Introduction
SET nuclear proto-oncogene (SET), also known as
protein phosphatase type 2A inhibitor (I2PP2A) or
template-activating factor-1β, is involved in a number of cell
processes, including the replication of DNA, chromatin remodeling,
gene transcription, differentiation, migration and cell cycle
regulation (1). The SET protein
was initially extracted from the bone marrow of a leukemia patient
(2). Originally identified as a
potent PP2A inhibitor, SET is overexpressed in several neoplasms
(3,4), particularly acute myeloid leukemia
(5). N-terminal and internal amino
acid sequencing indicated that I2PP2A was a truncated form of SET
(5) Clinical data have indicated
that patients with SET overexpression exhibit worse overall
survival and event-free survival, suggesting an oncogenic role of
SET in tumorigenesis (1).
In our previous proteomics study, the ATRA-resistant
acute promyelocytic leukemia (APL) NB4-R1 cell line was treated
with 25 μmol/l As4S4 and an
As4S4-induced differential protein expression
profile was obtained (6). There
was a significant decrease in the expression of the SET oncoprotein
after the treatment with As4S4 for 24 h,
which was completely abolished after treatment for 48 h compared
with the vector control; this indicated the importance of SET in
the regulation of signal transduction pathways and in the mechanism
of APL cell apoptosis. Next, we further investigated the possible
cellular and molecular mechanisms of As4S4
treatment and the role of SET in the apoptosis of NB4-R1 cells.
Consistent with previous studies, it was found that
As4S4-induced apoptosis was accompanied by
reduced mRNA and protein expression levels of SET in the NB4-R1
cells (Wang et al, unpublished data). These results
indicated that As4S4 may induce the apoptosis
of NB4-R1 cells through the downregulation of SET protein
expression.
In the present study the SET gene was selected for
further investigation. To avoid interference and possible
compensation by other genes, SET was transfected into human
embryonic kidney 293T cells and it was observed whether SET
expression led to a change in the expression of PP2A or the
apoptosis-related genes Bcl-2 and Bax in 293T cells treated with
As4S4.
Materials and methods
Cell culture and reagents
The human embryonic kidney 293T cell line and the
APL NB4-R1 cell line were stored in in the Oncology Research
Laboratory, The First Affiiated Hospital of Xi'an Jiaotong
University (Xi'an, China). The 293T cells were maintained in
Dulbecco's modified Eagle's medium (Gibco; Thermo Fisher Scientific
Inc., Waltham, MA, USA) at 37°C in a 5% CO2 atmosphere
in air. The growth medium was supplemented with 10%
heat-inactivated (at 56°C for 30 min) fetal bovine serum (FBS),
penicillin (100 U/ml) and streptomycin (100 μg/ml). The
NB4-R1 cells were cultured in RPMI-1640 medium (Gibco; Thermo
Fisher Scientific Inc.) supplemented with 10% FBS, 100 U/ml
penicillin and 100 μg/ml streptomycin, and maintained in a
5% CO2 humidified atmosphere at 37°C.
Extraction of total RNA and reverse
transcription-polymerase chain reaction (RT-PCR)
The total RNA was extracted from the NB4-R1 cells
using TRIzol reagent (Invitrogen; Thermo Fisher Scientific Inc.)
and was precisely quantified using an ultraviolet
spectrophotometer. The sequences of the primers for SET
amplification are listed below: Forward,
5′-CCGCTCGAGATGTCGGCGCCGGCGGCCAAAG-3′ (the underlined section
represents the XhoI cleavage site); and reverse,
5′-GGGGTACCGTGTCATCTTCTCCTTCATCC-3′ (the underlined section
indicates the KpnI cleavage site). The RT reaction system
included 2 μg RNA, 2 μl 5X PrimeScript RT Master Mix
(Takara Bio Inc., Otsu, Japan) and RNase-free dH2O to a
final volume of 10 μl. The reaction program was as follows:
37°C for 15 min, followed by 85°C for 5 sec. The PCR was performed
in a total volume of 20 μl, which included 1 μl (10
ng/μl) cDNA, 4 μl 5X Taq buffer; 1.6 μl (2.5
mM) dNTPs, 0.4 μl (10 μM) forward and reverse primer;
0.2 μl Taq polymerase; and 12.4 μl ddH2O.
