Introduction
Acute myocardial infarction (AMI) is a
life-threatening episode of coronary artery disease, and an as yet
unresolved clinical issue with high morbidity and mortality.
Insufficient blood supply and oxidative stress result in necrosis
of cardiac tissue, pathological remodeling and left ventricular
dysfunction (1–3). An early and accurate diagnosis of AMI
is essential for an optimal treatment outcome. Therefore, new
approaches that are able to complement and improve current
strategies for AMI diagnosis are urgently needed.
Recent discoveries have revealed the existence of
stable cardiomyocyte-enriched microRNAs (miRNAs) circulating in
human blood cells or plasma/serum (4,5).
miRNAs are small, non-coding RNA molecules, 20–25 nucleotides long,
which inhibit gene expression by promoting messenger RNA (mRNA)
degradation or inhibiting translation (6–8). It
is noteworthy that numerous studies have revealed that some
fraction of the circulating miRNAs is secreted from healthy or
damaged cells (9). The fact that
these miRNAs are able to be detected in peripheral blood and are
relatively stable in serum, plasma and other biofluids makes them
potentially useful in aiding diagnosis or guiding therapy through
rapid and easy tests eliminating the necessity of performing an
invasive procedure (10,11).
The present study aimed to compare the miRNA
profiles in plasma samples of patients on the first day of AMI
(admission) with those from the identical patients collected six
months after AMI (stable phase) in order to identify differentially
expressed miRNAs that could be potentially dysregulated in response
to early myocardial damage. The most promising miRNAs were
additionally studied using a set of AMI serum samples from a second
independent cohort and a control group of patients with a stable
coronary artery disease (CAD).
Materials and methods
Patients
Sixteen patients for the study group and fourteen
patients for the validation group, diagnosed with ST-segment
elevation myocardial infarction (STEMI), were randomly selected
from our previously described cohorts of patients admitted to the
Medical University of Warsaw and the Medical University of
Bialystok (12). The control group
comprised seven age- and sex-matched individuals selected from a
cohort of patients with a stable CAD and no history of myocardial
infarction (MI). The whole cohort of CAD patients has been
characterized in our previous study (12).
The design and conduct of this study complied with
the Declaration of Helsinki. The protocol of the study was approved
by the Ethics Committees of the Medical University of Warsaw and
the Medical University of Bialystok. Written informant consent was
obtained from all patients.
Plasma and serum collection, and
hemolysis assessment
Venous whole blood samples (4–8 ml) were drawn from
the patients diagnosed with STEMI at two time points: On the first
day of AMI (admission), and six months following AMI using standard
phlebotomy techniques. For the study group, plasma was isolated
using BD Vacutainer® CPT™ glass tubes with sodium
citrate (BD Biosciences, Franklin Lakes, NJ, USA), following the
manufacturer's protocol. For the validation and control groups,
blood samples were drawn into serum separator tubes (Profilab s.c.,
Warsaw, Poland) according to the manufacturer's instructions. The
plasma and serum were transferred into fresh tubes and stored at
−80°C prior to subsequent analysis.
Oxyhemoglobin was assayed in all plasma and serum
samples analyzed from the study, validation and control groups.
Absorbance at λ=414 nm was measured spectrophotometrically
(NanoDrop ND-1000; Thermo Fisher Scientific, Inc., Waltham, MA,
USA). Additionally, at the profiling stage, hemolysis in the plasma
samples was assessed using two miRNAs available on the Serum/Plasma
Focus miRNA Polymerase Chain Reaction (PCR) panel: miRNA-451a,
which is specific to erythrocytes, and miRNA-23a-3p, which is
unaffected by hemolysis. The ΔCp (crossing point) values for
(miR-23a-3p - miR-451a) were calculated. Samples with a ΔCp value
>7.0 were likely to have undergone hemolysis. Both the
absorbance measurements at λ=414 nm (data not shown) and the ΔCp
data (Fig. 1) indicated that no
serum and plasma sample was affected by hemolysis.
miR NA isolation, complementary DNA
(cDNA) synthesis and quality controls
Total RNA was extracted from 200 µl
plasma/serum using an miRCURY™ RNA Isolation kit - Biofluids
(Exiqon A/S, Vedbaek, Denmark). To improve the yield and
reproducibility between isolations, 1.25 µg/ml MS2
bacteriophage RNA carrier (Roche Diagnostics GmbH, Mannheim,
Germany) was added at the beginning of the procedure. To control
isolation efficiency and yield, three synthetic RNA spike-ins
(UniSp2, UniSp4 and UniSp5; Exiqon A/S) were added to the samples
at concentrations recommended by the manufacturer. Total RNA was
eluted with 50 µl ribonuclease-free water and stored at
−80°C prior to analysis. cDNA was synthesized from purified miRNA
using the miRCURY™ LNA™ microRNA PCR, Polyadenylation and cDNA
synthesis kit II from Exiqon A/S, according to the manufacturer's
protocol. During cDNA synthesis, two spike-ins (UniSp6 and
cel-miR-39; Exiqon A/S) were added to detect the presence of
potential inhibitors in the cDNA synthesis process or in reverse
transcription-quantitative PCR (RT-qPCR). The quality control
analysis was performed according to the protocol provided in the
manual (http://www.exiqon.com/ls/Documents/Scientific/QC-PCR-Panel-Manual.pdf).
