Introduction
Osteogenesis imperfecta (OI) is an inherited
connective tissue disorder characterized by brittle bone fractures
and short stature. In addition, other connective tissue disorders,
including blue or gray sclera, dentinogenesis imperfecta,
hyperlaxity of ligaments and skin, and progressive conductive
hearing loss, are common in OI patients (1). OI patients have been classified into
four types by Sillence and Rimoin (2) according to clinical characteristics
and patterns of inheritance. An expanded Sillence classification
was published by Rauch and Glorieux in 2004 (3). Forlino et al (4) classified OI into 11 types based on
the discovery of each novel gene defect. To date, a total of 19
genes have been identified as causing OI: Collagen, type I, alpha 1
(COL1A1; MIM 120150), collagen, type I, alpha 2
(COL1A2; MIM 120160), cartilage-associated protein
(CRTAP; MIM 605497), leucine- and praline-enriched
proteoglycan1 (LEPRE1; MIM 610339), peptidyl-prolyl
isomerase B (PPIB; MIM 123841), FK506 binding protein 10
(MIM 607063), serpin family H member 1 (MIM 600943), Wnt family
member 1 (MIM 164820), bone morphogenetic protein 1 (MIM 112264),
interferon induced transmembrane protein 5 (IFITM5; MIM
614757), plastin 3 (MIM 300131), procollagen-lysine, 2-oxoglutarate
5-dioxygenase 2 (MIM 601865), serpin peptidase inhibitor, clade F
(alpha-2 antiplasmin, pigment epithelium derived factor), member 1
(MIM 172860), Sp7 transcription factor (MIM 606633), transmembrane
protein 38B (MIM 611236), cAMP responsive element binding protein
3-like 1 (HGNC ID 18856; https://oi.gene.le.ac.uk/home.php?select_db=CREB3L1),
prolyl 4-hydroxylase, beta polypeptide (MIM 176790), SEC24 family
member D (MIM 607186) and Secreted protein, acidic, cysteine-rich
(SPARC; MIM 182120) (5–8).
Although numerous genetic causes of OI have been reported,
mutations in COL1A1 and COL1A2 are responsible for
~90% of OI cases (9). To date, the
COL1A1 and COL1A2 mutations in Chinese patients with
OI have been reported only in certain sporadic cases (10–14).
To investigate the pathogenic gene mutation spectrum and the
clinical manifestations of mutations in COL1A1 and
COL1A2 genes in Chinese patients with OI requires a large
sample size.
Our previous study (14) identified 56 heterozygous mutations
in COL1A1 and COL1A2, including 43 mutations in
COL1A1 and 13 mutations in COL1A2. Of those Chinese
patients, 56.9% had OI type I, 19.0% had type III and 20.7% had
type IV. In addition, the study identified 2 novel compound
heterozygous mutations in the LEPRE1 gene in two probands
with OI. Our other previous study identified mutations in the
IFITM5 gene in four Chinese families with OI type V
(15). To more accurately reflect
the Chinese OI clinical characteristics and pathogenic gene
mutations, the present study recruited a further 61 OI patients
from 2012 to 2015. Mutations in the COL1A1 and COL1A2
genes were identified in the 61 Chinese OI patients. Although the
exact incidence of OI in China remains unknown, this is a further
large report of Chinese OI cases. The aim of the present study was
to investigate the pathogenic gene mutation spectrum and clinical
manifestations of mutations of COL1A1 and COL1A2
genes in Chinese patients with OI. These data may be useful for
future clinical diagnosis and genetic counseling.
Materials and methods
Subjects
A total of 61 unrelated probands from 61 separate
families were recruited from the Department of Osteoporosis and
Bone Diseases, Shanghai Jiao Tong University Affiliated Sixth
People's Hospital (Shanghai, China) over a 2-year period
(2012–2015). The probands came from 16 provinces of China, with the
majority from Eastern cities. All patients were of Han ethnicity. A
total of 410 DNA samples were collected from the 61 probands, 99
family members and 250 healthy control donors. Samples from healthy
controls were sequenced to determine whether mutations occurred as
polymorphisms. Clinical characteristics, including scleral hue, the
presence of dentinogenesis imperfecta and hearing loss, a history
of fractures, height, and OI type, were recorded. The OI type was
classified in all probands according to the Sillence classification
(2). None of the patients belonged
to a consanguineous family or had received any treatment prior to
the present study. All family members were examined by a single
experienced clinician familiar with OI.
Ethics statement
The present study was approved by the Ethics
Committee of Shanghai Jiao Tong University Affiliated Sixth
People's Hospital. Written informed consent was obtained from all
adult participants prior to recruitment. In addition, written
informed consent was obtained from parents on behalf of pediatric
participants. This consent procedure was approved by the Ethics
Committee of the Shanghai Jiao Tong University Affiliated Sixth
People's Hospital.
