Introduction
The domestic goat (Capra hircus) is reared
throughout the world, particularly in China, India and other
developing countries (1), and
serves as an important source of fibers and pelts (2). The Laiwu black goat, of which there
are 230,000, is indigenous to Shandong and supplies cashmere, pelts
and meat. The advantages of this breed include the high-quality
cashmere produced, resistance to disease and a high reproductive
rate. With regards to cashmere, the fleece fiber diameter is <12
μm, cashmere length is up to 56 mm, and the maximum cashmere
yield is 300 g (3). The cashmere
growth in Laiwu black goats begins in late July, continuing to
March (3).
The identification of genes involved in cashmere
production may provide an opportunity to improve production
efficiency and product quality and diversity. This may be
accomplished through breeding programs, the development of
transgenic lines or via the manufacture of therapeutic agents that
improve fibers by altering gene expression (4). Previous studies have investigated the
effect of genetic polymorphisms (single nucleotide polymorphisms
and quantitative trait loci) on cashmere growth and regulation
(5,6). Goats have two types of hair
follicles: The primary hair follicle, which produces coarse coat
hair and the secondary hair follicle, which produces cashmere
(1). Comparative transcriptomic
analysis identified 51 genes that were differentially expressed
(DE) in primary and secondary hair follicles (1). In addition, microRNAs and protein
coding genes with potential roles in goat and sheep hair growth
have been identified (7–13). The present study used the Agilent
sheep gene expression array to identify genes and proteins
associated with wool follicle growth and cycling (14,15).
The Agilent sheep gene expression array has previously been
performed on goat samples to investigate the contribution of
mammary epithelial cells to the immune response during the early
stages of Staphylococcus aureus infection (16).
To date, to the best of our knowledge, no microarray
or other transcriptomic analysis has been conducted to investigate
hair and cashmere growth in various skin areas of adult goats. In
addition, only a limited number of studies have described the
molecular characteristics of cashmere growth in the Laiwu black
goat (11,12,17).
The aim of the present study was to analyze and compare gene
expression levels in body (hair and cashmere rich) and groin skin
(no hair or cashmere) during the cashmere growth period using
microarray technology. This may enable the identification of genes
potentially associated with hair and cashmere growth regulation in
the Laiwu black goat.
Materials and methods
Sampling
The protocol of the present study was approved by
the Ethics Committee of Qingdao Agricultural University (Qingdao,
China). The goats were obtained from Qingdao Aote Sheep Farm
(Qingdao, China), and housed in semi-closed sheep houses. Sampling
was performed as described previously (14). Sampling was performed in August,
during the cashmere growth period, on one ram and two ewes, aged 2
years. Full thickness skin from the body (cashmere/wool-bearing)
and groin (no cashmere/wool) were sampled from the three animals
under local anesthesia (2 ml 0.5% procaine hydrochloride; Harbin
Pharmaceutical Group Co., Ltd., Harbin, China) for complementary
DNA microarray experiments.
RNA preparation, microarray hybridization
and data analysis
Total RNA was isolated from the skin samples using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) according to the manufacturer's
instructions. RNA integrity was confirmed by denaturing agarose gel
electrophoresis and RNA was quantified using a NanoDrop®
ND-1000 Spectrophotometer (NanoDrop Technologies; Thermo Fisher
Scientific, Inc.). Total RNA (2.5 μg) from each sample was
doubled and transcribed into fluorescent complementary RNA using a
Quick Amp Labeling kit (Agilent Technologies, Inc., Santa Clara,
CA, USA) according to the manufacturer's instructions.
Subsequently, RNA samples were incubated with the Agilent Sheep
Gene Expression Microarray (Agilent Technologies, Inc.). The
microarray signals were scanned and analyzed as previously
described (14). Clustering
analysis of DE genes was performed using Cluster software version
3.0 (bonsai.hgc.jp/~mdehoon/software/cluster/software.htm)
(18,19) to analyze the similarity in the
expression patterns among different skin areas. The functional
annotation of DE genes was performed using the Database for
Annotation, Visualization and Integrated Discovery (DAVID) gene
annotation tool version 6.6 (david.abcc.ncifcrf.gov/) (20).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) verification
Equal amounts of RNA were reverse-transcribed using
Superscript III (Invitrogen; Thermo Fisher Scientific, Inc.). qPCR
was performed with SYBR Green Master Mix (Roche Applied Science,
Penzberg, Germany) and gene-specific primers. The cycling
conditions consisted of an initial, single cycle for 5 min at 95°C,
followed by 40 cycles of 20 sec at 95°C, 30 sec at 60°C and 30 sec
at 72°C. The primer sequences, melting temperatures and product
sizes are listed in Table I. GAPDH
served as the housekeeping gene, and results were normalized using
the ΔΔCq method (21).
 | Table IPrimers used for qPCR validation. |
Table I
Primers used for qPCR validation.