All samples passed the criteria and were included in further
studies.
miRNA profiling
For initial screening, quantification of the miRNA
levels in samples taken from patients with AMI [both on the first
day of AMI (admission) and six months following AMI] was performed
by using the Serum/Plasma Focus microRNA PCR panel, Version V3
(Exiqon A/S) in a 96-well format, which was designed to detect the
179 most expressed miRNAs in human serum/plasma. RT-qPCR reactions
were performed using ExiLENT SYBR® Green master mix
(Exiqon A/S) according to the protocol provided by the
manufacturer. Negative controls (no template) were performed and
profiled in an identical manner as for the samples. The
amplification was performed in a LightCycler® 480
Real-Time PCR system (Roche Diagnostics, Basel, Switzerland). The
amplification curves were analyzed using the Roche LC software
(version 1.5), both for determination of the Cp values (by the
second derivative method) and for melting curve analysis.
Quantification of individual miRNAs
Selected miRNAs whose levels were found to differ
between patients on admission and six months following AMI were
subjected to a subsequent validation step by RT-qPCR. The specific
microRNA LNA™ PCR primer sets and ExiLENT SYBR® Green
Master Mix (Exiqon A/S) were used to assess the presence of
individual miRNAs in serum samples according to the manufacturer's
protocol.
miRNA RT-qPCR data analysis
RT-qPCR results were analyzed using the GenEx
software, version 6.0 (MultiD Analyses AB, Göthenburg, Sweden).
Data obtained for the negative control plate were subtracted from
the data for the miRNA PCR panels. Only miRNA species with a Cp
value <37 and at least 5 points below the negative control Cp
value were included in the data analysis. For the profiling study,
the expression data were normalized to a global mean. A logarithmic
transformation (log2) was used to normalize the
expression data in the profiling stage. The geNorm and NormFinder
algorithms (Exiqon A/S software) were used to select the reference
gene for the validation studies. The data were normalized to
miR-19b-3p as a stable endogenous reference gene, and UniSp2 as a
stable exogenous reference gene.
Prediction and functional analysis of
miRNA targets
Ingenuity Pathway Analysis (IPA; www.ingenuity.com; Qiagen, Inc., Valencia, CA, USA)
was used to search mRNA targets for dysregulated miRNAs. To avoid
exceeding the maximum gene list size allowed by IPA, the miRNAs
were analyzed using the microRNA Target Filter limited to
experimentally validated miRNA-mRNA interactions. Target genes were
further analyzed for over-represented biological functions and
canonical pathways using the IPA database.
Statistical analysis
Statistical analyses were performed using R 3.1.2
software (The R Project for Statistical Computing; https://www.R-project.org). The Shapiro-Wilk test was
used to test for normal distribution of continuous variables, and
subsequently, continuous variables were expressed as the mean ±
standard deviation for normally distributed ones and the median
(first quartile - third quartile) for the variables that deviated
from a normal distribution. Categorical variables were presented as
frequencies and percentages. Student's t-test (for normally
distributed variables) and the Mann-Whitney test (for the variables
deviating from a normal distribution) were used to compare
continuous variables. Fisher's exact test was used to compare
categorical variables. Statistical significance between miRNAs that
were differentially expressed in study and validation groups was
determined using either a paired, two-tailed Student's t-test
(admission compared with six months following AMI) or an unpaired,
two-tailed Student's t-test (admission compared with the control
group). P<0.05 was taken to indicate a statistically significant
value. Principal component analysis (PCA) was performed using GenEx
software (version 6.0; MultiD Analyses AB). Receiver operating
characteristic (ROC) curve analysis and the area under the curve
(AUC) were used to estimate the ability of biomarkers to
distinguish the AMI group from the control group. The optimal
cut-off points for each miRNA were determined using the highest sum
of sensitivity and specificity.
Results
Patient characteristics
In the present study, patients with STEMI who were
treated with primary percutaneous revascularization were included.
The mean age of participants was 54.9±11.3 years for the study
group (n=16) and 58.2±11.1 years for the validation group (n=14).
