Introduction
Oral squamous cell carcinoma (OSCC) is the most
common malignancy of the oral and maxillofacial region, and the
sixth most common type of tumor worldwide (1). Although surgical treatment,
radiotherapy and chemotherapy have been regarded as mature
technologies, the prognosis of patients with OSCC remains poor and
the overall 5-year survival rates have remained at 50% (2). Therefore, further investigation into
the pathogenesis of OSCC continues to be important.
The majority of human cancer types are characterized
by genetic instabilities. Mitochondrial DNA (mtDNA) is the only
genetic material of the human genome, with the exception of nuclear
DNA (3–5). mtDNA somatic mutations have been
increasingly observed in human cancer, such as stomach, liver and
lung cancer (6–8), and have been proposed as important
oncological biomarkers. However, the biological significance of
mtDNA mutations in OSCC remains to be elucidated. The
displacement-loop (D-loop) region, the only non-coding region in
mtDNA, is considered to be important as it is the major control
site for mtDNA expression and it is also involved in mtDNA
replication. Mutations occur throughout the mitochondrial genome in
tumors, but are most frequently detected in the D-loop region
(6,7).
In the present study, gene mutations in the D-loop
region of mtDNA were investigated in thirty patients with OSCC in
order to examine the role of gene mutations of mtDNA in OSCC
tumorigenesis.
Materials and methods
Patients and samples
Tumor samples were obtained from resected specimens
of 30 patients with primary OSCC in the Department of Oral and
Maxillofacial Surgery, Affiliated Hospital Medical College Qingdao
University (Qingdao, China) between March, 2009 and June, 2010. The
study was approved by the ethics committee of the Affiliated
Hospital of Qingdao University (Qingdao, China). Written informed
consent was obtained from all patients or their families. Fresh
cancer tissues, paracancerous tissues and normal mucosal tissues
from the same patient were obtained and immediately put into 1.5 ml
frozen pipes for preservation in −196°C liquid nitrogen. The
patients consisted of 21 males and nine females, and ranged in age
between 29 and 72 years old (mean age, 56.5 years). The tumor sites
included 11 tongue cancers, six gingival cancers, four floor of the
mouth cancers, four soft palate cancers, two buccal mucosa cancers,
two oropharyngeal cancers and one lip cancer.
Extraction of mtDNA
A total of 30 mg tissue of the sample was extracted
as required and ground into a fine powder, then mtDNA was extracted
using an mtDNA extraction kit (Shanghai Genmed Pharmaceutical
Scientific Company, Shanghai, China). The purity and content of the
mtDNA were measured using an ultra trace spectrophotometer
(NanoDrop ND-2000C; Thermo Fisher Scientific, Waltham, MA, USA).
The value required was A280/A260>1.80, which meant 20 μl
(2 ng/μl) mtDNA was obtained.
Polymerase chain reaction (PCR)
amplification
The PCR amplification primer sequences were as
follows: P1, 5′-TGATGTGAGCCCGTCTAAAC-3′; P 2, 5′- GAA
TCGGAGGACAACCAGTA-3′; P3: 5′-TATCCCGCACAAGAGTGCTACTC-3′ and P4:
5′-CTCCAGCGTCTCGCAATGCTA-3′ which resulted in an amplification
product of 1,450 bp, including the D-loop region. Amplification was
accomplished with a 50 μl PCR reaction using 25 μl of
Power Taq PCR MasterMix polymerase (DR100A; Takara Bio, Inc., Otsu,
Japan) and 3 μl template DNA (including 15 μl of
upstream and 1.5 μl of downstream primers supplied by
Shanghai Sangon Biological Engineering Co., Ltd, Shanghai, China).
The PCR conditions were as follows: 94°C for 5 min, 35 cycles of
94°C for 1 min, 55°C for 1 min, 72°C for 2 min and then an
extension step at 72°C for 7 min followed by holding at 4°C. The
specificity of the reactions was confirmed using agarose gel
electrophoresis. The concentration of each purified PCR product was
measured using a Tanon 2500 gel imaging analysis system (Tanon
Science and Technology Co., Ltd., Shanghai, China).
Gene sequencing and analysis
All PCR products were purified and sequenced by
Shanghai Maipu Biotechnology Company according to the
manufacturer’s instructions. There were four primers as shown in
Table I. Chromas software
(2.3.0.0; Technelysium, Brisbane, Australia) and BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi)
were used to analyze the sequencing and to search mutations. The
sequences of D-loop region mtDNA in the present study were compared
with the Cambridge Standard sequence in GenBank (http://www.ncbi.nlm.nih.gov/genbank/).