| Gene | Primer sequence
(5′–3′)
| Tm, °Ca | Target size,
bp |
|---|
| Forward | Reverse |
|---|
| GAPDHb |
ACAGTCAAGGCAGAGAACGG |
CCAGCATCACCCCACTTGAT | 60 | 98 |
| AMP18 (GKN1) |
TCAAGCCCTTGGTATGCTGG |
TGAAGTCCGGCTTCTTGGTC | 60 | 198 |
| CYP1A1 |
TTTCACCCTCGCTCTGAAGG |
AAGTTCTGTGGCCGAGATGG | 60 | 190 |
| Connexin 43 |
TTGTACCCGGGAGGAGACAT |
CTGAGCCCCTCCAAAGACTG | 60 | 101 |
| C-jun |
GGATCAAGGCGGAGAGGAAG |
CTGCGTTAGCATGAGTTGGC | 60 | 224 |
| ITGB1 |
AGCACGGATGAGGTGAACAG |
ATCTCACAGGTTGGCCCTTG | 60 | 389 |
| C3 |
AACAAACGGGATCCCCTGAC |
GAGTTCCCCTGCGTGTTGTA | 60 | 113 |
Network analysis
Groups of genes were analyzed using the Meme
Software Suite version 4.10.2 (meme-suite.org/) to identify common motifs and
cis-regulatory elements (22). Common motifs and
cis-regulatory elements were searched by Patch in the
TRANSFAC® Public version 7.0 database (gene-regulation.com/pub/databases.html) to identify
transcription factor binding sites (23).
Statistical analysis
Comparisons between groups were performed using
Student's t-tests in Microsoft Excel 2007 (Microsoft
Corporation, Redmond, WA, USA). Data are expressed as the mean ±
standard deviation. P≤0.05 was considered to indicate a
statistically significant difference.
Results
Comparative transcriptome analysis
between body and groin skin
Skin samples collected from the two skin areas were
analyzed. A total of 15,008 transcripts (98.68% of all probe sets)
were detected in the two skin areas (data not shown). Following
normalization and statistical analyses, 655 genes were identified
as DE, with 217 upregulated and 438 downregulated in body compared
with groin skin, over the threshold of fold-change (>2.0; data
not shown).
To further investigate the similarity in expression
patterns of gene transcripts in the two skin areas, cluster
analysis was performed using the Cluster 3.0 tool. As presented in
Fig. 1, cluster analysis revealed
differences between body and groin skin.
Using DAVID, 22 of the DE genes were classified into
16 categories according to their functional correlation (data not
shown). A number of these genes could be classified into more than
one category. The majority of the genes associated with hair growth
regulation could be assigned into the intracellular, intracellular
organelle, membrane-bound vesicle, cytoplasmic vesicle, pattern
binding, heparin binding, polysaccharide binding, glycosaminoglycan
binding and cytoplasmic membrane-bound vesicle categories. Other DE
genes, not classified in DAVID, are discussed below.
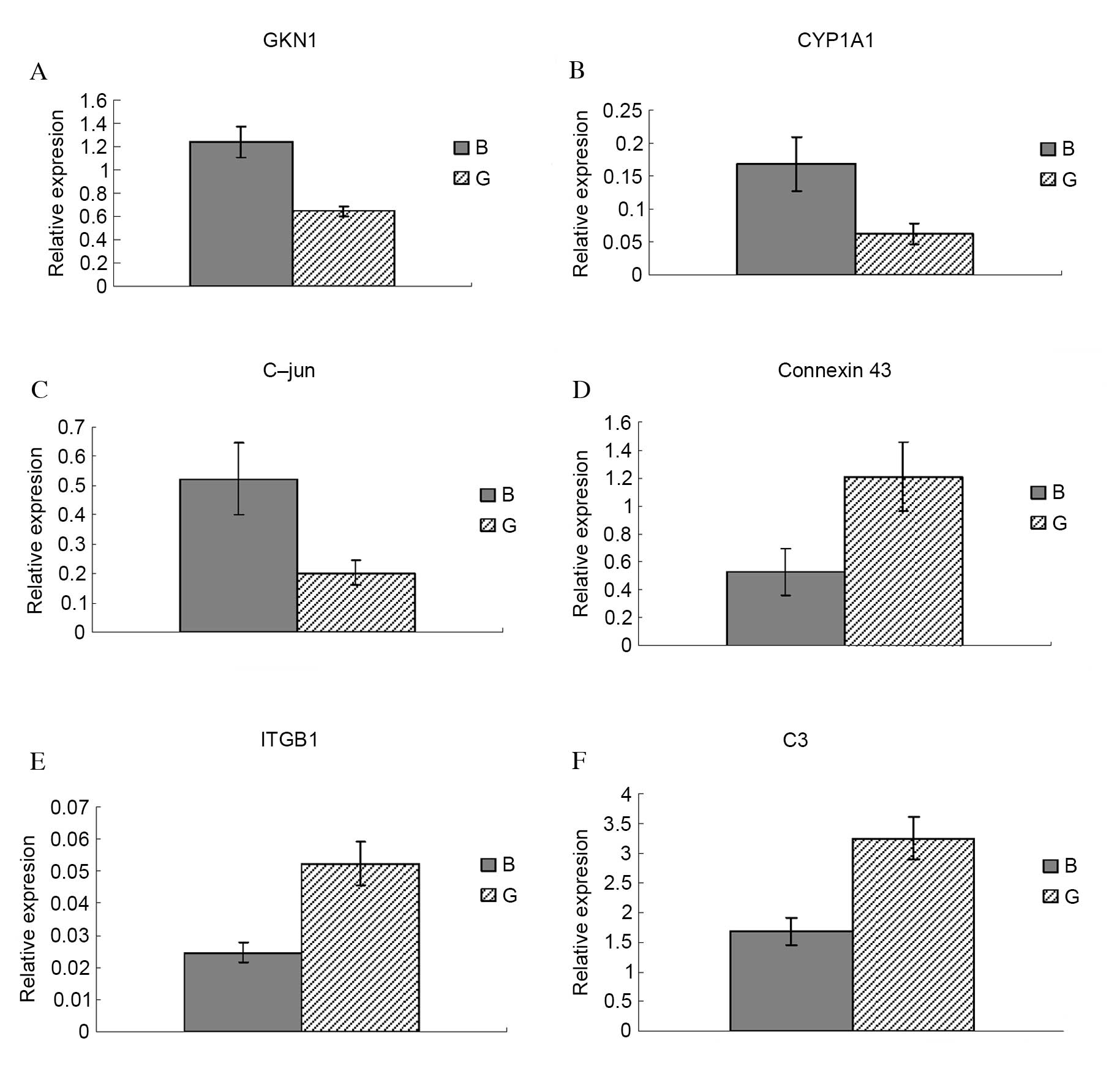
Confirmation of DE genes by RT-qPCR
To confirm the microarray data, six DE genes were
selected for assessment of their relative expression levels by
RT-qPCR: Antrum muscle protein 18 (gastrokine 1; GKN1; Fig. 2A), cytochrome P450 family 1
subfamily A member 1 (CYP1A1; Fig.
