Introduction
More than 90% of the genome is transcribed into
non-protein-coding RNAs, including micro- (mi), short interfering
(si) and small nuclear (sn) RNAs. Another class of non-coding (nc)
RNA, that does not fit into any of these categories, is >200 nt
in length and has been defined as long ncRNA (lncRNA) (1). Recently, lncRNAs have gained
widespread attention due to their notable regulatory roles in
various pathophysiological changes, including differentiation,
proliferation and apoptosis, as well as disease states such as
hepatocellular carcinoma and tuberculosis (2,3).
Thousands of lncRNA transcripts have been identified in humans,
mice and pigs (2,3). Previous studies have indicated that
lncRNAs are highly associated with cardiovascular disease,
particularly with regard to cardiac development and myocardial
fibrosis (2,3). In addition, certain studies have
employed the hypertensive rat model; however, the role of lncRNAs
in this type of model remains to be elucidated (4,5).
Hypertension is a cardiovascular disease associated
with high morbidity and is increasing in prevalence worldwide
(6). Multiple factors are
implicated in the pathogenesis of high blood pressure, including
the renin-angiotensin-aldosterone system, vascular smooth muscle
and vascular endothelial dysfunction, as well as impaired platelet
function and kidney-related factors. To further investigate
renal-associated hypertension, and taking advantage of recent
developments in microarray technology, the present study employed
lncRNA microarray to detect and compare differentially expressed
ncRNAs in the renal cortex of spontaneously hypertensive rats
(SHRs) and normotensive Wistar-Kyoto (WKY) rats.
Materials and methods
Tissue collection and RNA extraction
SHRs demonstrating increased blood pressure levels
at the age of 5–6 weeks, were first obtained by Okamoto and
colleagues by inbreeding WKY rats with high blood pressure
(7,8). Twelve-week-old SHR and WKY rats
(weight, 250–330 g) were obtained from the Experimental Animal
Research Center (Zhejiang Chinese Medical University, Hangzhou,
China) and maintained at 18–29°C, 40–70% humidity, in a 12/12 h
light/dark cycle, 3–5 rats/cage. Blood pressure levels were
monitored using tail cuffs until the rats were 15-weeks-old
(ALCOTT-automated non-invasive blood pressure rat sphygmomanometer;
Shanghai Alcott Biotech Co., Ltd., Shanghai, China). All rats were
anesthetized with an injection of pentobarbital sodium (25 mg/kg)
(Sigma-Aldrich, St. Louis, MO, USA) and perfusion with normal
saline prior to the removal of the renal cortex and the heart. All
surgical instruments, as well as the operating table, were
sterilized using high temperature and high pressure. To remove the
renal cortex, an abdominal incision was made, the intestines moved
aside and the kidney removed. The kidney was bisected to expose the
junction of the medulla and cortex, and the cortex was removed. All
of the specimens were flash-frozen in liquid nitrogen and stored in
freezing tubes (cat no. 430661; Corning Life Sciences, Corning, NY,
USA) at −80°C until use. Three pairs of rats with marked
differences in blood pressure were selected as the hypertension and
control groups. These were selected, from five pairs of rats with
marked differences in blood pressure, following total RNA
extraction, as the three selected pairs of samples exhibited the
greatest density and purification of RNA. The animal study was
approved by the animal welfare committee of Wenzhou Medical
University (Wenzhou, China), according to state and institutional
regulations.
Total RNA was extracted from the ipsilateral renal
cortex (left side) of SHR and WKY rats with TRIzol (Invitrogen Life
Technologies, Carlsbad, CA, USA) according to the manufacturer's
instructions. The purity and yield of the RNA was evaluated by the
ratio of absorbance at A260−A280 with a
NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Inc.,
Wilmington, DE, USA) and 2% agarose gel electrophoresis (120V, 15
min) using 2% agarose (Biowest, Hong Kong, China), a gel imaging
and analysis system (Bio-Rad, Hercules, CA, USA), and an
electrophoresis system (Bio-Rad).
