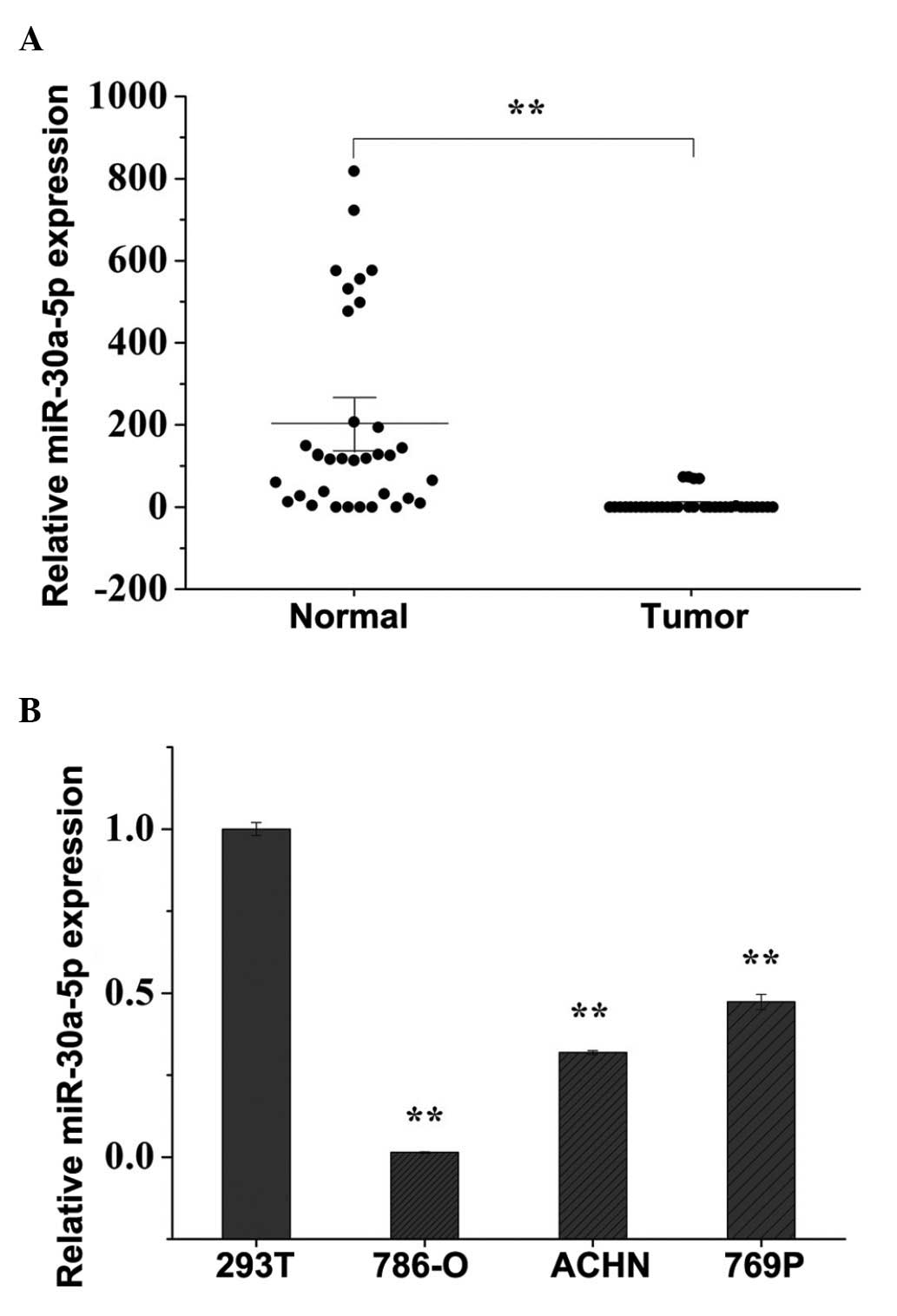
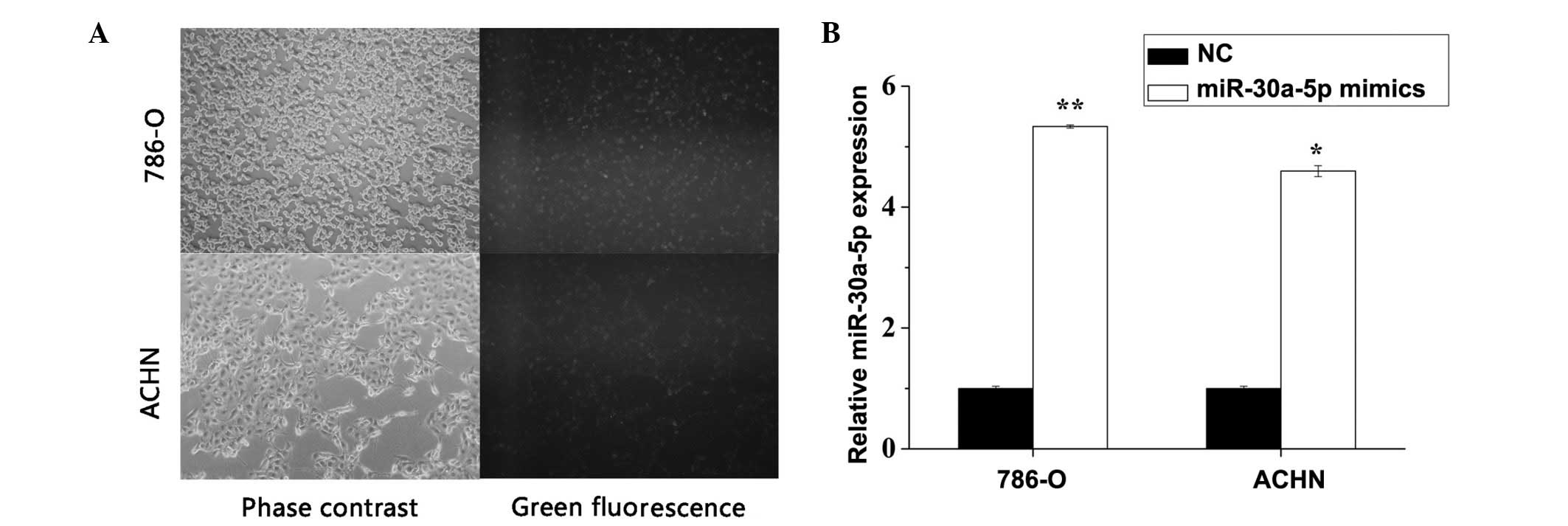
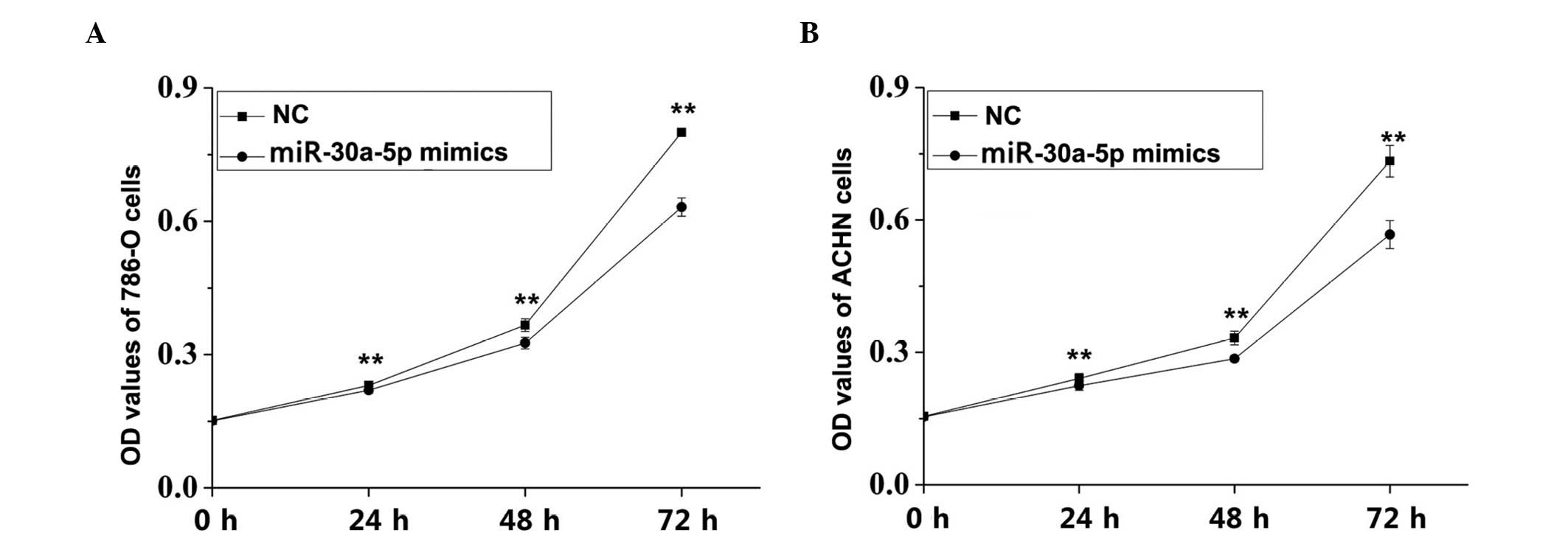
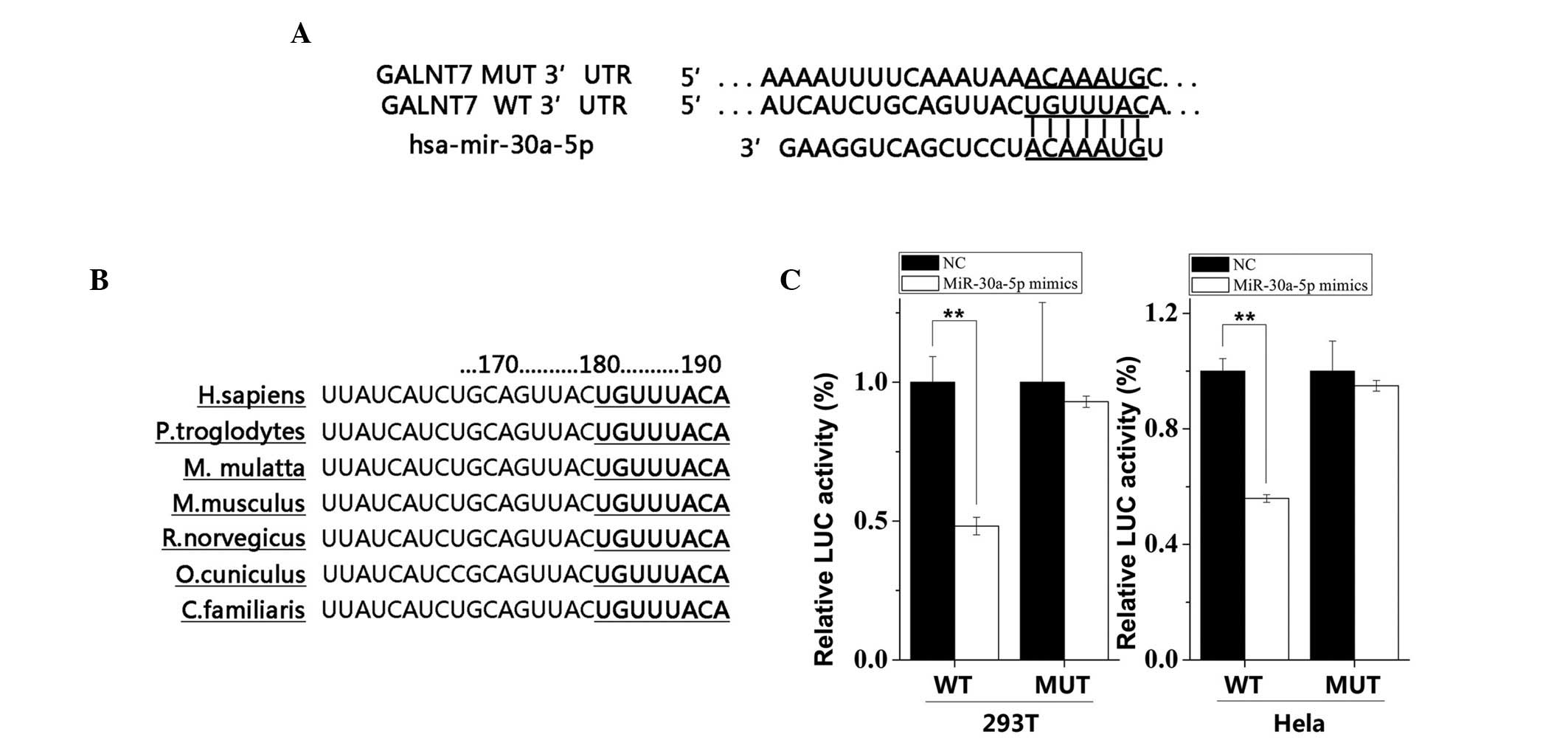
|
1
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rini BI, Campbell SC and Escudier B: Renal
cell carcinoma. Lancet. 373:1119–1132. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Motzer RJ, Bander NH and Nanus DM:
Renal-cell carcinoma. N Engl J Med. 335:865–875. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Janzen NK, Kim HL, Figlin RA and
Belldegrun AS: Surveillance after radical or partial nephrectomy
for localized renal cell carcinoma and management of recurrent
disease. Urol Clin North Am. 30:843–852. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rouvière O, Bouvier R, Négrier S, Badet L