2B), C-jun (Fig. 2C), Connexin
43 (Fig. 2D), integrin subunit β1
(ITGB1; Fig. 2E) and complement
component 3 (C3; Fig. 2F). GKN1,
CYP1A1 and C-jun were significantly upregulated in body compared
with groin skin (P=0.013, P=0.025 and P=0.005, respectively).
Connexin 43, ITGB1 and C3 were significantly downregulated in body
compared with groin skin (P=0.041, P=0.022 and P=0.038,
respectively). RT-qPCR results for all six genes were consistent
with the microarray data, therefore validating its reliability.
GKN1 (8.14-fold) was one of the most upregulated
genes in body compared with groin skin in the present study (data
not shown). The other two genes upregulated in body compared with
groin skin, CYP1A1 (2.04-fold) and C-jun (2.04-fold), are involved
in skin disease development and epidermal keratinocyte survival and
differentiation (24,25). Connexin 43 (−19.35-fold) and ITGB1
(−2.45-fold) are associated with differentiation of hair follicle
stem cells (26,27). The downregulated C3 gene
(−8.92-fold) is a core member of the complement system (28).
Analysis of the region 1,800 bp upstream
of the translation start site of DE genes
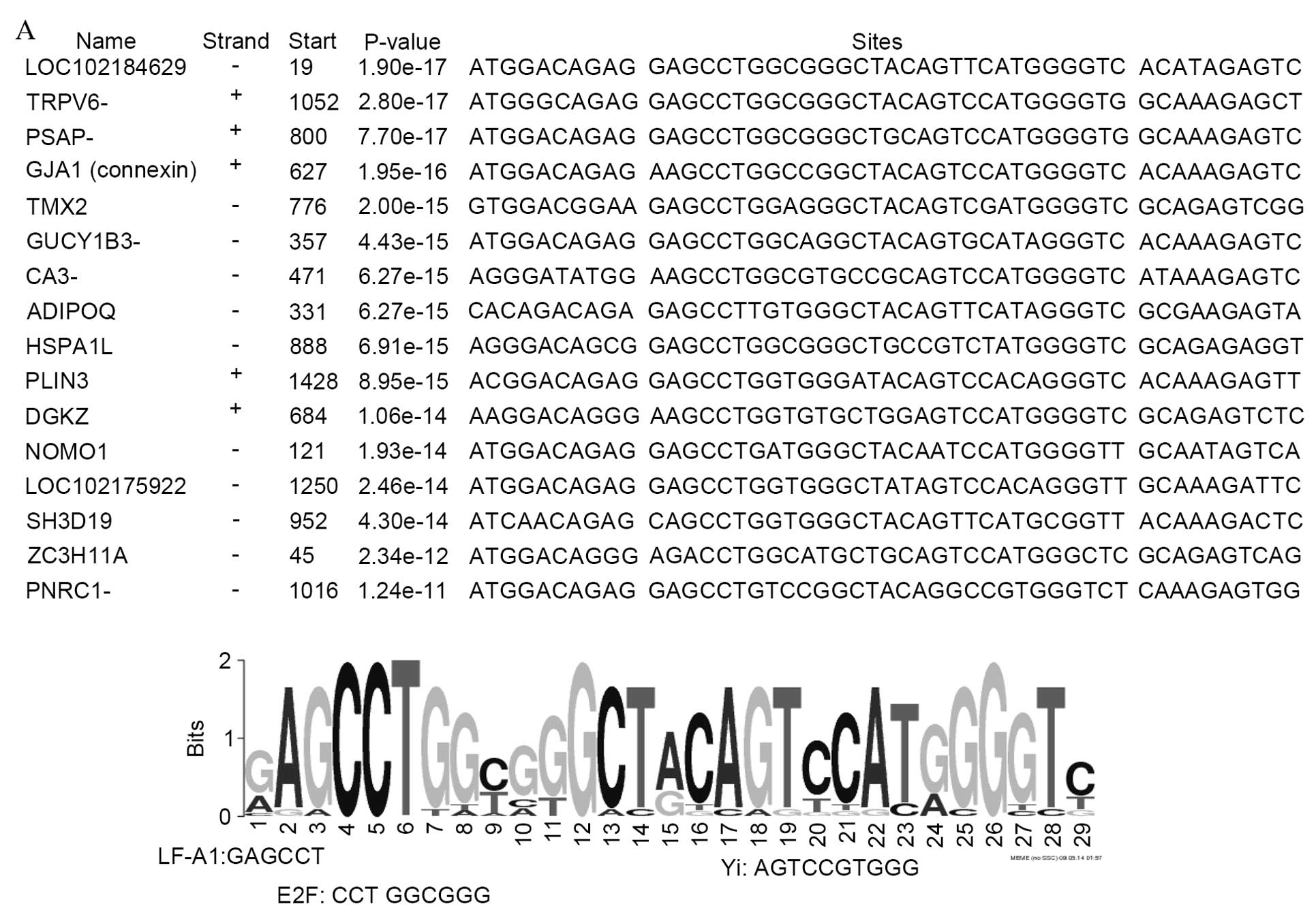
As presented in Fig.