Clinical characteristics of patients from the two groups are shown
in Table I.
 | Table IClinical characteristics of patients
from study and validation groups. |
Table I
Clinical characteristics of patients
from study and validation groups.
| Characteristic | Study group
(n=16) | Validation group
(n=14) | P-value |
|---|
| Gender
(female/male) | 3/13
(18.8%/81.2%) | 0/14
(0.0%/100.0%) | 0.200 |
| Age (years) | 54.9±11.3 | 58.2±11.1 | 0.422 |
| BMI
(kg/m2) | 26.8±2.3 | 28.0±4.3 | 0.345 |
| Smoking | 7 (43.8%) | 8 (57.1%) | 0.715 |
| Hypertension | 4 (25.0%) | 10 (71.4%) | 0.026 |
| Diabetes | 2 (12.5%) | 2 (14.3%) | >0.999 |
|
Hypercholesterolemia | 9 (56.2%) | 10 (71.4%) | 0.466 |
| Previous MI | 0 (0.0%) | 0 (0.0%) | NA |
| Anterior MI | 9 (60.0%) | 5 (35.7%) | 0.272 |
| Previous
revascularization | 0 (0.0%) | 0 (0.0%) | NA |
| Non-coronary
atherosclerosis | 0 (0.0%) | 1 (7.1%) | 0.467 |
| WBC
(×103/µl) | 12.2±3.3 | 13.0±3.6 | 0.506 |
| NT-proBNP
(pg/ml) | 1,052.4
(458.1–1,504.3) | 784.1
(514.5–1,640.0) | 0.861 |
| LVEF (%) | 49.9±11.3 | 41.0±10.0 | 0.039 |
| Medication |
| Aspirin | 16 (100.0%) | 14 (100.0%) | NA |
| Clopidogrel | 15 (93.8%) | 14 (100.0%) | >0.999 |
| Beta blockers | 16 (100.0%) | 13 (92.9%) | 0.467 |
| ACE inhibitors | 16 (100.0%) | 14 (100.0%) | NA |
| Statins | 16 (100.0%) | 14 (100.0%) | NA |
| Diuretics | 6 (37.5%) | 6 (42.9%) | >0.999 |
Identification of differentially
expressed miRNAs in the plasma of patients with AMI
miRNA profiling was performed on plasma samples
derived from patients on the first day of AMI (n=16), and on
samples from the identical patients collected six months following
AMI (n=16, stable phase), which reduced the impact of
inter-individual variability. Following data analysis, miRNA
candidates were selected on the basis of fulfillment of the
criterion of significance (P<0.05) in the comparison between
admission and six months following AMI. A total of 32 miRNAs (14
up- and 18 down-regulated) were differentially quantified in the
acute phase of MI compared with the stable phase following MI
(Table II).
 | Table IIDifferential miRNAs between the first
day of AMI and the stable phase following myocardial infarction in
the study group. |
Table II
Differential miRNAs between the first
day of AMI and the stable phase following myocardial infarction in
the study group.
| miRNA | Fold change | P-value |
|---|
| hsa-miR-133b | 45.764 | 9.0E-06 |
| hsa-miR-133a | 30.127 | 1.6E-04 |
| hsa-miR-208a | 27.074 | 3.0E-06 |
| hsa-miR-1 | 12.139 | 2.0E-03 |
| hsa-miR-30a-5p | 3.643 | 7.0E-03 |
| hsa-miR-629-5p | 2.573 | 4.5E-02 |
| hsa-miR-20b-5p | 2.570 | 2.4E-02 |
| hsa-miR-22-5p | 2.360 | 1.1E-02 |
| hsa-miR-145-5p | 1.776 | 9.0E-03 |
| hsa-miR-22-3p | 1.507 | 2.0E-02 |
| hsa-miR-486-5p | 1.402 | 3.8E-02 |
| hsa-miR-451a | 1.349 | 3.3E-02 |
| hsa-miR-92a-3p | 1.327 | 1.0E-02 |
| hsa-miR-93-5p | 1.133 | 3.1E-02 |
| hsa-let-7i-5p | −1.198 | 1.8E-02 |
|
hsa-miR-148b-3p | −1.226 | 3.4E-02 |
|
hsa-miR-103a-3p | −1.237 | 1.3E-02 |
| hsa-miR-223-3p | −1.237 | 1.7E-02 |
| hsa-miR-652-3p | −1.251 | 3.6E-02 |
| hsa-miR-26b-5p | −1.257 | 4.2E-02 |
| hsa-miR-107 | −1.272 | 7.0E-03 |
|
hsa-miR-199a-3p | −1.278 | 3.0E-02 |
|
hsa-miR-151a-5p | −1.326 | 9.0E-03 |
| hsa-miR-30b-5p | −1.414 | 3.0E-03 |
|
hsa-miR-181a-5p | −1.436 | 4.6E-02 |
| hsa-miR-142-3p | −1.693 | 3.6E-04 |
|
hsa-miR-374b-5p | −1.829 | 3.6E-04 |
| hsa-miR-335-5p | −2.384 | 1.5E-02 |
| hsa-miR-505-3p | −2.873 | 5.0E-03 |
| hsa-miR-885-5p | −2.898 | 4.0E-02 |
| hsa-miR-326 | −3.535 | 7.0E-03 |
|
hsa-miR-301a-3p | −4.147 | 7.0E-03 |
PCA was performed on the miRNA results from the
analyzed samples to determine how the 32 differentially expressed
miRNAs were distributed among the samples from the first day of
AMI, and those collected six months afterwards. As shown in
Fig. 2, the PCA clearly separated
the plasma samples on admission from those collected six months
following AMI. This suggests that the observed miRNA differences
are associated with the pathophysiology of MI, and these miRNAs
might constitute an early biomarker signature for AMI.