and Lyonnet D: Nonmetastatic renal-cell carcinoma: Is it really
possible to define rational guidelines for post-treatment
follow-up? Nat Clin Pract Oncol. 3:200–213. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Carthew RW and Sontheimer EJ: Origins and
mechanisms of miRNAs and siRNAs. Cell. 136:642–655. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Huntzinger E and Izaurralde E: Gene
silencing by microRNAs: Contributions of translational repression
and mRNA decay. Nat Rev Genet. 12:99–110. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Krol J, Loedige I and Filipowicz W: The
widespread regulation of microRNA biogenesis, function and decay.
Nat Rev Genet. 11:597–610. 2010.PubMed/NCBI
|
|
10
|
Ichimi T, Enokida H, Okuno Y, Kunimoto R,
Chiyomaru T, Kawamoto K, Kawahara K, Toki K, Kawakami K, Nishiyama
K, et al: Identification of novel microRNA targets based on
microRNA signatures in bladder cancer. Int J Cancer. 125:345–352.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rydzanicz M, Wrzesiński T, Bluyssen HA and
Wesoly J: Genomics and epigenomics of clear cell renal cell
carcinoma: Recent developments and potential applications. Cancer
Lett. 341:111–126. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fendler A, Stephan C, Yousef GM and Jung
K: MicroRNAs as regulators of signal transduction in urological
tumors. Clin Chem. 57:954–968. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Huang QB, Ma X, Zhang X, Liu SW, Ai Q, Shi
TP, Zhang Y, Gao Y, Fan Y, Ni D, et al: Down-Regulated miR-30a in
Clear Cell Renal Cell Carcinoma Correlated with Tumor Hematogenous
Metastasis by Targeting Angiogenesis-Specific DLL4. PLoS One.
82013.
|
|
14
|
Mathew LK, Lee SS, Skuli N, Rao S, Keith
B, Nathanson KL, Lal P and Simon MC: Restricted expression of
miR-30c-2-3p and miR-30a-3p in clear cell renal cell carcinomas
enhances HIF2alpha activity. Cancer Discov. 4:53–60. 2014.
View Article : Google Scholar :
|
|
15
|
Tan X, Qin W, Zhang L, Hang J, Li B, Zhang
C, Wan J, Zhou F, Shao K, Sun Y, et al: A 5-microRNA signature for
lung squamous cell carcinoma diagnosis and hsa-miR-31 for
prognosis. Clin Cancer Res. 17:6802–6811. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fuster O, Llop M, Dolz S, García P, Such
E, Ibáñez M, Luna I, Gómez I, López M, Cervera J, et al: Adverse
prognostic value of MYBL2 overexpression and association with
microRNA-30 family in acute myeloid leukemia patients. Leuk Res.
