Y, et al: The
transcription factor FoxH1 (FAST) mediates Nodal signaling during
anterior-posterior patterning and node formation in the mouse.
Genes Dev. 15:1242–1256. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen G, Nomura M, Morinaga H, et al:
Modulation of androgen receptor transactivation by FoxH1 a newly
identified androgen receptor corepressor. J Biol Chem.
280:36355–36363. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Escudier B, Eisen T, Stadler WM, et al:
Sorafenib in advanced clear-cell renal-cell carcinoma. N Engl J
Med. 356:125–134. 2007. View Article : Google Scholar : PubMed/NCBI
|