Validation of selected miRNAs in an
independent group of patients with AMI
miRNA candidates for validation were selected
following an extensive review of the literature on the basis of
their inferred relevance to cardiovascular disease. Additionally,
the potential candidates were filtered for highly expressed miRNAs
according to inspection of their raw Cp values in individual
samples. The validation was performed on serum samples of an
independent patient group on admission (n=14) and samples from the
identical patients collected six months following AMI (n=14), and a
control group (n=7). The levels of these miRNAs were quantified
using RT-qPCR for individual miRNAs. Three miRNAs were further
investigated: miR-133b, which is known to be associated with MI;
miR-374b-5p, which limited literature has suggested has an
involvement in MI; and miR-22-5p, which has not yet been reported
to be associated with MI, and therefore may be a possible novel
biomarker. Two of the miRNAs, miR-133b and miR-22-5p, demonstrated
significant differences in the comparison between admission and six
months following AMI, the direction and magnitude of the changes
reflecting reasonably well those found in the profiling stage
(Table III). These two miRNAs
were further investigated in the patients with AMI and the control
group. As shown in Table IV, the
expression levels of miR-133b and miR-22-5p were significantly
increased in patients with AMI compared with the control group.
 | Table IIIRT-qPCR quantification of selected
miRNAs on admission vs. six months following AMI in study and
validation groups. |
Table III
RT-qPCR quantification of selected
miRNAs on admission vs. six months following AMI in study and
validation groups.