The reaction program was performed in a PCR Thermocycle Instrument
(Bio-Rad Laboratories, Inc., Hercules, CA, USA) as follows: 94°C
for 5 min, and then 30 cycles of 94°C for 30 sec, 55°C for 30 sec,
72°C for 50 sec and an extension step of 72°C for 10 min. The
amplified PCR products were electrophoresed on a 1% agarose gel
(Beijing SBS Genetech Co., Ltd., Beijing, China) and were
visualized under ultraviolet light using a Chemidox XRS and
Quantity One gel image analysis system (Bio-Rad Laboratories,
Inc.). The length of the amplified PCR segment was 850 bp and the
experiment was replicated 3 times.
Construction of pEGFP-N1-SET plasmid
The gel-purified PCR segment was cut with
XhoI and KpnI restriction enzymes. The length of the
restriction enzyme digestion product was 835 bp. Next, the segment
was introduced into the pEGFP-N1 plasmid (Shanghai Genechem Co.,
Ltd., Shanghai, China). The pEGFP-N1-SET construct was transformed
into DH5α cells (stored in our laboratory), and then the correct
fragment was identified as a mini-plasmid by digestion with
XhoI and KpnI, and was verified as the correct clone
through sequence analysis.
Transfection of 293T cells
For transient transfection, the 293T cells were
plated at a density of 8×104/ml cells per well in 6-well
plates and transfected with 2 μg pEGFP-N1-SET or pEGFP-N1
(vector group) plasmids using Lipofectamine 2000 reagent
(Invitrogen; Thermo Fisher Scientific Inc.). After 48 h of
transfection, the cells were observed with an inverted fluorescence
microscope (Olympus Corporation, Tokyo, Japan) and used for further
assays.
SET and PP2A mRNA analyses
The total RNA was extracted from each sample of 293T
cells in TRIzol reagent. Each RNA sample (1 μg) was
reverse-transcribed into cDNA using a RevertAid First Strand cDNA
Synthesis kit (Thermo Fisher Scientific Inc.) according to the
manufacturer's protocols, and then the cDNA was used for
RT-quantitative PCR (RT-qPCR). The RT-qPCR amplification reaction
contained 12.5 μl 2X SYBR-Green I, 1 μl forward (0.4
μM) and 1 μl reverse (0.4 μM) primer and 10.5
μl cDNA diluted with RNase-free water. The RT-qPCR included
control samples with RNase-free water instead of the cDNA template.
All samples were analyzed in three independent biological
replicates. The PCR conditions were as follows: Initial
denaturation at 95°C for 30 sec, denaturing at 95°C for 5 sec, and
annealing at 60°C for 30 sec, where the last two steps of the
reaction were repeated for 40 cycles. The primer sequences used to
amplify the target genes are listed as follows: SET forward,
5′-CATCTTCGAAGTCCACCGAAATC-3′ and reverse;
5′-TGCATCAGAATGGTCAGTAAACCAG-3′; PP2A forward,
5′-GTAACCAAGCTGCAATCATGGAA-3′ and reverse,
5′-CTCTACGAGGTGCTGGGTCAAAC-3′; and glyceraldehyde 3-phosphate
dehydrogenase (GAPDH) forward, 5′-CACCCTGTTGCTGTAGCCAAA-3′ and
reverse, 5′-CACCCTGTTGCTGTAGCCAAA-3′. RT-qPCR was conducted using
an iCycler iQ Real-Time PCR Detection system (Bio-Rad Laboratories
Inc.) with SYBR-Green I fluorescent dye (Takara Bio Inc.). The
relative mRNA levels of the target genes were expressed in terms of
the rates that were based on the cycle threshold (Cq) (7). The mRNA expression levels of each
group were compared through 2−ΔΔCq. The GAPDH gene was
used for inter-sample normalization and three independent
experiments were performed in triplicate.
SET and PP2A protein analysis
The cultured cells were harvested and washed three
times with cold phosphate-buffered saline (PBS). Next, the cells
were solubilized in radioimmu-noprecipitation assay buffer
containing a protease inhibitor cocktail containing 104 mM AEBSF,
80 μM aprotinin, 4 mM bestatin, 1.4 mM E-64, 2 mM leupeptin
and 1.5 mM pepstatin A (Sigma-Aldrich, St. Louis, MO, USA).