lncRNA and mRNA microarray expression
profiling and quantitative reverse transcription-polymerase chain
reaction (RT-qPCR) validation
The microarray contained ~10,000 rat lncRNAs that
were derived from authoritative databases, including RefSeq
(http://www.ncbi.nlm.nih.gov/refseq/),
Ensemble and Ultra-conserved Region Encoding LncRNA (https://users.soe.ucsc.edu/~jill/ultra.html), lncRNAdb
(http://www.lncrnadb.org/), ncRNA (http://www.ncrna.org/), and sequencing data from the
Beijing Aerospace Control Center, which contained 30,367 mRNA
probes. Total RNA was reverse transcribed to cDNA using a RevertAid
First Strand cDNA Synthesis kit (Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) with random primers. Following fragmentation, the
samples were applied to a custom rat lncRNA microarray, using a
LowInput Quick-Amp Labeling kit, Gene Expression Wash kit, Gene
Expression Hybridization kit and a RNA Spike-In kit (Agilent
Technologies, Inc., Santa Clara, CA, USA). The cDNA fragments were
hybridized with the lncRNA microarray chip at 65°C for 17 h
according to the manufacturer's instructions. Slides were scanned
with the Agilent Microarray Scanner G2505C (Agilent Technologies,
Inc.). The data extraction was performed following selection of the
appropriate data to export to text, using Feature Extraction
software, version 11.0.1.1 (Agilent Technologies, Inc.).
In addition, Feature Extraction software (version
11.0.1.1) was used to analyze the acquired array images of the
results. Quantile normalization and subsequent data processing were
performed using the GeneSpring GX v12.0 software package (Agilent
Technologies, Inc.). Microarray profiling and analysis was
conducted by the OE Biotechnology Co. (Shanghai, China).
Six differentially expressed lncRNAs of interest
were selected for validation. RT-qPCR was performed using a
LightCycler® 480 Instrument II (Roche Diagnostics,
Basel, Switzerland) with 10 ml PCR reaction mixture (1 ml cDNA, 5
ml 2X LightCycler® 480 SYBR Green I Master mix (Roche
Diagnostics), 0.2 ml forward primer, 0.2 ml reverse primer and 3.6
ml nuclease-free water). Reactions were incubated in a 384-well
optical plate (Roche Diagnostics) at 95°C for 10 min, followed by
40 cycles of 95°C for 10 sec and 60°C for 30 sec. Each sample was
run in triplicate for analysis. Following the PCR cycles, melting
curve analysis was performed specifically to validate the
generation of the expected PCR product. The expression levels of
sequences were normalized to GAPDH and were calculated according to
the 2−ΔΔCt methodology (9). The primer sequences were designed in
the laboratory using Primer 6.0 and Oligo 5.0 and synthesized by
Generay Biotech (Shanghai) Co., Ltd. (Shanghai, China) based on the
ncRNA sequences obtained from the National Center for Biotechnology
Information database (http://www.ncbi.nlm.nih.gov/nucleotide/), as presented
in Table I.
 | Table IPrimer sequences. |
Table I
Primer sequences.
| Gene symbol | Primer sequence
(5′-3′) |
|---|
| XR_005438. 2 |
TGACTGTAGCTTCACAGGAAT |
|
TCCAGGACAGTTCAGGAT |
| TCONS_00016231 |
CAAAGTACCTCACCTTACCAG |
|
ACTTCCATGACTCTAGCCT |
| TCONS_00089902 |
TTATCGGGAGAGGCTCAAC |
|
GCTACATTGGATCATCTTGTCA |
| NR_038078. 1 |
CTGCTTTGTAAAAGCAAAGGT |
|
GCCAAAGACAGTACTAACAAC |
| XR_086023. 2 |
TGAGGTGAGGACCACTAGGAAGAC |
|
AGGTGCCATCGGAGGAATCATCT |
| Uc344+ |
CCAATATCCCTGCCTATAACA |
|
TAAGTCCAATCCGCCGTA |
Gene ontology (GO) analysis, and
functional predictions of mRNAs and lncRNAs
GO enrichment analysis was used to make preliminary
predictions of the biological functions of the sequences of
interest. GO is a common method for gene annotation and
predominantly includes three fields: Biological processes, cellular
components and molecular functions. Kyoto Encyclopedia of Genes and
Genomes (KEGG) pathway analysis (http://www.kegg.jp/kegg/docs/statistics.html) is
another synthetic pathway prediction tool that contains >200
pathways, and a collection of pathway maps representing information
based on the molecular interaction and reaction networks for
sequences (10).