Collagen type I mutation analysis
Genomic DNA was extracted from 2 ml peripheral blood
samples using the QuickGene DNA whole blood kit (Kurabo Industries
Ltd., Osaka, Japan) and a Nucleic Acid Isolation system
(QuickGene-610L; Autogen, Inc., Holliston, MA, USA). All exons of
the COL1A1 and COL1A2 genes, including the
exon-intron boundaries, were amplified by polymerase chain reaction
using the primers described in our previous study (14). The genomic (AF017178.2 and
AF004877.1) and mRNA (Z74615.1 and Z74616.1) sequences of the
COL1A1 and COL1A2 genes were used as reference
sequences. DNA mutations were numbered according to the cDNA
sequence and the A of the ATG codon was designated as nucleotide
+1. Novel mutations were identified according to the Type 1
Collagen Mutation Database (https://oi.gene.le.ac.uk/variants.php?select_db=COL1A1&action=view_all
and https://oi.gene.le.ac.uk/variants.php?select_db=COL1A2&action=view_all).
In addition, control alleles from 250 healthy individuals were
sequenced to determine whether novel mutations occurred as
polymorphisms.
To perform a cross-species analysis of the gene
sequences of the mutations that were not substitution mutations of
glycine, the Uniprot database (http://www.uniprot.org/uniprot/?query=COL1A1&sort=score
and http://www.uniprot.org/uniprot/?query=COL1A2&sort=score)
was used to align 10 protein sequences, and the Clustal Omega
program (http://www.ebi.ac.uk/Tools/msa/clustalo/) was used to
view their characteristics alongside each other. The 10
COL1A1 protein sequences compared were from human,
zebrafish, Japanese common newt, chicken, African clawed frog,
mouse, rat, cow, dog and cat. The 10 COL1A2 protein
sequences compared were human, zebrafish, green anole or horse,
chicken, African clawed frog, mouse, rat, cow, dog and cat, since
the protein sequence of Japanese common newt of COL1A2 was
not included in the database.
Bone densitometry
The bone mineral density (BMD; g/cm2) of
the lumbar spine (L1-L4) was measured using dual-energy X-ray
absorptiometry (DXA). Vertebrae were excluded from analysis if they
were affected by fractures. All subjects were assessed using Lunar
Prodigy equipment (GE Healthcare Life Sciences, Chalfont, UK). The
Lunar Prodigy device was calibrated daily. The coefficient of
variability of the lumbar spine DXA measurements was 1.39%
(16). All DXA scans were
conducted by the same trained specialist. Lumber spine BMD results
were converted to age- and gender-specific Z-scores as previously
described (17).
Statistical analysis
Statistical analyses were performed in SPSS software
version 11.0 (SPSS, Inc., Chicago, IL, USA). Group differences in
enumeration data were analyzed for significance using the
chi-square test. Data with normal distribution was tested for
significance using independent-samples t-test and data with
non-normal distribution using the Mann-Whitney U test. All tests
were two-sided. P<0.05 was considered to indicate a
statistically significant difference.
Results
Clinical characteristics of the 61
patients
A total of 44 probands had a mutation in the
COL1A1 gene, and 17 had a mutation in COL1A2
(Table I). Of the patients, 35
were male and 26 female, with a median age of 11.0 years (range,
0.4–60 years). Family history was positive for OI in 33 probands
(54%), negative for OI in 27 probands (44%) and unknown in 1
proband (2%). All probands had suffered fractures, and 13.2% of
probands with COL1A1 mutations had suffered >10 fractures
(Table I). A total of 49 probands
presented with blue sclerae (80.3%), 20 probands suffered from
dentinogenesis imperfecta (32.8%) and 1 patient had hearing loss
(1.6%).
 | Table I.Clinical findings of OI in the 61
patients. |
Table I.
Clinical findings of OI in the 61
patients.
| Mutation | Sex (M/F) | Age (years) | Fr >10 | Blue sclerae | DI | Hearing loss | Height Z score | Weight Z score | LS BMD Z score |
|---|
| COL1A1
(n=44) | 28/16 | 11.0 | 6 | 37 | 15 | 1 | −0.8 | −0.5 | −1.0±1.4 |
|
|
| (4.5–22.5) | (13.6%) | (84.1%) | (34.1%) | (2.3%) | (−2.5, 0.6) | (−1.2, 2.1) |
|
| COL1A2
(n=17) | 7/10 | 9.0 | 0 | 12 | 5 | 0 | −1.2 | 0.2 | −2.7±0.6 |
|
|
| (7.0–20.0) |
| (70.6%) | (29.4%) |
| (−2.5, −0.7) | (−1.1, 0.9) |
|
COL1A1 and COL1A2 mutations
A total of 44 patients had a mutation within the
COL1A1 gene, and 17 had a mutation in COL1A2 (Table I). These mutations included 33
missense mutations, 8 nonsense mutations, 7 splicing variants and
13 frameshift mutations (Table
II). Almost half the probands (42.6%; 26/61) had a substitution
mutation of the glycine within the Gly-X-Y triplet domain of the
triple helix, of which 13 were in COL1A1 and 13 were in
COL1A2. Serine substitutions were the most common in the
present study (42.3%).
 | Table II.Types of COL1A1 and
COL1A2 mutations in the present study. |
Table II.