Subsequent to being maintained on ice for 10 min, the cell
suspension was centrifuged for protein at 15,500 × g for 15 min at
4°C. The protein (30 μg) was separated on 10% sodium dodecyl
sulfate polyacrylamide gel electrophoresis gels and transferred to
a nitrocellulose membrane at 110 V for 2 h. The non-specific
binding sites on the membranes were blocked with 5% (w/v) skimmed
milk in Tris-buffered saline [TBS: 20 mM Tris-HCl and 200 mM NaCl
(pH 7.6)] for 2 h under gentle rocking at room temperature. Next,
the membranes were incubated with the following primary antibodies:
Rabbit polyclonal anti-SET antibody (cat. no. ab92872) and rabbit
monoclonal anti-PP2A (cat. no. ab32141) antibodies (1:10,000;
Abcam, Cambridge, MA, USA); and mouse monoclonal anti-GAPDH
antibody (1:10,000; cat. no. sc-47724; Santa Cruz Biotechnology
Inc., Dallas, TX, USA) directed against the protein or enzyme of
interest for 1 h at room temperature and then at 4°C overnight. The
membranes were then incubated with goat anti-rabbit (cat. no.
sc-2004) or goat anti-mouse (cat. no. sc-2005) horseradish
peroxidase-conjugated secondary antibody (Santa Cruz Biotechnology,
Inc.) for 1 h at room temperature. Subsequent to being washed
extensively with TBS plus Tween 20, the membranes were incubated
under chemiluminescence and wrapped in clear plastic wrap for film
exposure. The bands on the immunoblots were quantified using
Quantity One version 4.6.2 software (Bio-Rad Laboratories Inc.).
The protein expression of each sample was internally normalized to
GAPDH, and the quantity was compared with the expression of the
transfected control groups.
Cell cycle analysis
The cell cycle distribution was analyzed via flow
cytometry (Becton Dickinson FACSCalibur double laser flow
cytometer; BD Biosciences, Franklin Lakes, NJ, USA). The 293T cells
(1×106/ml) were harvested and washed twice with ice-cold
PBS. The cells were suspended gently in 70% chilled ethanol at
−20°C overnight. After washing with PBS, the cells were
re-suspended in 500 μl of PBS containing propidium iodide
(50 μg/ml) and RNase (50 μg/ml), and incubated for 30
min at 37°C in the dark. The cell cycle phase distribution of each
experiment was analyzed using 10,000 cells per sample. The
proportion of cells in the G0/G1, S and
G2/M phases were represented as DNA histograms.
Bcl-2 and Bax protein analyses
At 48 h post-transfection, the transfected cell
medium was changed to new medium that contained 5 μmol/l
As4S4 for 24 h. The method was similar to
that used for western blotting analysis. The relevant primary
antibodies were mouse monoclonal anti-Bcl-2 (cat. no. sc-7382) or
rabbit polyclonal anti-Bax (cat. no. sc-493) antibodies (1:1,000
dilutions; Santa Cruz Biotechnology Inc.). The protein expression
of each sample was internally normalized to GAPDH.
Statistical analysis
Experiments were performed in duplicates or
triplicates over three or more independent experiments and the
results are presented as the mean ± standard deviation. Statistical
significance was calculated via a t-test using SPSS software,
version 19.0 (IBM SPSS, Armonk, NY, USA). P<0.05 was used to
indicate a statistically significant difference.
Results
Construction and package of plasmid
Using PCR-based techniques, an 850-bp human SET gene
product of the correct size was generated, and then the
gel-purified PCR segment was cut with the XhoI and
KpnI restriction enzymes. The length of the restriction
enzyme digestion product was 835 bp. Next, the segment was inserted
into the pEGFP-N1 expression vector.
The SET gene product and the positive recombinant
clones were identified with PCR (Fig.
1). DNA sequencing analysis confirmed that the sequences were
correct.