3, three common motifs [41 bp (Fig. 3A), 50 bp (Fig. 3B) and 50 bp (Fig. 3C)] were identified in the upstream
sequences of genes upregulated in body compared with groin skin.
Notably, nuclear factor 1A (NF-1A), Yi, E2 factor (E2F) and cyclic
adenosine monophosphate response element binding (CREB)/CREBβ
binding sites are present in the promoters of a number of
upregulated genes. The promoter regions of genes downregulated in
body compared with groin skin demonstrate 3 common motif sequences
of 29 bp (Fig. 4A), 39 bp
(Fig. 4B) and 50 bp (Fig. 4C). In these regions, binding sites
were identified for the transcription factors LF-A1, Yi, E2F,
Collier/Olfactory-1/early B-cell factor 1 (COE1) and peroxisome
proliferator-activated receptor α (PPARα). In addition, a binding
site (the U site) was identified from the promoter regions of 12
downregulated genes, which has been previously demonstrated to be
present in the olfactory cyclic nucleotide gated channel and type
III cyclase promoters (29).
Discussion
In the present study, the molecular events
associated with wool and cashmere growth control in the Laiwu black
goat were investigated using microarray technology. Statistical
analysis revealed hundreds of genes DE (>2-fold-change) between
the two sampled skin areas. RT-qPCR confirmed the reliability of
the microarray results.
Heat shock protein 70.1 (Hsp70.1) is necessary and
sufficient to accelerate depigmentation in vitiligo-prone Pmel-1
mice (30), while at least three
different mutations of Agouti signaling peptide (ASIP) were
hypothesized to cause the recessive black coat color pattern in
sheep (31–33). A gene duplication affecting
expression of the ovine ASIP gene is responsible for white and
black sheep (32). In addition,
Hsp70.1 was downregulated in body compared with groin skin of the
Aohan fine-wool sheep (34). In
the present study, Hsp70.1 in body skin may facilitate the
regulation of the depigmentation of wool fiber. Upregulation of
ASIP may have a similar role.
Overexpression of Hsp70.1 alone significantly
inhibits aminoglycoside-induced hair cell death in mice (35). However, in the present study
Hsp70.1 was downregulated in body skin of the Laiwu black goat. The
reason for this contradiction remains to be elucidated.
Matrix metalloproteinase 2 (MMP2) activity is
associated with the disappearance of collagen VII during the
invasion of epithelial cords of hair follicles and sweat glands in
human skin (36). MMP2 is involved
in hair growth-associated extracellular matrix remodeling and cell
migration, and may be a downstream effector, through which thymosin
β4 exerts its effect on hair growth (37). However, in the present study MMP2
was significantly downregulated in body compared with groin skin. A
similar result was observed in the Aohan fine wool sheep (38), suggesting that MMP2 functions may
differ in goat and sheep.
Integrins, including α3, α11 and β3, have been
demonstrated to be DE between primary and secondary hair follicles
(11). In keratinocytes there was
revealed to be a log-linear association between the relative
expression levels of β1 integrins (ITGB1) on the cell surface and
proliferative capacity (39). In
addition, ITGB1-mediated signaling is important in human hair
growth control (40). Skin and
hair follicle integrity is crucially dependent on ITGB1 expression
in keratinocytes (41). However,
ITGB1 expression in body skin of the Aohan fine wool sheep was
downregulated >10 fold (38).
The results of the present study were consistent with this,
although the fold change was reduced at −2.45. The underlying
mechanism requires further investigation.
Rowe et al (24) identified the primary CYP1A1
location as the sebaceous gland surrounding the hair shaft.
Consistent with this, in the present study CYP1A1 expression was
upregulated in body skin according to the microarray and RT-qPCR
data.
Connexin 43 is a gap junction protein expressed in
the dermal papilla (26). The G60S
connexin 43 mutant regulates hair growth and hair fiber morphology
in a mouse model of human oculodentodigital dysplasia (27). However, connexin 43 was
downregulated in body skin in the microarray and RT-qPCR data of
the present study. A similar pattern was observed in the Aohan fine
wool sheep (38).
Fibroblast growth factor (FGF) 2 and −10 were
upregulated and downregulated respectively in body compared with
groin skin. FGF signaling is crucial for hair growth regulation
(42,43). FGF5, expressed in the outer root
sheath, has been demonstrated to control anagen-catagen transition
(44) via the inhibition of hair
elongation (45). FGF21 and −22,
which promote the transition of catagen (46), were reportedly down-regulated in
primary compared with secondary hair follicles of the Yunnan black
goats (1). FGF expression varied
throughout the hair follicle cycle, reflecting the role of FGFs in
regulatory and developmental processes, which include patterning,
morphogenesis, proliferation, differentiation and migration of
cells (42). Expression analysis
suggested an important role for FGF7 and −10, as well as their
cognate receptor FGFR2-IIIb, in the proliferation and
differentiation of the mature hair follicle (46).