The final gene list was submitted to the FunNet
database (http://www.funnet.ws/) for in-depth
analysis of the mRNA or associated lncRNA in the microarray. FunNet
is described as an exploratory tool that performs relevance-based
searching in transcriptional coexpression networks; the information
in FunNet was extracted from genomic databases, converted to a
comprehensive exploratory framework and then updated to the website
(11). The results provide values
to describe the association between the submitted genes and the
terms in the database.
Results
lncRNAs are differentially expressed
between SHRs and WKY rats
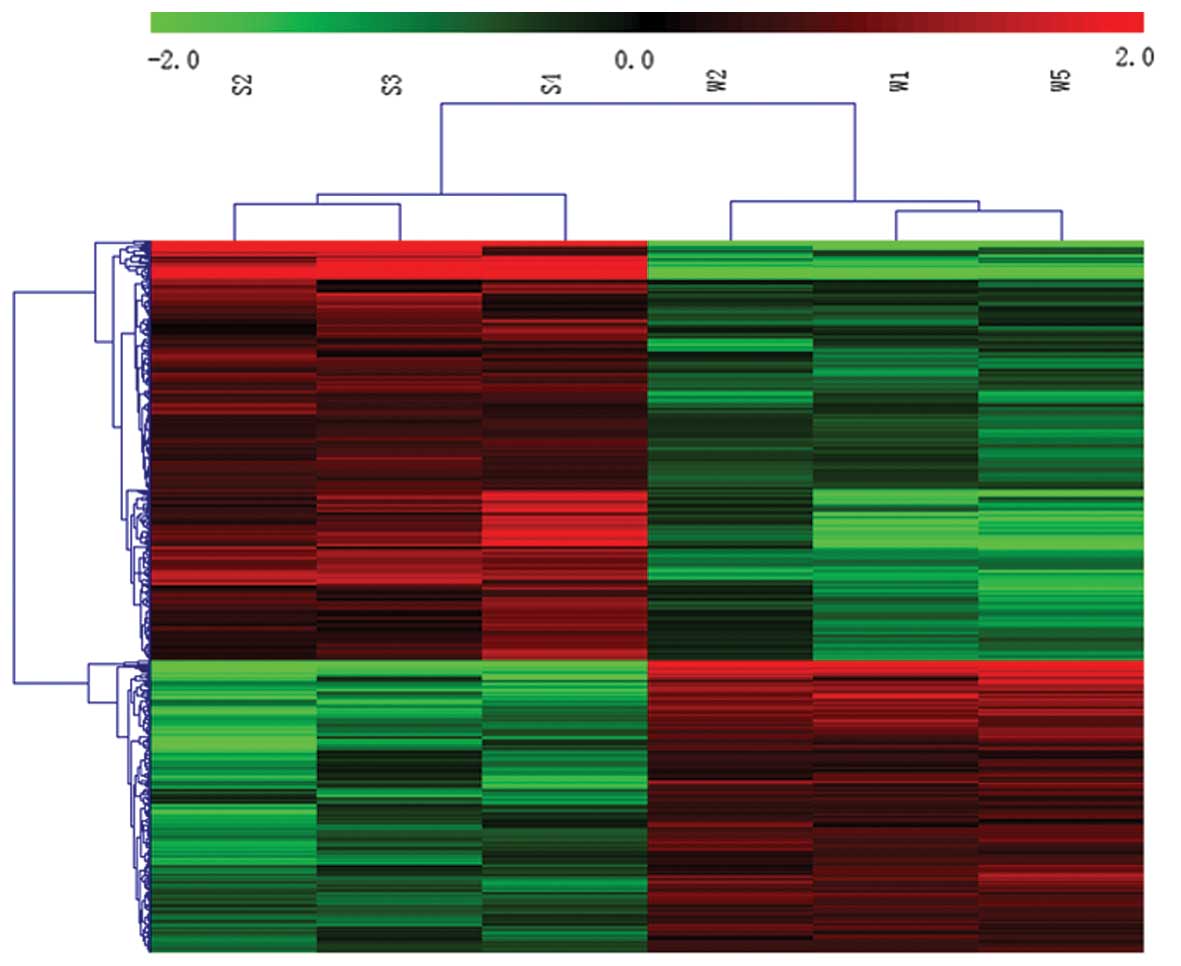
Microarray analysis indicated that 145 of 9,991
ncRNAs were differentially expressed in SHRs compared with WKY
rats; of these, 93 ncRNAs were upregulated and 52 were
downregulated (≥2-fold; Fig. 1).