 | Figure 1Gels showing products of (A) PCR and
(B) restriction enzyme analysis. Lanes: M, DNA marker (5,000,
3,000, 2,000, 1,500, 1,000, 750, 500, 250 and 100 bp); 1,
PCR-amplified SET gene; 2, PCR-amplified product of
double-digestion by XhoI and KpnI. (C) Gel
electrophoretic analysis of vectors. Lanes: 1, double-distilled
H2O; 2, recombinant negative vector pEGFP-N1; 3, GAPDH;
4, DNA marker (as above); 5–12, recombinant positive vector of
pEGFP-N1-SET. PCR, polymerase chain reaction; EGFP, enhanced green
fluorescent protein; GAPDH, glyceraldehyde 3-phosphate
dehydrogenase. |
SET expression analysis
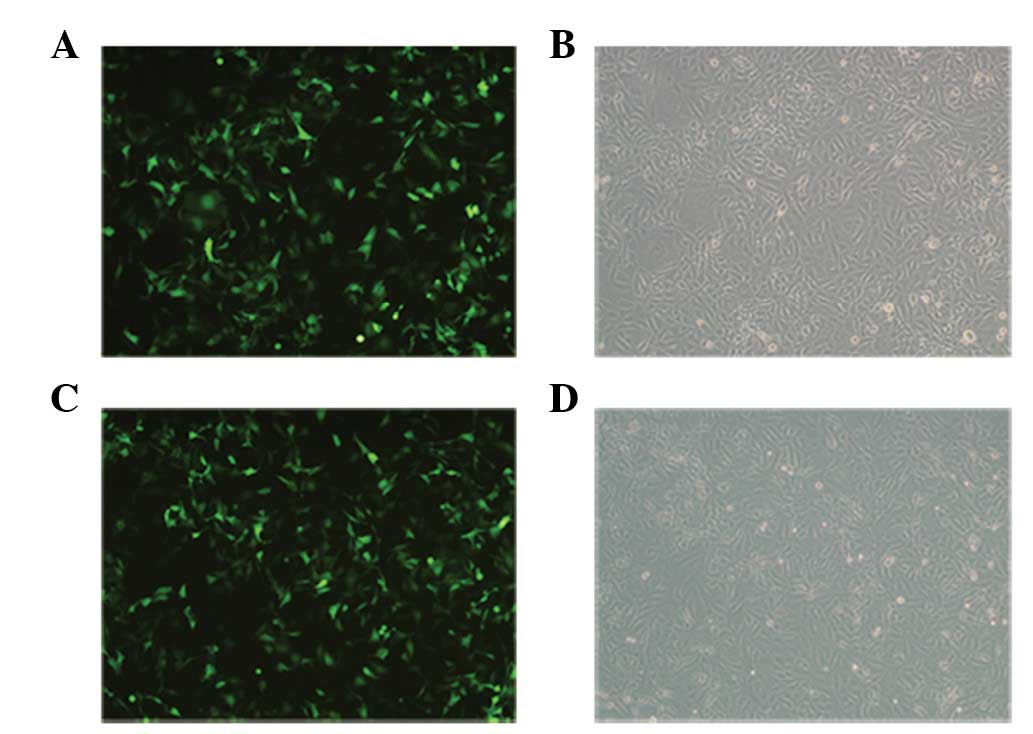
As the recombinant plasmid pEGFP-N1-SET was labeled
with enhanced green florescent protein (EGFP), EGFP expression
could be observed by laser confocal microscopy, indirectly
reflecting the transfection rate of the SET gene. At 48 h
post-transfection, the pEGFP-N1 empty vector and pEGFP-N1-SET
construct transfection yielded efficiencies of >90%, as
visualized by fluorescence microscopy for EGFP expression (Fig. 2).
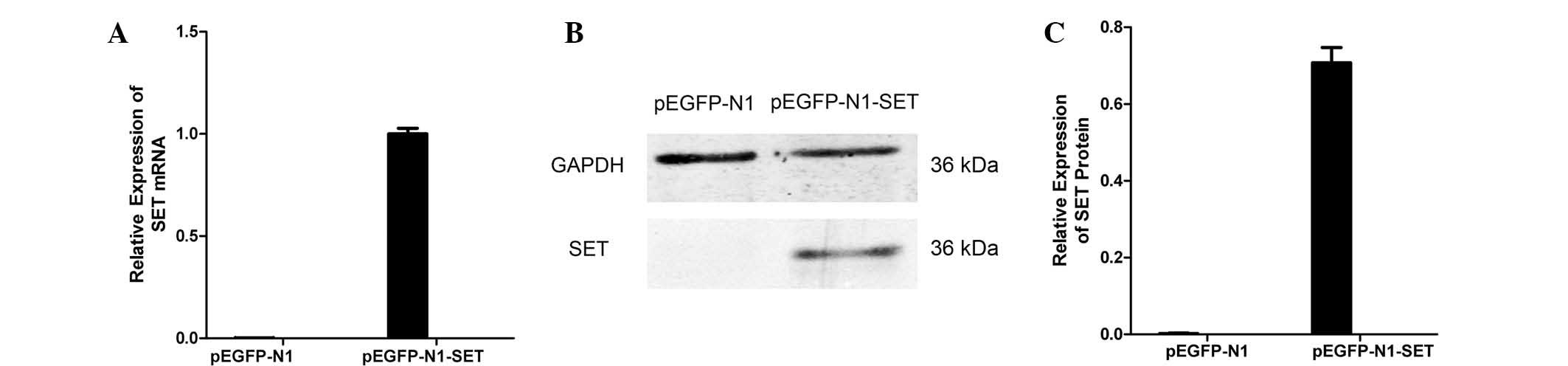
To confirm that the gene had been transfected into
the cells, RT-qPCR and western blotting was used to detect SET
expression in the recombinant cells. The results showed that the
SET expression of the construct transfection group was markedly
increased at the mRNA and protein levels compared with that of the
empty vector group. Both real-time quantitative PCR (P=2.1×10 −7)
and western blotting (P=0.001) demonstrated that pEGFP-N1-SET were
effective for SET overexpression. These results demonstrated that
the target gene had been successfully transfected (Fig. 3).