| miRNA | Admission vs. six
months
|
|---|
Study group
| Validation group
|
|---|
| Fold change | P-value | Fold change | P-value |
|---|
| hsa-miR-133b | 45.764 | <0.001 | 5.449 | <0.05 |
| hsa-miR-22-5p | 2.360 | <0.05 | 4.872 | <0.01 |
|
hsa-miR-374b-5p | −1.829 | <0.001 | 1.642 | NS |
 | Table IVRT-qPCR quantification of the two
selected miRNAs on admission vs. the control group in the
validation group. |
Table IV
RT-qPCR quantification of the two
selected miRNAs on admission vs. the control group in the
validation group.
| miRNA | Admission vs.
control group
|
|---|
Validation group
|
|---|
| Fold change | P-value |
|---|
| hsa-miR-133b | 7.273 | <0.05 |
| hsa-miR-22-5p | 4.505 | <0.01 |
Diagnostic accuracy of selected
circulating miRNAs
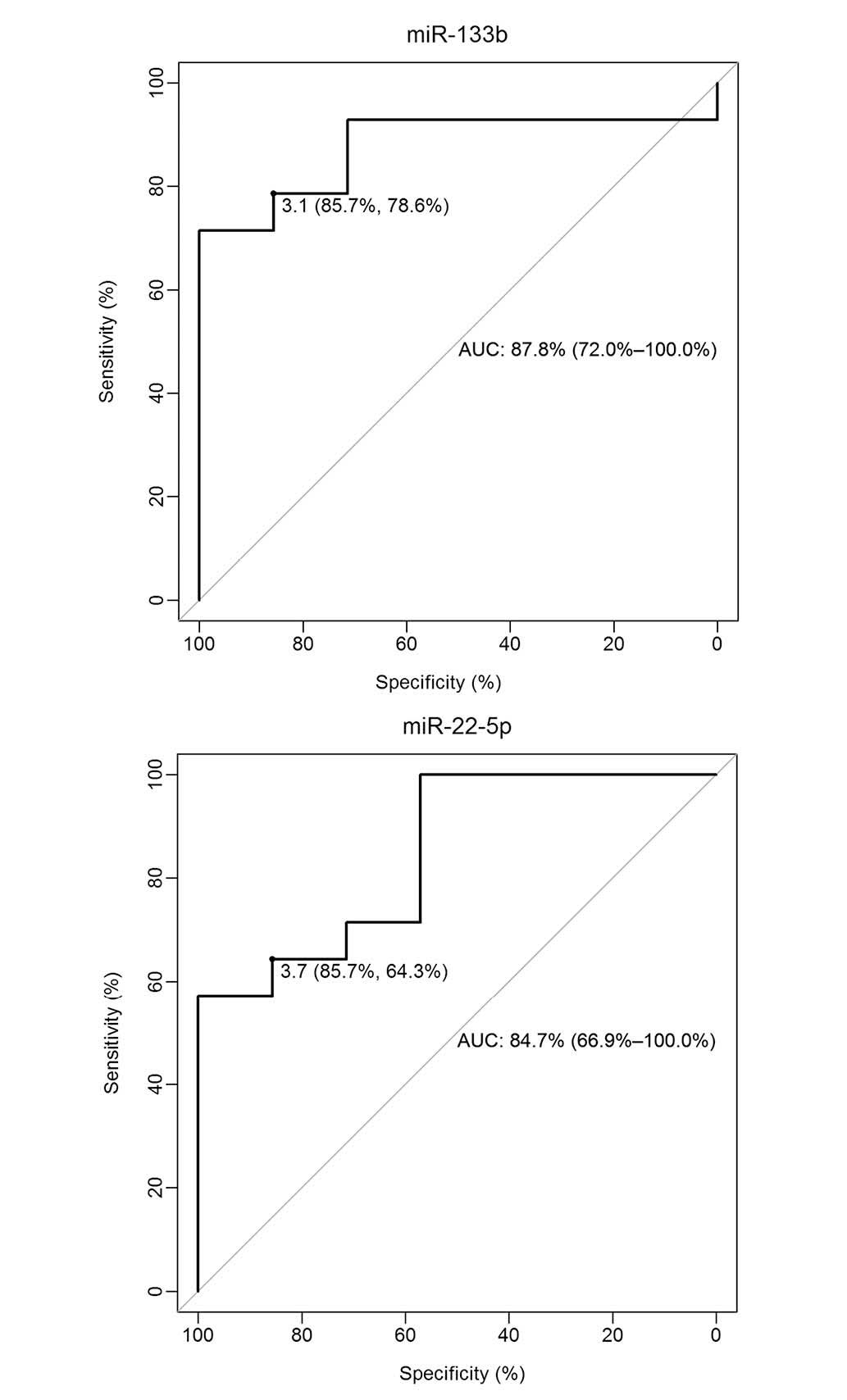
To evaluate the diagnostic value of miR-133b and
miR-22-5p as potential biomarkers of AMI, ROC curve analysis,
together with calculation of the AUC, was performed. As shown in
Fig. 3, the ROC curves of miR-133b
and miR-22-5p reflected a good separation between the patients with
AMI and the control group, with AUC measurements of 87.8% [95%
confidence interval (CI): 72.0–100.0] and 84.7% (95% CI:
66.9–100.0), respectively. ROC curves yielded an optimal cut-off
value of 3.1 for miR-133b, with a sensitivity of 78.6% and a
specificity of 85.7%, and an optimal cut-off value of 3.7 for
miR-22-5p, with a sensitivity of 64.3% and a specificity of 85.7%.
These results suggest that miR-133b and miR-22-5p are of good
diagnostic value for patients with AMI.
Discussion
Previous studies have revealed that heart-specific
miRNAs are released into the circulation during AMI, and therefore
may be used to detect and monitor myocardial injury (13,14).