The included sequences were demonstrated to have functions that
predominantly begin with 'NR', such as NR_038078.1, also referred
to as small nucleolar RNA host gene 4 (SNHG4), a gene which may be
associated with the bystander effect in radiation biology (12). The included sequences were also
functionally associated with miscellaneous RNAs, pseudogenes,
ultraconserved elements and unclassified sequences (partial data
are listed in Table II). Using
the same criteria, 383 differentially coexpressed messenger RNAs
were identified.
 | Table IIPartial differentially expressed
lncRNAs (abs ≥ 3-fold) between SHRs and control WKY rats. |
Table II
Partial differentially expressed
lncRNAs (abs ≥ 3-fold) between SHRs and control WKY rats.
| Target_id | Chr | Regulation | FC (abs) |
|---|
| TCONS_00016233 | 1 | Up | 3.03 |
| TCONS_00054808 | 16 | Up | 3.11 |
| XR_085734. 1 | | Up | 3.12 |
| XR_086159. 2 | | Up | 3.13 |
| TCONS_00054032 | 16 | Up | 3.17 |
| TCONS_00110995 | 6 | Up | 3.23 |
| TCONS_00045352 | 14 | Up | 3.25 |
| TCONS_00108373 | 6 | Down | 3.26 |
| TCONS_00058983 | 17 | Up | 3.26 |
| uc. 107+ | 3 | Down | 3.30 |
| NR_038078. 1
(Snhg4) | 18 | Down | 3.35 |
| XR_086329. 2 | | Up | 3.35 |
| TCONS_00005805 | 1 | Up | 3.39 |
| TCONS_00045737 | 14 | Down | 3.39 |
| TCONS_00089821 | 3 | Down | 3.41 |
| TCONS_00066490 | 2 | Up | 3.45 |
| XR_146926. 1 | | Down | 3.49 |
| TCONS_00016688 | 1 | Up | 3.53 |
| TCONS_00135006 | 9 | Up | 3.55 |
| TCONS_00045364 | 14 | Up | 3.70 |
| TCONS_00054225 | 16 | Up | 3.75 |
| XR_085687. 1 | | Up | 3.76 |
| XR_147157. 1 | | Up | 3.98 |
| TCONS_00077817 | 20 | Up | 4.01 |
| TCONS_00016690 | 1 | Up | 4.06 |
| TCONS_00043934 | 14 | Up | 4.07 |
| TCONS_00053182 | 16 | Up | 4.16 |
| TCONS_00003762 | 1 | Up | 4.25 |
| TCONS_00066377 | 19 | Down | 4.30 |
| XR_085881. 1 | | Down | 4.38 |
| TCONS_00134998 | 9 | Up | 4.40 |
| TCONS_00073324 | 2 | Down | 4.41 |
| TCONS_00079171 | 20 | Up | 4.49 |
| TCONS_00041050 | 14 | Up | 4.68 |
| TCONS_00119346 | 7 | Down | 4.85 |
| TCONS_00063145 | 19 | Up | 4.88 |
| TCONS_00017712 | 10 | Down | 4.93 |
|
ENSRNOT00000010509 | 7 | Down | 4.94 |
| TCONS_00016231 | 1 | Up | 5.00 |
| XR_086204. 2 | | Up | 5.03 |
| uc. 101+ | 3 | Up | 5.43 |
| TCONS_00134930 | 9 | Up | 5.47 |
| TCONS_00058134 | 17 | Up | 6.20 |
| TCONS_00089902 | 3 | Up | 6.36 |
|
ENSRNOT00000036466 | X | Down | 6.39 |
| TCONS_00045074 | 14 | Up | 6.61 |
| TCONS_00045348 | 14 | Up | 6.97 |
| XR_085575. 1 | | Down | 7.38 |
| NR_003722. 1
(Gapdh-ps1) | 3 | Up | 9.81 |
| XR_006589. 2 | | Down | 10.49 |
| XR_006589. 2 | | Down | 10.49 |
| TCONS_00106261 | 5 | Down | 11.38 |
| TCONS_00016693 | 1 | Up | 17.88 |
| XR_006738. 3 | | Up | 18.33 |
| TCONS_00139993 | X | Down | 27.31 |
| TCONS_00031872 | 11 | Down | 29.43 |
| TCONS_00098342 | 4 | Down | 62.10 |
RT-qPCR validation of the differential
expression of partial sequences
Six differentially expressed sequences were selected
to validate the results of the microarray using RT-qPCR. Partial
sequences indicated greater differences, however were not selected
due to disparities between groups, for example, TCONS_00031872
demonstrated virtually no expression in the SHR group. The result
of RT-qPCR validation is presented in Fig. 2.
lncRNAs may serve roles in numerous
biological processes and pathophysiological alterations
GO enrichment analysis is based on the knowledge of
various biological elements, and KEGG annotations contain
information on >200 pathways. Certain notable or high-scoring
terms in the GO and KEGG pathway analyses were collected in a
summary and are listed in Fig. 3.