PP2A expression analyses
PP2A expression was detected by RT-qPCR and western
blotting to ascertain if the PP2A gene was involved in the study.
PP2A expression was decreased by ~92% (mRNA; P=0.0002) and ~47%
(protein; P=0.002) in the pEGFP-N1-SET-transfected 293T cells
compared with the pEGFP-N1-transfected control group (Fig. 4).
SET increases the percentage of
transfected cells in S and G2/M phases
To monitor the stimulatory effect of SET on cell
growth, the effect of SET overexpression on cell cycle progression
was investigated by flow cytometric analysis. The result
demonstrated that the overexpression of SET significantly increased
the cell percentage in the S (P=0.025) and G2/M
(P=0.045) phases in the 293T cells compared with that in the
control transfectants (Fig.
5).
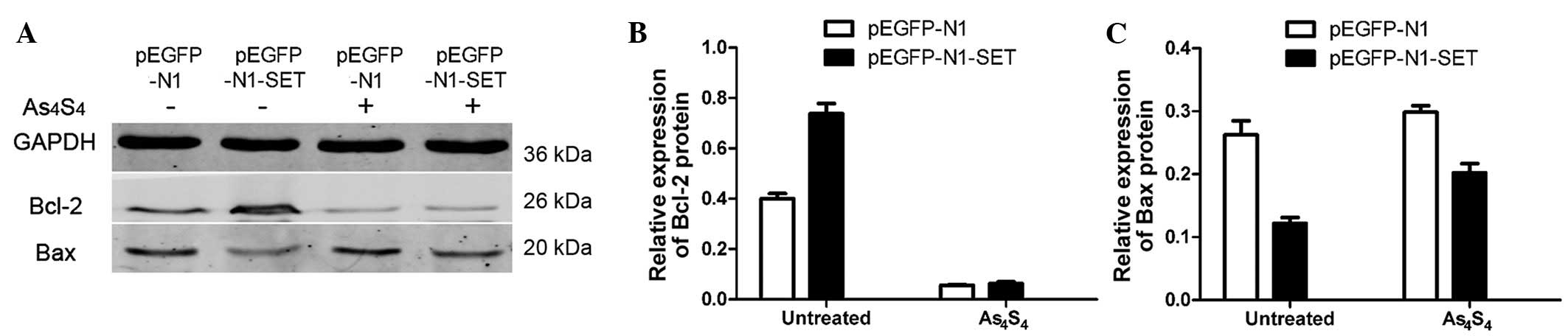
Bcl-2 and Bax expression analyses
Anti-apoptotic Bcl-2 expression and pro-apoptotic
Bax expression was detected by western blotting to ascertain if
Bcl-2 and Bax were involved in the apoptosis in this study. Bcl-2
expression was increased by ~84% (P=0.001) in the
pEGFP-N1-SET-transfected 293T cells compared with the
pEGFP-N1-transfected control group. The incubation with 5
μmol/l As4S4 for 24 h decreased the
relative Bcl-2 expression level by 86% (P=0.001) in the
pEGFP-N1-transfected 293T cells and by 91% (P=0.001) in the
pEGFP-N1-SET-transfected cells, when compared with the untreated
control cells (Fig. 6).