If the sequences in normal tissue, adjacent tissue and cancer
tissue were the identical, but differed to the Cambridge Standard
sequence provided in GenBank, they were identified as single
nucleotide polymorphisms. Mutations were identified if the sequence
in normal tissue was identical to that of adjacent tissue, but
differed to that of cancer tissue.
 | Table ISequences of primers. |
Table I
Sequences of primers.
| Primer | Sequence |
|---|
| P1 |
5′-TGATGTGAGCCCGTCTAAAC-3′ |
| P2 |
5′-GAATCGGAGGACAACCAGTA-3′ |
| P3 |
5′-TATCCCGCACAAGAGTGCTACTC-3′ |
| P4 |
5′-CTCCAGCGTCTCGCAATGCTA-3′ |
Statistical analysis
Statistical analysis using the χ2 test
and identification of significant mutations were performed using
SPSS 16.0 software (SPSS, Inc., Chicago, IL, USA).
Results
PCR amplification and sequencing
All PCR amplification products of the thirty cases
with OSCC exhibited a 1,450-bp fragment of the mtDNA D-loop region,
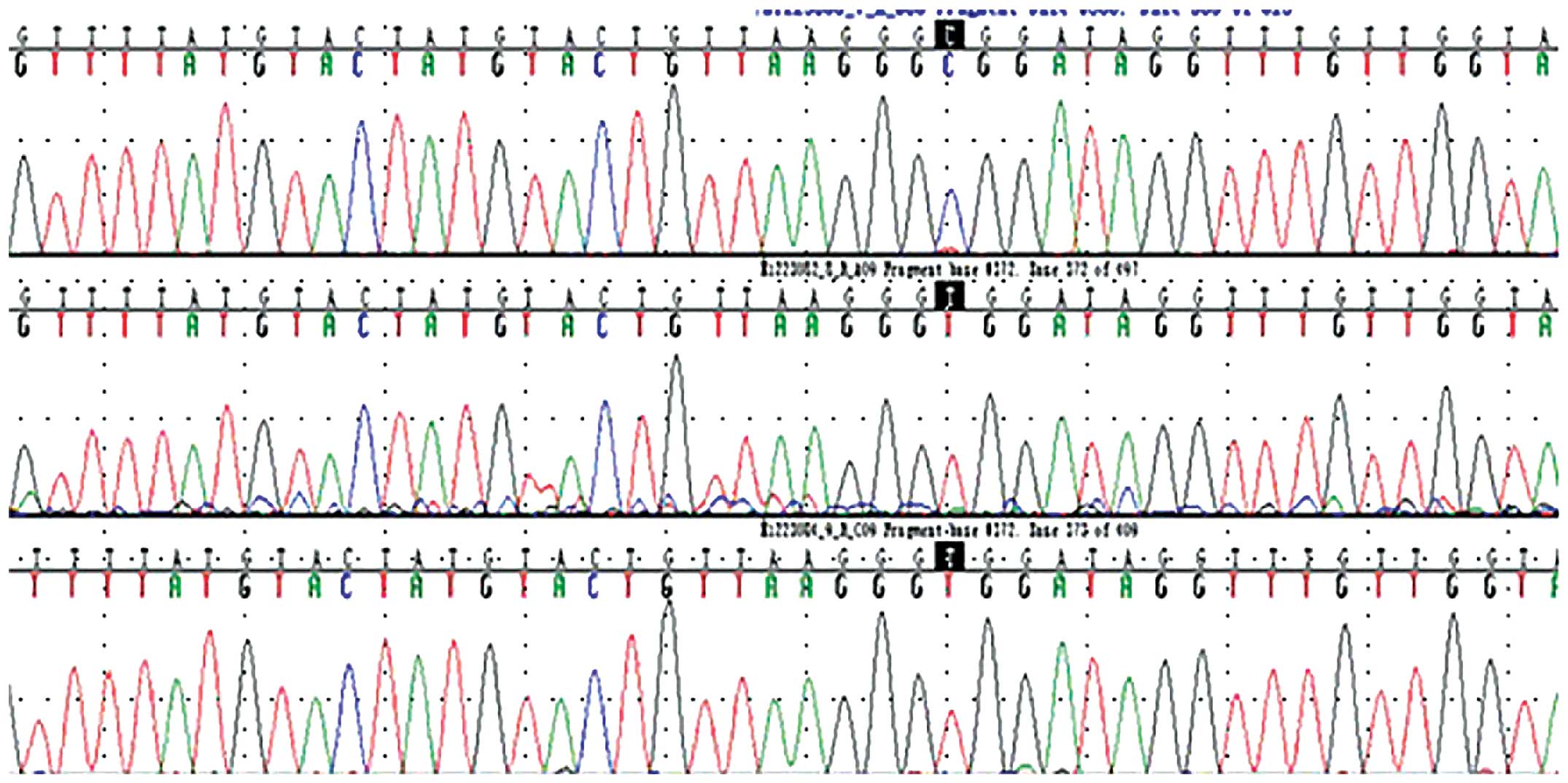
(Fig. 1). The sequencing results
demonstrated clear peaks (Fig. 2).