Homeobox (Hox) A7 and HoxC6 were downregulated in
body compared with groin skin in the present study. The first Hox
gene demonstrated to have a universal role in hair follicle
development was Hoxc13, as Hoxc13-deficient and -overexpressing
mice exhibit severe hair growth and patterning defects (47). Members of the Hox family may
perform essential and functionally diverse roles in hair that
require complex transcriptional control mechanisms to ensure
correct spatio-temporal patterns of Hox gene expression at
homeostatic levels (47,48). HOXA7 silences
differentiation-specific genes during keratinocyte proliferation
(49), which is consistent with
the downregulation of HOXA7 in body skin compared with groin skin
in the present study.
Growth hormone (GH) and GH receptor (GHR) were
upregulated in body compared with groin skin, consistent with
results in Aohan fine wool sheep (38). A number of growth factors have been
demonstrated to be critical for hair growth, including FGFs,
transforming growth factors, insulin-like growth factors (IGFs),
epidermal growth factors and hepatocyte growth factor (50). The hair cycle in the dorsal skin of
male GH-deficient rats enters a long-lasting telogen phase at 8
weeks of age; however, depilation induces a transient hair cycle
(51). Therefore, GH and GHR may
be involved in wool and cashmere growth regulation.
IGF2 is DE in anagen, catagen and telogen stages in
the Shaanbei White cashmere goat (9). IGF binding protein (IGFBP) 5-mediated
FGFR2-IIIb signaling is a critical regulator of the structure of
the hair shaft medulla (46). The
expression of IGFBP3 and IGFBP5 was decreased in early anagen and
anagen phases (IGFBP3) or in catagen and telogen phases (IGFBP5)
during a depilation-induced hair cycle (51). Downregulation of FGFR2-IIIb, an
effect associated with increased expression levels of IGFBP5, was
demonstrated to decrease the thickness of the hair shaft and to
reduce the cellularity of the hair shaft medulla (46). In accordance with these results,
IGFBP2 was downregulated in body compared with groin skin in the
microarray data of the present study.
In addition, the present study identified NF-1A, Yi,
E2F and CREB/CREBβ binding sites in the promoter region of a number
of upregulated genes. NF-1A negatively regulates target gene
expression via binding to a silencer element (52); therefore, the reduced expression
level of the gene subset may be caused by NF-1A upregulation or
activation. CREB is a transcription factor important for
keratinocyte proliferation (53–56).
Adenosine stimulates growth of hair follicles by triggering
phosphorylation of CREB (25).
Combined recruitment of CREB, CCAAT-enhancer-binding protein β and
c-jun determines the activation of promoters upon keratinocyte
differentiation (25). In the
present study, no differential expression was observed in CREB
between the two skin regions. Therefore, CREB may have important
additional roles in wool and cashmere growth, potentially via
phosphorylation or other modifications.
The promoter regions of downregulated genes
demonstrated three common motif sequences of 29, 39 and 50 bp. In
these regions, binding sites were identified for the transcription
factors LF-A1, Yi, E2F, COE1 and PPARα. The transcription factor
LF-A1 is required for the cell-specific expression of the human
α1-antitrypsin gene in hepatocytes (57). Binding sites of LF-A1 are present
in the promoter regions of various genes expressed in the liver
(α1-antitrypsin, apolipoproteins A1, B1 and A4, and pyruvate
kinase). In the present study, thioredoxin-related transmembrane
protein 2, adiponectin, C1Q and collagen domain containing and
perilipin 3 may associate with apolipoproteins. COE1 is essential
for B-cell differentiation (58).
Therefore, COE1 may be involved in the immune privilege process as
described previously (14).
Furthermore, PPARα has been reported to contribute to hair growth
and epidermal healing (59).
Notably, putative Yi and E2F sites were side-by-side
in the upstream regions of upregulated and downregulated genes. Yi
is an inducible DNA binding activity (60), consistent with its involvement in
upregulated and downregulated genes. E2F has important roles in
hair follicle growth, differentiation and survival (61,62).
E2F1-deficient mice have a high incidence of spontaneous epidermal
tumors of hair follicle origin (63). However, the reason for the
existence of putative E2F sites in upregulated and downregulated
genes requires further investigation. The U site was present in the
promoter regions of 12 downregulated genes, of which 11 contained
COE1 binding sites, consistent with a previous study (29).
A previous study in sheep revealed that the number
and fold-change of DE genes in December were markedly reduced
compared with August (21), which
is consistent with the decreased activity of hair follicles in
winter. Di et al (64)
investigated DE genes of skin tissue in fine wool sheep with
various fiber diameters using the Agilent sheep gene expression
microarray. Therefore, the present study validates the use of
Agilent Sheep Gene Expression Microarray in wool follicle
investigations and provides a strong rationale for future
studies.
Brenaut et al (16) investigated the contribution of
mammary epithelial cells to the early stages of an immune response
against Staphylococcus aureus in goats using the Agilent
sheep gene expression microarray. Goyal et al (65) conducted microarray with advanced
bioinformatic analysis on carotid arteries from the normoxic
near-term ovine fetus at sea level and high altitude. These studies
identified genes and networks involved in the investigated
processes. Therefore, microarray technology may be used to examine
various aspects of the sheep and goat species.
The present study used microarray methodologies to
investigate hair and cashmere growth in the Laiwu black goat. Gene
transcripts expressed in skin cells from hair-rich and hairless
areas were compared, and hundreds of DE genes were identified.
However, no keratin or keratin-associated protein genes were DE,
which contradicts the results of a previous study (34). Whether this problem is the result
of species differences in probe sets remains to be elucidated.