Bax expression was decreased significantly by ~54%
(P=0.004) in the pEGFP-N1-SET-transfected group compared with the
pEGFP-N1-transfected control group.
As4S4-treatment increased Bax expression by
14% (P>0.05) in the pEGFP-N1-transfected cells. In comparison,
As4S4 addition increased Bax expression by
66% (P=0.009) in the pEGFP-N1-SET-transfected cells (Fig. 6).
Discussion
Abnormally expressed genes associated with signal
transduction play a role in the development of carcinoma. The
investigation of the abnormal expression of genes in cancer can not
only enhance our understanding of the pathogenesis of the tumor,
but can also provide reliable therapeutic targets. Our previous
proteomics study showed a total of 21 proteins that were
differentially regulated after treatment with
As4S4 in NB4-R1 cells, including 5
downregulated proteins and 16 upregulated proteins (6). It was suggested that either the loss
of tumor suppressor genes or oncogene overexpression would be
involved in the neoplastic cell transcription of normal cells.
Among these differential displayed proteins, the expression of the
SET oncoprotein showed a significant decrease after the treatment
with As4S4, which indicated the importance of
SET in the regulation of signal transduction pathways (6).
SET oncoprotein expression is often associated with
cell proliferation and tumorigenesis (8,9).
Therefore, in the present study, a eukaryotic cell expression
plasmid was constructed and transfected into human 293T cells by
lipofection. Transient cell transfection resulted in a high level
of SET oncoprotein expression, which was not regulated by natural
promoters. A fluorescence marker was included in the expression
cassette in order to indicate the expression and localization of
the target protein in the transfected cells. SET expression in the
pEGFP-N1-SET-transfected group compared with the control group was
monitored by RT-qPCR and western blotting. EGFP was expressed in
the pEGFP-N1- and pEGFP-N1-SET-transfected groups, but only
pEGFP-N1-SET effectively exhibited SET overexpression. This result
demonstrated that the target gene had been successfully
transfected.
As a nuclear protein, SET has been identified to
play a key role in the inhibition of normal histone acetylation and
the demethylation of ectopically methylated DNA, ultimately leading
to the inhibition of gene silencing and transcriptional activation
(4). It has been reported that SET
is overexpressed in chronic myeloid leukemia and acute myeloid
leukemia, in which it is correlated with BCR/ABL expression and
activity, resulting in the inhibition of PP2A (5,10).
In addition, clinical data have indicated that the increasing
expression of the SET gene corresponds to higher degrees of tumor
malignancy (11), suggesting an
oncogenic role of SET in tumorigenesis. In in vitro
experiments, SET was identified to promote the progression of the
cell cycle by modulating the activities of CDKs, including CDK1,
CDK2 and CDK5, by interacting with the activators and/or inhibitors
of the CDKs (12). In the present
study, the functional activity of SET was shown to be present in
the 293T cell line. The accumulation of cells in the S and
G2/M phases was observed for the 293T cells, which may
be associated with the anti-apoptosis function of SET, suggesting
that the overexpression of SET may exert its carcinogenesis through
cell cycle arrest. Therefore, SET may be a potential target of gene
therapy for leukemia and other cancers in the future.
It has been demonstrated that SET overexpression is
a key contributing mechanism involved in the PP2A inhibition
observed in tumors (1). Therefore,
the change in PP2A level was detected in the following experiments
of the present study. The relative PP2A mRNA and protein expression
levels in the pEGFP-N1-SET-transfected 293T cells were
down-regulated compared with those in the pEGFP-N1-transfected
cells. The results showed that SET mediated the decreased
expression of PP2A may be of greater importance in light of the
potential role of PP2A in leukemogenesis and other cancers. PP2A is
a major serine/threonine phosphatase in eukaryotic cells, which
consists of a 36-kD catalytic subunit C (PP2Ac), a regulatory
subunit A with a molecular mass of 65 kD, and a third regulatory
subunit B (13). As a tumor
suppressor, PP2A plays an irreplaceable role in a number of
cellular processes, such as cellular metabolism, DNA replication,
transcription, RNA splicing, translation, cell cycle progression
and cell transformation (14).
Furthermore, PP2A exhibits a positive regulatory function in
apoptosis, mediating the dephosphorylation and inactivation of the
anti-apoptotic Bcl-2 and activation of the pro-apoptotic factor Bad
(15). As a result, PP2A has been
noted as a potential therapeutic target in leukemia patients
(9,16,17).