In the present study it has been confirmed that the well-known
'cardiac miRNAs' of ongoing early myocardial damage, miR-1,
miR-133a, miR-133b and miR-208a, are significantly up-regulated in
AMI. An additional 28 differentially expressed miRNAs that were
apparently associated with AMI were also identified. To determine
the biological significance of miRNAs dysregulated in AMI, in
silico target prediction was performed using the IPA software
(Qiagen, Inc.). Targeting information was available for 22 out of
the 32 miRNAs in the database, resulting in a total of 412
experimentally validated target mRNAs for up-regulated miRNAs, and
a total of 304 experimentally validated mRNAs for down-regulated
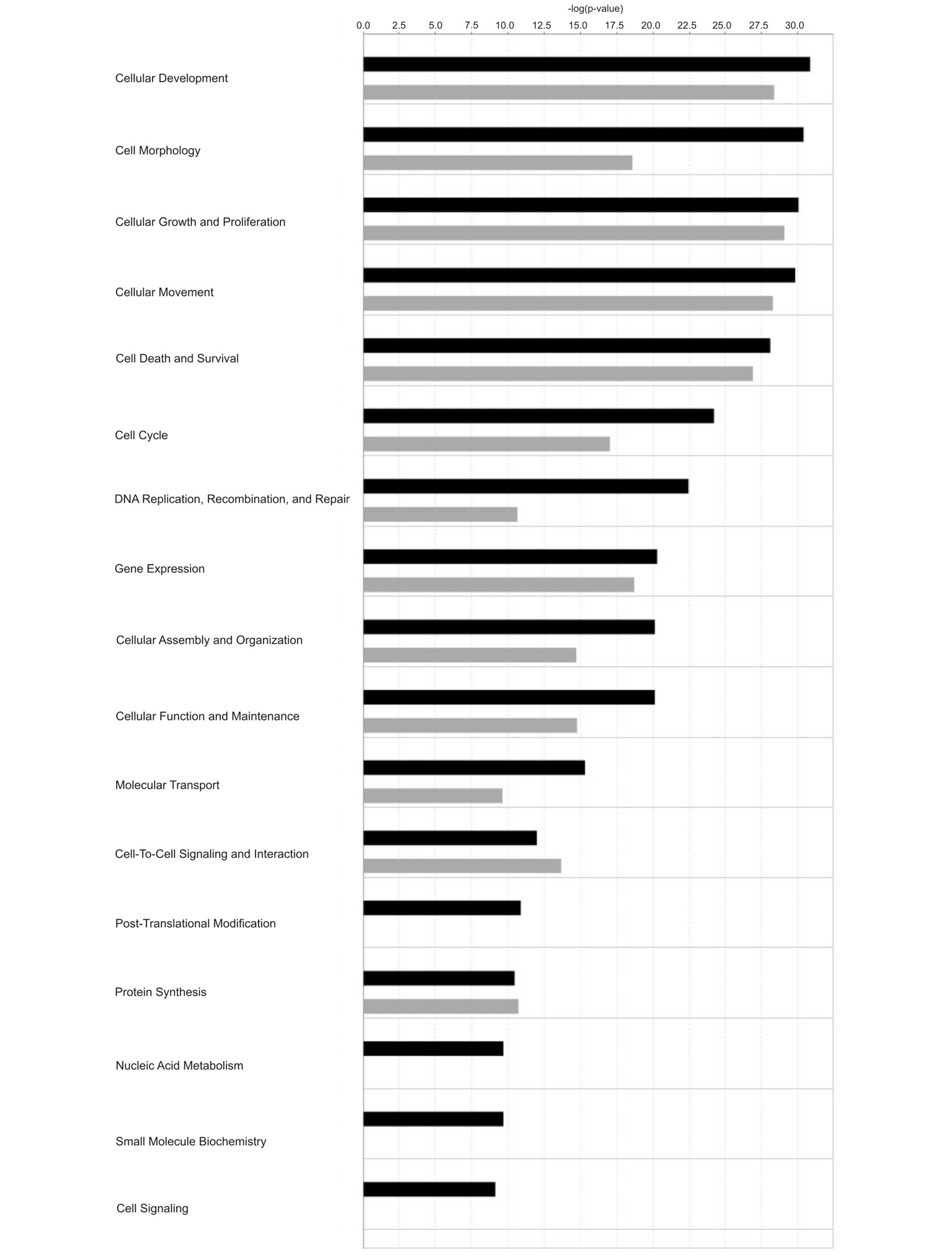
miRNAs. Functional analysis revealed that targets for both up- and
down-regulated miRNAs were generally involved in identical
molecular and cellular functions (Fig.
4). Only post-translational modification, nucleic acid
metabolism, small molecule biochemistry and cell signaling were
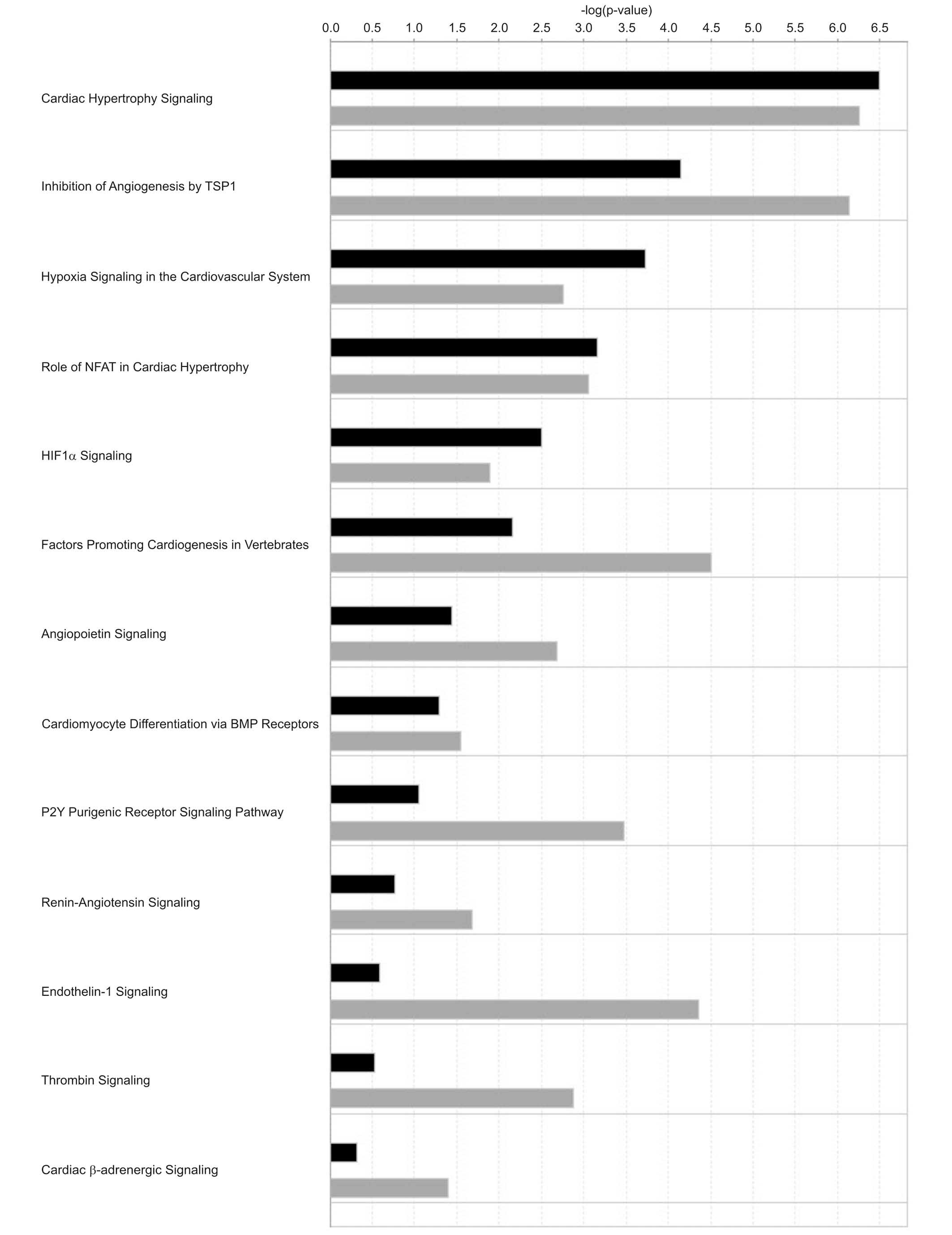
predicted to be associated with up-regulated miRNAs. In addition,
several pathways involved in cardiovascular signaling were revealed
to be associated with the canonical pathway analysis, the most
important being cardiac hypertrophy signaling, inhibition of
angiogenesis by thrombospondin 1 and hypoxia signaling in the
cardiovascular system (Fig. 5).
These findings reveal that the identified miRNAs could have a role
in the pathogenesis of MI through their ability to negatively
regulate the expression of genes that govern processes important
for myocardial function.
Numerous biochemical biomarkers of MI are commonly
used in clinical practice [e.g. cardiac troponins (Tn) I and T,
creatinine kinase isoenzyme MB, N-terminal pro B-type natriuretic