The GO functions identified were predominantly associated with
isoprenoid biosynthetic processes, fatty acid metabolic processes,
responses to cyclic AMP, cell migration in biological processes,
the apical plasma membrane, microsomes, the external side of the
plasma membrane, the perinuclear region of the cytoplasm
(identified during cellular component analysis), glycoprotein
binding, phosphatase activity and isopentenyl-pyrophosphate
δ-isomerase activity (identified during molecular function
analysis). The KEGG pathway analysis demonstrated that lncRNA
coexpressed genes predominantly mapped to three pathways, including
the cytochrome p450-associated process, extracellular
matrix-receptor interaction and viral myocarditis.
Discussion
These preliminary data indicate that numerous
lncRNAs are differentially expressed between SHRs and normal (WKY)
rats. These results were expected as high blood pressure
(considered to be a multifactorial disease) involves complicated
biological networks.
According to the GO and pathway analyses, numerous
genes are associated with a variety of membrane functions,
particularly including cell membrane permeability, transmembrane
receptors and membrane potential. However, this finding contributes
little guidance for follow-up experiments, as these physiological
elements are involved in almost all biological processes. Although
type I diabetes mellitus and viral myocarditis are associated with
blood pressure or are involved in the cardiovascular system and
received high scores in the pathway analysis, it is challenging to
conduct further research on pathways with such a direct association
to blood pressure.
One notable finding of the present study was the
observation of altered cell migration and ion channel activity,
which are two pathogenic factors associated with hypertension.
Excessive activation or deactivation of ion channels may have a
marked effect on vasomotion or blood pressure, and the target genes
of lncRNAs may be associated with the
Na+/K+ATP pump or Ca+ channel
(13,14). Changes in vascular smooth muscle
cell proliferation and migration are considered to be pathogenic
factors leading to high blood pressure, as these processes
significantly contribute to angiogenesis and microcirculation blood
volume (15).
A fundamental limitation of the present study was
the disorganized database and unofficial analysis tool used for the
identification of animal lncRNAs, including the use of trans-
and/or cis-regulatory network analysis. Although this is a common
method to analyze associated protein-coding genes and transcription
factors, as well as to predict biological pathways, these databases
are based on the human genome and therefore cannot be used to
determine sequence information from rats or mice. The
above-mentioned GO and KEGG pathway analyses were used as an
effective prediction tool for mRNAs; however, with this approach,
the user is capable of predicting only the biological function of
coexpressed lncRNAs indirectly. Furthermore, the number of studies
and the database integrity for rat lncRNAs are markedly lower than
those for human and mouse models. Other biological tools, such as
ncFANS Function Annotation (http://www.bioinfo.org/ncfans/), are primarily based
on data from humans and mice.
Additional limitations of the present study, as well
as other ncRNA studies, are the result of the following factors.
First, a number of ncRNAs are shorter than 200 nt, are functional,
and cannot be grouped into miRNAs and structural RNA groups
(16); the microarray in the
present study contained only lncRNA probes longer than 200 nt. In
addition, many ncRNAs are too similar to other mRNAs with regards
to sequence. For example, the lncRNA, NR_003722.1 was upregulated
in the SHR group in the current results; however, this represents a
pseudogene of GAPDH and there is little difference in base sequence
composition. Therefore, one of the main obstacles in investigating
lncRNAs is designing a primer or siRNA that cannot be associated
with similar mRNAs. Furthermore, siRNAs typically interfere with
the expression of their target mRNA in the cytoplasm, whereas
numerous lncRNAs have been suggested to be located intranuclearly,
where the majority of siRNAs have difficulty entering (17). Further to design and synthesize,
ensuring the function of the siRNA presents another challenge.