37:1690–1696. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu Z, Tu K and Liu Q: Effects of
microRNA-30a on migration, invasion and prognosis of hepatocellular
carcinoma. FEBS Lett. 588:3089–3097. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang HY, Li YY, Fu S, Wang XP, Huang MY,
Zhang X, Shao Q, Deng L, Zeng MS, Zeng YX and Shao JY: MicroRNA-30a
promotes invasiveness and metastasis in vitro and in vivo through
epithelial-mesenchymal transition and results in poor survival of
nasopharyngeal carcinoma patients. Exp Biol Med. 239:891–898. 2014.
View Article : Google Scholar
|
|
19
|
Huang Q, Jiang Z, Meng T, Yin H, Wang J,
Wan W, Cheng M, Yan W, Liu T, Song D, et al: MiR-30a inhibits
osteolysis by targeting RunX2 in giant cell tumor of bone. Biochem
Biophys Res Commun. 453:160–165. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fu J, Xu X, Kang L, Zhou L, Wang S, Lu J,
Cheng L, Fan Z, Yuan B, Tian P, et al: miR-30a suppresses breast
cancer cell proliferation and migration by targeting Eya2. Biochem
Biophys Res Commun. 445:314–319. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jiang X, Du L, Wang L, Li J, Liu Y, Zheng
G, Qu A, Zhang X, Pan H, Yang Y and Wang C: Serum microRNA
expression signatures identified from genome-wide microRNA
profiling serve as novel noninvasive biomarkers for diagnosis and
recurrence of bladder cancer. Int J Cancer. 136:854–862. 2015.
View Article : Google Scholar
|
|
22
|
Jia Z, Wang K, Wang G, Zhang A and Pu P:
MiR-30a-5p antisense oligonucleotide suppresses glioma cell growth
by targeting SEPT7. PLoS One. 8:e550082013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang K, Jia Z, Zou J, Zhang A, Wang G, Hao
J, Wang Y, Yang S and Pu P: Analysis of hsa-miR-30a-5p expression
in human gliomas. Pathol Oncol Res. 19:405–411. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu H, Brannon AR, Reddy AR, Alexe G,
Seiler MW, Arreola A, Oza JH, Yao M, Juan D, Liou LS, et al:
Identifying mRNA targets of microRNA dysregulated in cancer: With
application to clear cell renal cell carcinoma. BMC Syst Biol.
4:512010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Juan D, Alexe G, Antes T, Liu H,
Madabhushi A, Delisi C, Ganesan S, Bhanot G and Liou LS:
Identification of a microRNA panel for clear-cell kidney cancer.
Urology. 75:835–841. 2010. View Article : Google Scholar
|
|
26
|
Yi Z, Fu Y, Zhao S, Zhang X and Ma C:
Differential expression of miRNA patterns in renal cell carcinoma
and nontumorous tissues. J Cancer Res Clin Oncol. 136:855–862.
2010. View Article : Google Scholar
|
|
27
|
White NM, Khella HW, Grigull J, Adzovic S,
Youssef YM, Honey RJ, Stewart R, Pace KT, Bjarnason GA, Jewett MA,
et al: miRNA profiling in metastatic renal cell carcinoma reveals a
tumour-suppressor effect for miR-215. Br J Cancer. 105:1741–1749.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Müller S and Nowak K: Exploring the
miRNA-mRNA regulatory network in clear cell renal cell carcinomas
by next-generation sequencing expression profiles. Biomed Res Int.