peptide and B-type natriuretic peptide] (15). However, it should be noted that an
increase in the levels of these biochemical biomarkers above
reference values may also occur in various other disease states not
necessarily associated with MI (16). Therefore, selected miRNAs or miRNA
sets, particularly when combined with clinical parameters, are
likely to be more specific biomarkers of MI. Additionally, studying
their mechanism of action should provide an improved understanding
of the changes that occur in the myocardium, and determine the
potential role of extracellular miRNAs as paracrine signaling
molecules.
The up-regulation of miR-133b and miR-22-5p in two
independent patient groups using serum or plasma confirmed the high
diagnostic value of these miRNAs. The ROC curve analysis revealed
that the AUCs of miR-133b and miR-22-5p were 87.8 and 84.7%,
indicating that they may be clinically practicable biomarkers for
AMI diagnosis. The major novel finding reported in the present
study is the up-regulation of miR-22-5p in the acute phase of
STEMI. To date, mir-22-3p originating from the same hairpin has
been studied in depth to elucidate its role in cardiovascular
remodeling (17) and heart failure
(18). To the best of our
knowledge, no previous data regarding a role for miR-22-5p in
cardiovascular diseases is available, albeit a recent study has
demonstrated that up-regulation of mmu-miR-22-5p may prevent
myocardium regeneration in 7-day-old mice (19).
In conclusion, the present study has reported an
altered miRNA expression profile associated with AMI. A group of 32
circulating miRNAs that are significantly up- or down-regulated in
AMI compared with the stable phase of the disease has been
described. The circulating miRNA, miR-22-5p, has been identified as
a novel diagnostic biomarker of AMI.