Recently, an RNA sequencing study reported that
lncRNAs were differentially altered between the Brown Norway rat
and the Dahl salt-sensitive rat, which is recog-nized as another
classical animal model of hypertension. However, Wang et al
(4) stated that one limitation of
investigating lncRNAs in hypertension is the lack of systematic
lncRNA characterization in rats. In another renal lncRNA study that
performed sequencing and lncRNA transcriptome analysis, >3,000
transcripts were identified as rat lncRNAs; Gopalakrishnan et
al (5) reported numerous
lncRNAs that were differentially expressed between Dahl
salt-resistant, and Dahl salt-sensitive rats and SHRs. The study
also identified that certain mRNAs were coexpressed with lncRNAs
and that the majority of lncRNAs were not predicted to have target
genes (5). These studies
contribute important information to the role of lncRNAs in
hypertension in rats.
Previous studies have identified that genes
associated with hypertension exert significant influences on
lncRNAs. For example, angiotensin II (AngII), a classic
hypertensive factor, has been demonstrated to regulate the lncRNA
termed Lnc-Ang362 in vascular smooth muscle cells. Following
treatment with AngII, the expression level of more than three
lncRNAs was dynamically altered; specifically, upregulated lncRNAs
increased and downregulated lncRNAs decreased at an early
time-point following treatment. However, these levels all returned
to baseline within 24 h (18). The
growth of vascular endothelial cells was also indicated to be
regulated by the recently described lncRNA, metastasis associated
lung adenocarcinoma transcript 1 (MALAT1); in particular, silencing
MALAT1 using siRNA was observed to induce various changes in
endothelial cells with respect to angiogenesis (19).
Numerous studies have identified miRNAs as highly
associated with blood pressure (13,20),
and recent studies have highlighted the competing endogenous RNA
reaction between miRNAs and lncRNAs. This reaction may represent a
novel pathological process, as numerous miRNA-lncRNA or miRNA-mRNA
interactions may be implicated in the multi-layered
lncRNA-mRNA/protein crosstalk (21). Furthermore, these mechanisms have
been repeatedly reported for the cardiovascular system (22). In a previous study, two miRNAs
(miR-221 and miR-222) were verified to be associated with lncRNA in
AngII-treated vascular smooth muscle cells, indicating that lncRNAs
may serve as host genes for miRNAs (18). Recently, another study demonstrated
the importance of the tripartite axis of lncRNA-miRNA-mRNA in the
regulation of cardiovascular disease, which may present as a novel
research direction in future (23).
It is not considered to be practical to investigate
lncRNAs or miRNAs for the diagnosis of hypertension, as blood
pressure measurements may be more readily obtained; however, RNAs
may serve as valuable diagnostic tools for individuals with early
genetic alterations that do not demonstrate any disease symptoms.
For healthy individuals who live with anxiety, psychological
stress, or poor living habits and who may be asymptomatic genetic
carriers of the hypertensive trait, blood pressure measurements may
not indicate abnormalities, whereas molecular biology may provide
information on the likelihood of disease. Furthermore, a large
number of studies have indicated that miRNAs are closely associated
with hypertensive therapy (24).
However, the development of essential therapeutic agents utilizing
ncRNAs may require significant further research. In conclusion,
with the identifi-cation of the first miRNA-targeted therapeutic
agent and its application in volunteers (25), lncRNAs may present as a novel
target for the treatment of hypertension and may potentially serve
as molecular therapeutic strategies.
Acknowledgments
The present study was supported by the National
Natural Science Foundation of China (grant no. 81270230) and
Ministry of Health of the People's Republic of China Science
Foundation (grant no. WKJ-ZJ2014-020).
Abbreviations:
|
miRNAs
|
microRNAs
|
|
siRNAs
|
short interfering RNAs
|
|
snRNAs
|
small nuclear RNAs
|
|
ncRNA
|
non-coding RNA
|
|
lncRNA
|
long non-coding RNA
|
|
GO
|
gene ontology
|
|
SHR
|
spontaneous hypertensive rat
|
|
WKY
|
normotensive Wistar-Kyoto rat
|
References
|
1
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Schonrock N, Harvey RP and Mattick JS:
Long noncoding RNAs in cardiac development and pathophysiology.