SET may promote the suppression of PP2A by binding directly to the
catalytic subunits of PP2A and causing hyperphosphorylation of its
τ unit (18). Another study has
shown that SET could activate the mitogen-activated protein kinases
(MAPK) kinase/extracellular signal-regulated kinases/MAPK signaling
pathway, which is involved with cell spreading, motility and death
receptor-elicited apoptosis. Moreover, ERK has interactions with
PP2A, suggesting that cell migration may be regulated through a
SET-PP2A-ERK signaling complex (19). Therefore, the overexpression of the
SET oncoprotein may result in the inactivation of PP2A and thus the
deactivation of PP2A target gene dephosphorylation, leading to
apoptotic resistance in the tumor cells.
Our previous study showed that
As4S4-induced apoptosis was associated with a
reduced level of SET mRNA and protein expression in NB4-R1 cells
(Wang et al, unpublished data). Therefore, the present study
analyzed Bcl-2 (an anti-apoptotic protein) and Bax (a proapoptotic
protein) expression in transfected 293T cells in order to determine
if these Bcl-2 family members played a role in
As4S4-induced apoptosis. The Bcl-2 expression
level was increased in the pEGFP-N1-SET-transfected 293T cells
compared with the expression level in the pEGFP-N1-transfected
control group. Additionally, the Bcl-2 expression level was
decreased in the pEGFP-N1-transfected 293T cells treated with
As4S4 and was further decreased in the
pEGFP-N1-SET-transfected cells. Bax expression was increased
slightly in the pEGFP-N1-transfected 293T cells treated with
As4S4 and was further increased in the
pEGFP-N1-SET-transfected 293T cells. These results demonstrated
that transfection with SET led to upregulated Bcl-2 expression and
down-regulated Bax expression; these changes in expression may be
associated with the SET carcinogenic effect observed in previous
studies (8,9). The susceptibility of cells to
apoptosis may be determined by the ratio between anti-apoptotic and
proapoptotic members of the Bcl-2 family. The decrease in the
Bcl-2/Bax ratio leads to the translocation of Bax from the
cytoplasm to the mitochondria, promoting the release of cytochrome
c and the activation of caspases (20,21).
The present results also suggested that SET has suppressive effects
on apoptosis. However, further studied are required.
In conclusion, the present results showed that the
SET gene could lead to the promotion of 293T cell proliferation and
the inhibition of PP2A expression. Overexpression of SET increased
the protein expression of Bcl-2 and decreased the expression of Bax
in the cells, and their susceptibility to
As4S4-induced apoptosis was decreased. As a
key factor in carcinogenesis, this study identified SET as a
critical gene in As4S4-induced apoptosis and
a potential novel effective therapeutic target for leukemia and
other cancers. Further studies will be focused on defining the
mechanism through which SET regulates PP2A levels and the role of
such regulation in cell differentiation, growth and
carcinogenesis.
Acknowledgments
This study was supported by the Natural Science
Foundation of China (grant no. 81000218). The author would like to
express gratitude to Dr. Xinyang Wang for providing access to the
Oncology Research Laboratory, Key Laboratory of Environment and
Genes Related to Diseases, Ministry of Education (Xi'an, China) to
complete the experiment.
References
|
1
|
Cristóbal I, Garcia-Orti L, Cirauqui C,
Cortes-Lavaud X, García-Sánchez MA, Calasanz MJ and Odero MD:
Overexpression of SET is a recurrent event associated with poor
outcome and contributes to protein phosphatase 2A inhibition in
acute myeloid leukemia. Haematologica. 97:543–550. 2012. View Article : Google Scholar :
|
|
2
|
von Lindern M, van Baal S, Wiegant J, Raap
A, Hagemeijer A and Grosveld G: Can, a putative oncogene associated
with myeloid leukemogenesis, may be activated by fusion of its 3′
half to different genes: Characterization of the set gene. Mol Cell
Biol. 12:3346–3355. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Li M, Guo H and Damuni Z: Purification and
characterization of two potent heat-stable protein inhibitors of
protein phosphatase 2A from bovine kidney. Biochemistry.