Abbreviations:
|
AMI
|
acute myocardial infarction
|
|
BNP
|
B-type natriuretic peptide
|
|
CAD
|
coronary artery disease
|
|
CI
|
confidence interval
|
|
IPA
|
ingenuity pathway analysis
|
|
miRNA
|
microRNA
|
|
NT-proBNP
|
N-terminal pro B-type natriuretic
peptide
|
|
PCA
|
principal component analysis
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
STEMI
|
ST-segment elevation myocardial
infarction
|
Acknowledgments
We would like to thank the patients for their
participation in this study. We thank Katarzyna Rawa for technical
assistance in the miRNA profiling analysis. This work was supported
by the National Science Centre, Poland (grant no.
2014/13/N/NZ5/01403) and The National Centre for Research and
Development, Poland (grant no. N R13 0001 06).
References
|
1
|
Hori M and Nishida K: Oxidative stress and
left ventricular remodeling after myocardial infarction. Cardiovasc
Res. 81:457–464. 2009. View Article : Google Scholar
|
|
2
|
Sutton MG and Sharpe N: Left ventricular
remodeling after myocardial infarction: Pathophysiology and
therapy. Circulation. 101:2981–2988. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
White HD and Chew DP: Acute myocardial
infarction. Lancet. 372:570–584. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gilad S, Meiri E, Yogev Y, Benjamin S,
Lebanony D, Yerushalmi N, Benjamin H, Kushnir M, Cholakh H, Melamed
N, et al: Serum microRNAs are promising novel biomarkers. PLoS One.
3:e31482008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K,
Guo J, Zhang Y, Chen J, Guo X, et al: Characterization of microRNAs
in serum: A novel class of biomarkers for diagnosis of cancer and
other diseases. Cell Res. 18:997–1006. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bartel DP: microRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Inui M, Martello G and Piccolo S: MicroRNA
control of signal transduction. Nat Rev Mol Cell Biol. 11:252–263.
2010. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xu J, Zhao J, Evan G, Xiao C, Cheng Y and
Xiao J: Circulating microRNAs: Novel biomarkers for cardiovascular
diseases. J Mol Med (Berl). 90:865–875. 2012. View Article : Google Scholar
|
|
10
|
Oliveira-Carvalho V, Carvalho VO, Silva
MM, Guimarães GV and Bocch EA: MicroRNAs: A new paradigm in the
treatment and diagnosis of heart failure? Arq Bras Cardiol.
98:362–369. 2012.In English, Portuguese, Spanish. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Katoh M: Therapeutics targeting
angiogenesis: Genetics and epigenetics, extracellular miRNAs and
signaling networks (Review). Int J Mol Med. 32:763–767.
2013.PubMed/NCBI
|
|
12
|
Maciejak A, Kiliszek M, Michalak M, Tulacz
D, Opolski G, Matlak K, Dobrzycki S, Segiet A, Gora M and Burzynska
B: Gene expression profiling reveals potential prognostic
biomarkers associated with the progression of heart failure. Genome
Med. 7:262015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang GK, Zhu JQ, Zhang JT, Li Q, Li Y, He
J, Qin YW and Jing Q: Circulating microRNA: A novel potential
biomarker for early diagnosis of acute myocardial infarction in
humans. Eur Heart J. 31:659–666. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Białek S, Górko D, Zajkowska A, Kołtowski
Ł, Grabowski M, Stachurska A, Kochman J, Sygitowicz G, Małecki M,
Opolski G and Sitkiewicz D: Release kinetics of circulating
miRNA-208a in the early phase of myocardial infarction. Kardiol
Pol. 73:613–619. 2015. View Article : Google Scholar
|
|
15
|
Lindahl B: Acute coronary syndrome-the
present and future role of biomarkers. Clin Chem Lab Med.
51:1699–1706. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Iqbal N, Wentworth B, Choudhary R, Landa
Ade L, Kipper B, Fard A and Maisel AS: Cardiac biomarkers: New
tools for heart failure management. Cardiovasc Diagn Ther.
2:147–164. 2012.PubMed/NCBI
|
|
17
|
Huang ZP and Wang DZ: miR-22 in cardiac
remodeling and disease. Trends Cardiovasc Med. 24:267–272. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Goren Y, Kushnir M, Zafrir B, Tabak S,
Lewis BS and Amir O: Serum levels of microRNAs in patients with
heart failure. Eur J Heart Fail. 14:147–154. 2012. View Article : Google Scholar
|
|
19
|
Liu HL, Zhu JG, Liu YQ, Fan ZG, Zhu C and
Qian LM: Identification of the microRNA expression profile in the
regenerative neonatal mouse heart by deep sequencing. Cell Biochem
Biophys. 70:635–642. 2014. View Article : Google Scholar : PubMed/NCBI
|