Circulation Res. 111:1349–1362. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Thum T: Noncoding RNAs and myocardial
fibrosis. Nat Rev Cardiol. 11:655–663. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang F, Li L, Xu H, Liu Y, Yang C, Cowley
AW Jr, Wang N, Liu P and Liang M: Characteristics of long
non-coding RNAs in the Brown Norway rat and alterations in the Dahl
salt-sensitive rat. Sci Rep. 4:71462014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gopalakrishnan K, Kumarasamy S, Mell B and
Joe B: Genome-wide identification of long noncoding RNAs in rat
models of cardiovascular and renal disease. Hypertension.
65:200–210. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
James PA, Oparil S, Car ter BL, Cushman
WC, Dennison-Himmelfarb C, Handler J, Lackland DT, LeFevre ML,
MacKenzie TD, Ogedegbe O, et al: 2014 evidence-based guideline for
the management of high blood pressure in adults: Report from the
panel members appointed to the Eighth Joint National Committee (JNC
8). JAMA. 311:507–520. 2014. View Article : Google Scholar
|
|
7
|
Pinto YM, Paul M and Ganten D: Lessons
from rat models of hypertension: From Goldblatt to genetic
engineering. Cardiovasc Res. 39:77–88. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Okamoto K and Aoki K: Development of a
strain of spontaneously hypertensive rats. Jpn Circ J. 27:282–293.
1963. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
10
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar
|
|
11
|
Prifti E, Zucker JD, Clement K and Henegar
C: FunNet: An integrative tool for exploring transcriptional
interactions. Bioinformatics. 24:2636–2638. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chaudhry MA: Small nucleolar RNA host
genes and long non-coding RNA responses in directly irradiated and
bystander cells. Cancer Biother Radiopharm. 29:135–141. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Haddy FJ and Pamnani MB: The vascular
Na+-K+ pump in low renin hypertension. J
Cardiovasc Pharmacol. 6(Suppl 1): S61–S74. 1984. View Article : Google Scholar
|
|
14
|
Bátkai S and Thum T: MicroRNAs in
hypertension: Mechanisms and Therapeutic targets. Curr Hypertens
Rep. 14:79–87. 2012. View Article : Google Scholar
|
|
15
|
Qiu J, Zheng Y, Hu J, Liao D, Gregersen H,
Deng X, Fan Y and Wang G: Biomechanical regulation of vascular
smooth muscle cell functions: From in vitro to in vivo
understanding. J R Soc Interface. 11:201308522014. View Article : Google Scholar :
|
|
16
|
Spizzo R, Almeida MI, Colombatti A and
Calin GA: Long non-coding RNAs and cancer: A new frontier of
translational research? Oncogene. 31:4577–4587. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bagasra O and Prilliman KR: RNA
interference: the molecular immune system. J Mol Histol.
35:545–553. 2004.PubMed/NCBI
|
|
18
|
Leung A, Trac C, Jin W, Lanting L, Akbany
A, Sætrom P, Schones DE and Natarajan R: Novel long noncoding RNAs
are regulated by angiotensin II in vascular smooth muscle cells.
Circ Res. 113:266–278. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Michalik KM, You X, Manavski Y,
Doddaballapur A, Zörning M, Braun T, John D, Ponomareva Y, Chen W,
Uchida S, et al: Long noncoding RNA MALAT1 regulates endothelial
cell function and vessel growth. Circ Res. 114:1389–1397. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Synetos A, Toutouzas K, Stathogiannis K,
Latsios G, Tsiamis E, Tousoulis D and Stefanadis C: MicroRNAs in
arterial hypertension. Curr Top Med Chem. 13:1527–1532. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tay Y, Rinn J and Pandolfi PP: The
multilayered complexity of ceRNA crosstalk and competition. Nature.
505:344–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ge D, Han L, Huang S, Peng N, Wang P,
Jiang Z, Zhao J, Su L, Zhang S, Zhang Y, et al: Identification of a
novel MTOR activator and discovery of a competing endogenous RNA
regulating autophagy in vascular endothelial cells. Autophagy.
10:957–971. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wu C and Arora P: Long noncoding
RNA-microRNA-mRNA: A novel tripartite axis in the regulation of
cardiac hypertrophy. Circ Cardiovasc Genet. 7:729–731. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Neves VJ, Fernandes T, Roque FR, Soci UP,
Melo SF and de Oliveira EM: Exercise training in hypertension: Role
of microRNAs. World J Cardiol. 6:713–727. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lindow M and Kauppinen S: Discovering the
first microRNA-targeted drug. J Cell Biol. 199:407–412. 2012.
View Article : Google Scholar : PubMed/NCBI
|