The first line presents the cancer tissue sequence, the second
presents the adjacent tissue sequence and the third presents normal
tissue. Black markers indicate the mutation in cancer tissue, which
was identified as blue peak C differed to red peak T at the same
position in the adjacent and normal tissues.
Analysis of sequencing
There were multiple single nucleotides polymorphisms
in each case of OSCC when compared with the mtDNA Cambridge
sequence in GenBank (http://www.ncbi.nlm.nih.gov/genbank/) as shown in
Fig. 3. Mutations in the D-loop
region were identified in the cancer tissue samples of 8/30 cases
with OSCC, with a mutation rate of 27%. There were nine mutations
in total, including one point mutation, two base deletions, three
insertion mutations and three heterozygous mutations. In these
mutations, base deletions were different from each other and
heterozygous mutations did not have the same mutation form;
however, the three insertion mutations were the same, consisting of
an insertion of a C base. One case contained a T/A heterozygous
mutation as well as a base insertion of C (Table II).
 | Table IIMutations in the displacement-loop
region of mitochondrial DNA in oral squamous cell carcinoma. |
Table II
Mutations in the displacement-loop
region of mitochondrial DNA in oral squamous cell carcinoma.
| Sample no. | Age | Gender | Pathology | Mutation site | Seq. normal | Seq. adjacent | Sequence of cancer
tissue |
|---|
| 1 | 57 | Male | Well-differentiated
SCC | 249 | A | A | Deletion mutants
A |
| 2 | 67 | Male |
Moderately-differentiated SCC | 16289 | T | T | Replacement mutation
C |
| 3 | 29 | Female | Well-differentiated
SCC | 519 | T,G | T,G | Deletion mutants
T,G |
| 4 | 72 | Male | Well-differentiated
SCC | 313 | None | None | Insertional
mutagenesis C |
| 5 | 67 | Male |
Moderately-differentiated SCC | 16088 | G | G | Heterozygous mutation
G,A |
| 6 | 61 | Male |
Moderately-differentiated SCC | 313 | None | None | Insertional
mutagenesis C |
| 7 | 55 | Male | Well-differentiated
SCC | 16263 | C | C | Heterozygous mutation
C,T |
| 8 | 31 | Male | Well-differentiated
SCC | 313
16522 | None T | None T | Insertional
mutagenesis C, Heterozygous mutation T,A |
Clinical significance
The eight cases with a mutation in the D-loop region
consisted of three tongue cancers, two soft palate cancers, one
floor of the mouth cancer, one oropharyngeal cancer and one lip
cancer.
Discussion
mtDNA is the only genetic material of the human
genome not contained in the nucleus. It is present in a closed
double-stranded state, with a total length of 16,569 bp, containing
37 genes, including the structural genes encoding the thirteen
protein subunits, 12S rRNA, 16S rRNA and 2S tRNA, that make up the
oxidative phosphorylation system. The D-loop region, the only
non-coding region in mtDNA, is located at basepairs 1,602-577,
accounting for 6% of the total mtDNA. The D-loop region is the
major control site in the regulation of mtDNA transcription and is
also involved in mtDNA replication (3–5). In
recent years, somatic mutations in the mtDNA have been increasingly
observed in human cancers, such as stomach (6), liver (7), lung (8), ovarian (9), breast (10) and pancreatic cancer (11). Mutations occur throughout the
mitochondrial genome in tumors, but are most frequently detected in
the D-loop region. At present, studies investigating the mtDNA
mutations in head and neck tumors remain rare.
In our previous small sample study, it was
demonstrated that there were mutation sites in the D-loop region of
mtDNA in three out of seven OSCC cases and numerous single
nucleotide polymorphisms in the D-loop region in all cases. In the
present study, a larger sample study, the D-loop region of mtDNA in
thirty patients with OSCC was screened and the rate of gene
mutation in the D-loop region was 27%, which was similar to other
studies in which the mutation rates in human cancers were 20–78%
(12–14).
Excluding single nucleotide polymorphisms, nine
mutation sites were found in the D-loop region of mtDNA in the
present study and insertional mutagenesis C at site 313 was also
observed, which was similar to that identified in liver cancer and
colorectal cancer (13–14). Site 313 mutations may affect the
regulation of mtDNA transcription and may be an important area of
mtDNA investigation in the future.