34:1988–1996. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cervoni N, Detich N, Seo SB, Chakravarti D
and Szyf M: The oncoprotein Set/TAF-1beta, an inhibitor of histone
acetyltransferase, inhibits active demethylation of DNA,
integrating DNA methylation and transcriptional silencing. J Biol
Chem. 277:25026–25031. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li M, Makkinje A and Damuni Z: The myeloid
leukemia-associated protein SET is a potent inhibitor of protein
phosphatase 2A. J Biol Chem. 271:11059–11062. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jun Qi, He P, Chen W, Wang H, Wang X and
Zhang M: Comparative proteome study of apoptosis induced by
As4S4 in retinoid acid resistant human acute
promyelocytic leukemia NB4-R1 cells. Leuk Res. 34:1506–1516. 2010.
View Article : Google Scholar
|
|
7
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
8
|
Lee SG, Park TS, Cho SY, Lim G, Park GJ,
Oh SH, Cho EH, Chong SY and Huh JY: T-cell acute lymphoblastic
leukemia associated with complex karyotype and SET-NUP214
rearrangement: A case study and review of the literature. Ann Clin
Lab Sci. 41:267–272. 2011.PubMed/NCBI
|
|
9
|
Adler HT, Nallaseth FS, Walter G and
Tkachuk DC: HRX leukemic fusion proteins form a heterocomplex with
the leukemia-associated protein SET and protein phosphatase 2A. J
Biol Chem. 272:28407–28414. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Neviani P, Santhanam R, Trotta R, Notari
M, Blaser BW, Liu S, Mao H, Chang JS, Galietta A, Uttam A, et al:
The tumor suppressor PP2A is functionally inactivated in blast
crisis CML through the inhibitory activity of the BCR/ABL-regulated
SET protein. Cancer Cell. 8:355–368. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ouellet V, Le Page C, Guyot MC, Lussier C,
Tonin PN, Provencher DM and Mes-Masson AM: SET complex in serous
epithelial ovarian cancer. Int J Cancer. 119:2119–2126. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kumar RN, Radhakrishnan R, Ha JH and
Dhanasekaran N: Proteome analysis of NIH3T3 cells transformed by
activated Galpha12: Regulation of leukemia-associated protein SET.
J Proteome Res. 3:1177–1183. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Janssens V and Goris J: Protein
phosphatase 2A: A highly regulated family of serine/threonine
phosphatases implicated in cell growth and signalling. J Biol Chem.
353:417–439. 2001.
|
|
14
|
Westermarck J and Hahn WC: Multiple
pathways regulated by the tumor suppressor PP2A in transformation.
Trends Mol Med. 14:152–160. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Van Hoof C and Goris J: Phosphatases in
apoptosis: To be or not to be, PP2A is in the heart of the
question. Biochim Biophys Acta. 1640:97–104. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Neviani P, Santhanam R, Oaks JJ, Eiring
AM, Notari M, Blaser BW, Liu S, Trotta R, Muthusamy N,
Gambacorti-Passerini C, et al: FTY720, a new alternative for
treating blast crisis chronic myelogenous leukemia and Philadelphia
chromosome-positive acute lymphocytic leukemia. J Clin Invest.
117:2408–2421. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu Q, Zhao X, Frissora F, Ma Y, Santhanam
R, Jarjoura D, Lehman A, Perrotti D, Chen CS, Dalton JT, et al:
FTY720 demonstrates promising preclinical activity for chronic
lymphocytic leukemia and lymphoblastic leukemia/lymphoma. Blood.
111:275–284. 2008. View Article : Google Scholar
|
|
18
|
Arnaud L, Chen S, Liu F, Li B, Khatoon S,
Grundke-Iqbal I and Iqbal K: Mechanism of inhibition of PP2A
activity and abnormal hyperphosphorylation of tau by I2 (PP2A)/SET.
FEBS Lett. 585:2653–2659. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lam BD, Anthony EC and Hordijk PL:
Cytoplasmic targeting of the proto-oncogene SET promotes cell
spreading and migration. FEBS Lett. 587:111–119. 2013. View Article : Google Scholar
|
|
20
|
Rong Y and Distelhorst CW: Bcl-2 protein
family members: Versatile regulators of calcium signaling in cell
survival and apoptosis. Annu Rev Physiol. 70:73–91. 2008.
View Article : Google Scholar
|
|
21
|
Martinou JC and Youle RJ: Mitochondria in
apoptosis: Bcl-2 family members and mitochondrial dynamics. Dev
Cell. 21:92–101. 2011. View Article : Google Scholar : PubMed/NCBI
|