Gene annotation is incomplete and imperfect (certain
DE genes remain to be identified), which may explain why certain
factors, including keratin and keratin-associated proteins, were
not identified in the present study. However, the repertoire of
gene probes used (8×15K) confirmed the involvement of a number of
growth factors and HOX genes. The application of an original
algorithm to construct gene networks of temporal regulation of hair
growth was not performed due to limited DE genes. The present study
identified molecules in the cashmere-bearing skin area of the Laiwu
black goat, which may contribute to hair and cashmere traits.
Acknowledgments
The present study was funded by the National Hair
Sheep Industry Technology System (grant no. CARS-40), Projects of
Qingdao People's Livelihood Science and Technology (grant nos.
13-1-3-88-nsh and 14-2-3-45-nsh) and the National Natural Science
Foundation of China (grant no. 31301936).
References
|
1
|
Dong Y, Xie M, Jiang Y, Xiao N, Du X,
Zhang W, Tosser-Klopp G, Wang J, Yang S, Liang J, et al: Sequencing
and automated whole-genome optical mapping of the genome of a
domestic goat (Capra hircus). Nat Biotechnol. 31:135–141. 2013.
View Article : Google Scholar
|
|
2
|
Horner ME, Parkinson KE, Kaye V and Lynch
PJ: Dowling-Degos disease involving the vulva and back: Case report
and review of the literature. Dermatol Online J.
17:12011.PubMed/NCBI
|
|
3
|
Lueking A, Huber O, Wirths C, Schulte K,
Stieler KM, Blume-Peytavi U, Kowald A, Hensel-Wiegel K, Tauber R,
Lehrach H, et al: Profiling of alopecia areata autoantigens based
on protein microarray technology. Mol Cell Proteomics. 4:1382–1390.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Purvis IW and Franklin IR: Major genes and
QTL influencing wool production and quality: A review. Genet Sel
Evol. 37(Suppl 1): S97–S107. 2005. View Article : Google Scholar
|
|
5
|
Cano EM, Marrube G, Roldan DL, Bidinost F,
Abad M, Allain D, Vaiman D, Taddeo H and Poli MA: QTL affecting
fleece traits in Angora goats. Small Ruminant Research. 71:158–164.
2007. View Article : Google Scholar
|
|
6
|
Adams N and Cronjé P: A review of the
biology linking fibre diameter with fleece weight, liveweight, and
reproduction in Merino sheep. Australian Journal of Agricultural
Research. 54:1–10. 2003. View
Article : Google Scholar
|
|
7
|
Wenguang Z, Jianghong W, Jinquan L and
Yashizawa M: A subset of skin-expressed microRNAs with possible
roles in goat and sheep hair growth based on expression profiling
of mammalian microRNAs. OMICS. 11:385–396. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rufaut NW, Pearson AJ, Nixon AJ, Wheeler
TT and Wilkins RJ: Identification of differentially expressed genes
during a wool follicle growth cycle induced by prolactin. J Invest
Dermatol. 113:865–872. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang HR, Feng ZC, Du M, Ren JK and Li HR:
Initial research for seasonal variation of wool growth of Aohan
fine wool sheep. Inner Mong Anim Sci. 3:1–3. 1994.In Chinese.
|
|
10
|
McElwee KJ and Sinclair R: Hair Physiology
and its disorders. Drug discovery today: Disease Mechanisms.
5:e163–e171. 2008. View Article : Google Scholar
|
|
11
|
Zhu B, Xu T, Yuan J, Guo X and Liu D:
Transcriptome sequencing reveals differences between primary and
secondary hair follicle-derived dermal papilla cells of the
Cashmere goat (Capra hircus). PLoS One. 8:e762822013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhu B, Xu T, Zhang Z, Ta N, Gao X, Hui L,
Guo X and Liu D: Transcriptome sequencing reveals differences
between anagen and telogen secondary hair follicle-derived dermal
papilla cells of the Cashmere goat (Capra hircus). Physiol
Genomics. 46:104–111. 2014. View Article : Google Scholar
|
|
13
|
Wu Z, Fu Y, Cao J, Yu M, Tang X and Zhao
S: Identification of differentially expressed miRNAs between white
and black hair follicles by RNA-sequencing in the goat (Capra
hircus). Int J Mol Sci. 15:9531–9545. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu N, Li H, Liu K, Yu J, Cheng M, De W,
Liu J, Shi S, He Y and Zhao J: Differential expression of genes and
proteins associated with wool follicle cycling. Mol Biol Rep.
41:5343–5349. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Menzies M, Stockwell S, Brownlee A, Cam G
and Ingham A: Gene expression profiles of BMP4, FGF10 and cognate
inhibitors, in the skin of foetal Merino sheep, at the time of
secondary follicle branching. Exp Dermatol. 18:877–879. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Brenaut P, Lefèvre L, Rau A, Laloë D,
Pisoni G, Moroni P, Bevilacqua C and Martin P: Contribution of
mammary epithelial cells to the immune response during early stages
of a bacterial infection to Staphylococcus aureus. Vet Res.