2014:9484082014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Edge SB, Byrd DR, Compton CC, Fritz AG,
Greene FL and Trotti A: AJCC Cancer Staging Manual. 7th ed. New
York, NY: Springer; pp. 479–489. 2010
|
|
30
|
Duan HF, Li XQ, Hu HY, Li YC, Cai Z, Mei
XS, Yu P, Nie LP, Zhang W, Yu ZD and Nie GH: Functional elucidation
of miR-494 in the tumorigenesis of nasopharyngeal carcinoma. Tumour
Biol. 36:6679–6689. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shenouda SK and Alahari SK: MicroRNA
function in cancer: Oncogene or a tumor suppressor? Cancer
Metastasis Rev. 28:369–378. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Garzon R, Calin GA and Croce CM: MicroRNAs
in Cancer. Annu Rev Med. 60:167–179. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ortega M, Bhatnagar H, Lin AP, Wang L,
Aster JC, Sill H and Aguiar RC: A microRNA-mediated regulatory loop
modulates NOTCH and MYC oncogenic signals in B-and T-cell
malignancies. Leukemia. 29:968–976. 2015. View Article : Google Scholar :
|
|
34
|
Liu Y, Song Y, Ma W, Zheng W and Yin H:
Decreased microRNA-30a levels are associated with enhanced ABL1 and
BCR-ABL1 expression in chronic myeloid leukemia. Leuk Res.
37:349–356. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
González-Gugel E, Villa-Morales M, Santos
J, Bueno MJ, Malumbres M, Rodríguez-Pinilla SM, Piris MÁ and
Fernández-Piqueras J: Down-regulation of specific miRNAs enhances
the expression of the gene Smoothened and contributes to T-cell
lymphoblastic lymphoma development. Carcinogenesis. 34:902–908.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Perrine CL, Ganguli A, Wu P, Bertozzi CR,
Fritz TA, Raman J, Tabak LA and Gerken TA: Glycopeptide-preferring
polypeptide GalNAc transferase 10 (ppGalNAc T10), involved in
mucin-type O-glycosylation, has a unique GalNAc-O-Ser/Thr-binding
site in its catalytic domain not found in ppGalNAc T1 or T2. J Biol
Chem. 284:20387–20397. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Pegram MD, Borges VF, Ibrahim N, Fuloria
J, Shapiro C, Perez S, Wang K, Schaedli Stark F and Courtenay Luck
N: Phase I dose escalation pharmacokinetic assessment of
intravenous humanized anti-MUC1 antibody AS1402 in patients with
advanced breast cancer. Breast Cancer Res. 11:R732009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hollingsworth MA and Swanson BJ: Mucins in
cancer: Protection and control of the cell surface. Nat Rev Cancer.
4:45–60. 2004. View Article : Google Scholar
|
|
39
|
Peng RQ, Wan HY, Li HF, Liu M, Li X and
Tang H: MicroRNA-214 suppresses growth and invasiveness of cervical
cancer cells by targeting UDP-N-acetyl-α-D-galactosamine:
Polypeptide N-acetylgalactosaminyltransferase 7. J Biol Chem.
287:14301–14309. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Taniuchi K, Cerny RL, Tanouchi A, Kohno K,
Kotani N, Honke K, Saibara T and Hollingsworth MA: Overexpression
of GalNAc-transferase GalNAc-T3 promotes pancreatic cancer cell
growth. Oncogene. 30:4843–4854. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li W, Ma H and Sun J: MicroRNA34a/c
function as tumor suppressors in Hep2 laryngeal carcinoma cells and
may reduce GALNT7 expression. Mol Med Rep. 9:1293–1298.
2014.PubMed/NCBI
|
|
42
|
Gaziel-Sovran A, Segura MF, Di Micco R,
Collins MK, Hanniford D, Vega-Saenz de Miera E, Rakus JF, Dankert
JF, Shang S, Kerbel RS, et al: miR-30b/30d regulation of GalNAc
transferases enhances invasion and immunosuppression during
metastasis. Cancer Cell. 20:104–118. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Abend M, Pfeiffer RM, Ruf C, Hatch M,
Bogdanova TI, Tronko MD, Riecke A, Hartmann J, Meineke V, Boukheris
H, et al: Iodine-131 dose dependent gene expression in thyroid
cancers and corresponding normal tissues following the Chernobyl
accident. PLoS One. 7:e391032012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kim VN: MicroRNA biogenesis: Coordinated
cropping and dicing. Nat Rev Mol Cell Biol. 6:376–385. 2005.
View Article : Google Scholar : PubMed/NCBI
|