Bragoszewski et al (15) investigated mtDNA in ovarian cancer
and identified heterozygous mutations, such as C/T replacement at
site 16,193 and site 16,218 and G/A replacement at site 16,391,
which were seldom reported in the literature. In the present study,
there were similar mutations in the D-loop region of mtDNA which,
as a regulatory area, may be able to reduce mtDNA replication and
ND6 transcription. It was hypothesized that these heterozygous
mutations were rare mutation sites and require further
investigation.
Certain gene mutation sites in mtDNA, which are
closely associated with tumorigenesis and progression, have already
been identified (16). However,
the functional significance of mutations in the D-loop region of
mtDNA and its role in tumorigenesis remain to be elucidated and
require further investigation at the protein level and in larger
samples. These mutations may in the future provide novel markers
for early clinical diagnosis and novel targets for
chemotherapy.
Acknowledgments
The present study was supported by a grant from
Shanghai Science and Technology Committee (grant nos. 08DZ2271100
and S30206-kf13).
References
|
1
|
Ng SH, Yen TC, Liao CT, Chang JT, Chan SC,
Ko SF, Wang HM and Wong HF: 18F-FDG PET and CT/MRI in oral cavity
squamous cell carcinoma: a prospective study of 124 patients with
histologic correlation. J Nucl Med. 46:1136–1143. 2005.PubMed/NCBI
|
|
2
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Penta JS, Johnson FM, Wachsman JT and
Copeland WC: Mitochondrial DNA in human malignancy. Mutat Res.
488:119–133. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
McFarland R, Taylor RW and Turnbull DM:
Mitochondrial disease - its impact, etiology and pathology. Curr
Top Dev Biol. 77:113–155. 2007. View Article : Google Scholar
|
|
5
|
Stewart JB, Freyer C, Elson JL and Larsson
NG: Purifying selection of mtDNA and its implications for
understanding evolution and mitochondrial disease. Nat Rev Genet.
9:657–662. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hung WY, Wu CW, Yin PH, Chang CJ, Li AF,
Chi CW, Wei YH and Lee HC: Somatic mutations in mitochondrial
genome and their potential roles in the progression of human
gastric cancer. Biochim Biophys Acta. 1800:264–270. 2010.
View Article : Google Scholar
|
|
7
|
Wang C, Zhang F, Fan H, Peng L, Zhang R,
Liu S and Guo Z: Sequence polymorphisms of mitochondrial D-loop and
hepatocellular carcinoma outcome. Biochem Biophys Res Commun.
406:493–496. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ding C, Li R, Wang P, Jin P, Li S and Guo
Z: Identification of sequence polymorphisms in the D-loop region of
mitochondrial DNA as a risk factor for lung cancer. Mitochondrial
DNA. 23:251–254. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Guerra F, Kurelac I, Magini P, Cormio A,
Santini D, Ceccarelli C and Gasparre G: Mitochondrial DNA
genotyping reveals synchronous nature of simultaneously detected
endometrial and ovarian cancers. Gynecol Oncol. 122:457–458. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cai FF, Kohler C, Zhang B, Chen WJ,
Barekati Z, Garritsen HS, Lenner P, Toniolo P, Zhang JJ and Zhong
XY: Mutations of mitochondrial DNA as potential biomarkers in
breast cancer. Anticancer Res. 31:4267–4271. 2011.PubMed/NCBI
|
|
11
|
Lam ET, Bracci PM, Holly EA, Chu C, Poon
A, Wan E, White K, Kwok PY, Pawlikowska L and Tranah GJ:
Mitochondrial DNA sequence variation and risk of pancreatic cancer.
Cancer Res. 72:686–695. 2012. View Article : Google Scholar :
|
|
12
|
Czarnecka AM, Gammazza AM, Di Felice V,
Zummo G and Cappello F: Cancer as a ‘Mitochondriopathy’. J Cancer
Mol. 3:71–79. 2007.
|
|
13
|
Wong LJ, Tan DJ, Bai RK, Yehbackground KT
and Chang J: Molecular alterations in mitochondrial DNA of
hepatocellular carcinomas: is there a correlation with
clinicopathological profile? J Med Genet. 41:e652004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Boland CR, Thibodeau SN, Hamilton SR,
Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA,
Fodde R, Ranzani GN and Srivastava S: A national cancer institute
workshop on microsatellite instability for cancer detection and
familial predisposition: development of international criteria for
the determination of microsatellite instability in colorectal
cancer. Cancer Res. 58:5248–5257. 1998.PubMed/NCBI
|
|
15
|
Bragoszewski P, Kupryjanczyk J, Bartnik E,
Rachinger A and Ostrowski J: Limited clinical relevance of
mitochondrial DNA mutation and gene expression analyses in ovarian
cancer. BMC Cancer. 8:2922008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Brandon M, Baldi P and Wallace DC:
Mitochondrial mutations in cancer. Oncogene. 25:4647–4662. 2006.
View Article : Google Scholar : PubMed/NCBI
|