45:162014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Geng R, Yuan C and Chen Y: Exploring
differentially expressed genes by RNA-Seq in cashmere goat (Capra
hircus) skin during hair follicle development and cycling. PLoS
One. 8:e627042103. View Article : Google Scholar
|
|
18
|
Nacht M, Dracheva T, Gao Y, Fujii T, Chen
Y, Player A, Akmaev V, Cook B, Dufault M, Zhang M, et al: Molecular
characteristics of non-small cell lung cancer. Proc Natl Acad Sci
USA. 98:15203–15208. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tang Z, Li Y, Wan P, Li X, Zhao S, Liu B,
Fan B, Zhu M, Yu M and Li K: LongSAGE analysis of skeletal muscle
at three prenatal stages in Tongcheng and Landrace pigs. Genome
Biol. 8:R1152007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Dennis G Jr, Sherman BT, Hosack DA, Yang
J, Gao W, Lane HC and Lempicki RA: DAVID: Database for annotation,
visualization, and integrated discovery. Genome Biol. 4:P32003.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu N, Li H, Liu K, Yu J, Bu R, Cheng M,
De W, Liu J, He G and Zhao J: Identification of skin-expressed
genes possibly associated with wool growth regulation of Aohan fine
wool sheep. BMC Genet. 15:1442014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bailey TL, Boden M, Buske FA, Frith M,
Grant CE, Clementi L, Ren J, Li WW and Noble WS: MEME SUITE: Tools
for motif discovery and searching. Nucleic Acids Res. 37:W202–W208.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Matys V, Kel-Margoulis OV, Fricke E,
Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M,
Hornischer K, et al: TRANSFAC and its module TRANSCompel:
Transcriptional gene regulation in eukaryotes. Nucleic Acids Res.
34(Database Issue): D108–D110. 2006. View Article : Google Scholar
|
|
24
|
Rowe JM, Welsh C, Pena RN, Wolf CR, Brown
K and Whitelaw CB: Illuminating role of CYP1A1 in skin function. J
Invest Dermatol. 128:1866–1868. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Maimaiti A: Genetic polymorphism of five
KAP gene and their associations with wool quality Traitsin Chinese
merino sheep (Xinjiang type) group. PhD dissertation. Xinjiang
Agricultural University. Globe Thesis. 2013
|
|
26
|
Higgins CA, Richardson GD, Ferdinando D,
Westgate GE and Jahoda CA: Modelling the hair follicle dermal
papilla using spheroid cell cultures. Exp Dermatol. 19:546–548.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Churko JM, Chan J, Shao Q and Laird DW:
The G60S connexin43 mutant regulates hair growth and hair fiber
morphology in a mouse model of human oculodentodigital dysplasia. J
Invest Dermatol. 131:2197–2204. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Alcorlo M, López-Perrote A, Delgado S,
Yébenes H, Subías M, Rodríguez-Gallego C, Rodríguez de Córdoba S
and Llorca O: Structural insights on complement activation. FEBS J.
282:3883–3891. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chang CH, Jiang TX, Lin CM, Burrus LW,
Chuong CM and Widelitz R: Distinct Wnt members regulate the
hierarchical morphogenesis of skin regions (spinal tract) and
individual feathers. Mech Dev. 121:157–171. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mosenson JA, Zloza A, Klarquist J, Barfuss
AJ, Guevara-Patino JA and Poole IC: HSP70i is a critical component
of the immune response leading to vitiligo. Pigment Cell Melanoma
Res. 25:88–98. 2012. View Article : Google Scholar
|
|
31
|
Peñagaricano F, Zorrilla P, Naya H,
Robello C and Urioste JI: Gene expression analysis identifies new
candidate genes associated with the development of black skin spots
in Corriedale sheep. J Appl Genet. 53:99–106. 2012. View Article : Google Scholar
|
|
32
|
Norris BJ and Whan VA: A gene duplication
affecting expression of the ovine ASIP gene is responsible for
white and black sheep. Genome Res. 18:1282–1293. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hammond NL, Headon DJ and Dixon MJ: The
cell cycle regulator protein 14-3-3σ is essential for hair follicle
integrity and epidermal homeostasis. J Invest Dermatol.
132:1543–1553. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhao J, Li H, Liu K, Liu K, Liu Zuo and Li
J: Differential expression of immune genes between body side skin
and groin skin of Aohan fine wool sheep. Agric Sci Technol.
12:2475–2479. 2012.
|
|
35
|
Taleb M, Brandon CS, Lee FS, Lomax MI,
Dillmann WH and Cunningham LL: Hsp70 inhibits
aminoglycoside-induced hair cell death and is necessary for the
protective effect of heat shock. J Assoc Res Otolaryngol.
9:277–289. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Karelina TV, Bannikov GA and Eisen AZ:
Basement membrane zone remodeling during appendageal development in
human fetal skin. The absence of type VII collagen is associated
with gelatinase-A (MMP2) activity. J Invest Dermatol. 114:371–375.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Philp D, Nguyen M, Scheremeta B, St-Surin
S, Villa AM, Orgel A, Kleinman HK and Elkin M: Thymosin beta4
increases hair growth by activation of hair follicle stem cells.
FASEB J. 18:385–387. 2004.
|
|
38
|
Zhao J, Liu N, Liu K, He J, Yu J, Bu R,
Cheng M, De W, Liu J and Li H: Identification of genes and proteins
associated with anagen wool growth. Animal Genetics. In Press.
|
|
39
|
Jones PH and Watt FM: Separation of human
epidermal stem cells from transit amplifying cells on the basis of
differences in integrin function and expression. Cell. 73:713–724.
1993. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kloepper JE, Hendrix S, Bodó E, Tiede S,
Humphries MJ, Philpott MP, Fässler R and Paus R: Functional role of
beta 1 integrin-mediated signalling in the human hair follicle. Exp
Cell Res. 314:498–508. 2008. View Article : Google Scholar
|
|
41
|
Brakebusch C, Grose R, Quondamatteo F,
Ramirez A, Jorcano JL, Pirro A, Svensson M, Herken R, Sasaki T,
Timpl R, et al: Skin and hair follicle integrity is crucially
dependent on beta 1 integrin expression on keratinocytes. EMBO J.
19:3990–4003. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Galbraith H: Fundamental hair follicle
biology and fine fibre production in animals. Animal. 4:1490–1509.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Rosenquist TA and Martin GR: Fibroblast
growth factor signalling in the hair growth cycle: Expression of
the fibroblast growth factor receptor and ligand genes in the
murine hair follicle. Dev Dyn. 205:379–386. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Botchkarev VA and Paus R: Molecular
biology of hair morphogenesis: Development and cycling. J Exp Zool
B Mol Dev Evol. 298:164–180. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Hebert JM, Rosenquist T, Götz J and Martin
GR: FGF5 as a regulator of the hair growth cycle: Evidence from
targeted and spontaneous mutations. Cell. 78:1017–1025. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Schlake T: FGF signals specifically
regulate the structure of hair shaft medulla via IGF-binding
protein 5. Development. 132:2981–2990. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Awgulewitsch A: Hox in hair growth and
development. Naturwissenschaften. 90:193–211. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Stelnicki EJ, Kömüves LG, Kwong AO, Holmes
D, Klein P, Rozenfeld S, Lawrence HJ, Adzick NS, Harrison M and
Largman C: HOX homeobox genes exhibit spatial and temporal changes
in expression during human skin development. J Invest Dermatol.
110:110–115. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
La Celle PT and Polakowska RR: Human
homeobox HOXA7 regulates keratinocyte transglutaminase type 1 and
inhibits differentiation. J Biol Chem. 276:32844–32853. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Stenn KS and Paus R: Controls of hair
follicle cycling. Physiol Rev. 81:449–494. 2001.PubMed/NCBI
|
|
51
|
Umeda-Ikawa A, Shimokawa I and Doi K:
Time-course expression profiles of hair cycle-associated genes in
male mini rats after depilation of telogen-phase hairs. Int J Mol
Sci. 10:1967–1977. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Lee JS, Xiao J, Patel P, Schade J, Wang J,
Deneen B, Erdreich-Epstein A and Song HR: A novel tumor-promoting
role for nuclear factor IA in glioblastomas is mediated through
negative regulation of p53, p21, and PAI1. Neuro Oncol. 16:191–203.
2014. View Article : Google Scholar :
|
|
53
|
Jang SI and Steinert PM: Loricrin
expression in cultured human keratinocytes is controlled by a
complex interplay between transcription factors of the Sp1, CREB,
AP1, and AP2 families. J Biol Chem. 277:42268–42279. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Sander GR and Powell BC: Structure and
expression of the ovine Hoxc-13 gene. Gene. 327:107–116. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Tanaka S, Miura I, Yoshiki A, Kato Y,
Yokoyama H, Shinogi A, Masuya H, Wakana S, Tamura M and Shiroishi
T: Mutations in the helix termination motif of mouse type I IRS
keratin genes impair the assembly of keratin intermediate filament.
Genomics. 90:703–711. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Ansari KM, Rundhaug JE and Fischer SM:
Multiple signaling pathways are responsible for prostaglandin
E2-induced murine keratinocyte proliferation. Mol Cancer Res.
6:1003–1016. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Langbein L, Rogers MA, Praetzel-Wunder S,
Helmke B, Schirmacher P and Schweizer J: K25 (K25irs1), K26
(K25irs2), K27 (K25irs3), and K28 (K25irs4) represent the type I
inner root sheath keratins of the human hair follicle. J Invest
Dermatol. 126:2377–2386. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Lin H and Grosschedl R: Failure of B-cell
differentiation in mice lacking the transcription factor EBF.
Nature. 376:263–267. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Liu G, Liu R, Li Q, Tang X, Yu M, Li X,
Cao J and Zhao S: Identification of microRNAs in wool follicles
during anagen, catagen, and telogen phases in Tibetan sheep. PLoS
One. 8:e778012013. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Jiang Y, Xie M, Chen W, Talbot R, Maddox
JF, Faraut T, Wu C, Muzny DM, Li Y, Zhang W, et al: The sheep
genome illuminates biology of the rumen and lipid metabolism.
Science. 344:1168–1173. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Yu ZD, Bawden CS, Henderson HV, Nixon AJ,
Gordon SW and Pearson AJ: Micro-arrays as a discovery tool for wool
genomics. Proceedings of the New Zealand Society of Animal
Production. 66:129–133. 2006.
|
|
62
|
Xu T, Guo X, Wang H, Du X, Gao X and Liu
D: De novo transcriptome assembly and differential gene expression
profiling of three Capra hircus skin types during Anagen of the
hair growth cycle. Int J Genomics. 2013:2691912013. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Yu ZD, Gordon SW, Pearson AJ, Henderson
HV, Craven AJ and Nixon AJ: Gene expression profiling of wool
follicle growth cycles by cDNA microarray. Proceedings of the New
Zealand Society of Animal Production. 68:39–42. 2008.
|
|
64
|
Di J, La Z, Xu X, Zhang Y, Tian K, Tian Y,
Yu L and Ha N: Genome array on differentially expressed genes of
skin tissue in fine wool sheep with different fiber diameter. Acta
Veterinaria Et Zootechnica Sinica. 5:681–689. 2013.In Chinese.
|
|
65
|
Goyal R, Van Wickle J, Goyal D, Matei N
and Longo LD: Antenatal maternal long-term hypoxia: Acclimatization
responses with altered gene expression in ovine fetal carotid
arteries. PLoS One. 8:e822002013. View Article : Google Scholar : PubMed/NCBI
|