Types of COL1A1 and
COL1A2 mutations in the present study.
| Mutation | Missense | Nonsense | Splicing | Frameshift |
|---|
| COL1A1 | 17 | 7 | 7 | 13 |
| COL1A2 | 16 | 1 | 0 | 0 |
In total, 25 of the mutations of 26 probands (2
probands had the same novel mutation in COL1A2) identified were
novel: 18 in COL1A1 (Table
III) and 7 in COL1A2 (Table IV). Novel mutations were
identified according to the Type 1 Collagen Mutation Database. In
addition, these novel mutations were not present in the 250 healthy
controls. Of the 25 novel mutations, 7 missense mutations, 7
frameshift mutations and 4 nonsense mutations were identified in
COL1A1, and 6 missense mutations and 1 nonsense mutation
were identified in COL1A2. Of these mutations only 6 were
not substitution mutations of glycine. Therefore, the gene
sequences of these 6 mutations were compared across species.
Although the p.Ala508Thr missense mutation of COL1A1 is not
at a highly conserved position, other vertebrate species including
cow, dog and cat all contained alanine (Fig. 1A). The proband was classified as OI
type III and COL1A2, CRTAP and LEPRE1 gene mutations
were excluded. The mutation is potentially functionally damaging as
alanine is a nonpolar and threonine a polar amino acid. The
p.Thr766Ser (Fig. 1B) and
p.Thr1298Ile (Fig. 1C) missense
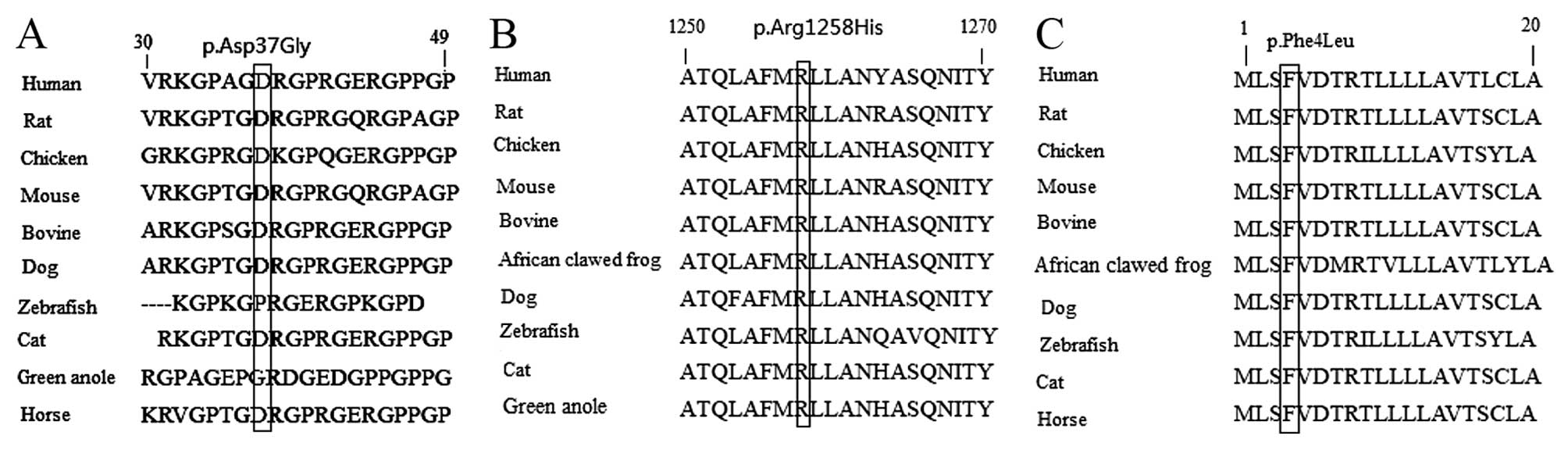
mutations of COL1A1, and the p.Asp37Gly (Fig. 2A), p.Arg1258His (Fig. 2B) and p.Phe4Leu (Fig. 2C) missense mutations of
COL1A2 are at highly conserved positions. In addition, these
missense variants were searched for in the large Exome Aggregation
Consortium (ExAC) database (exac.broadinstitute.org), and the frequencies of
p.Ala508Thr of COL1A1, p.Arg1258His and p.Phe4Leu missense
mutations of COL1A2 were revealed to be
2.471×10−5, 2.471×10−5 and
8.236×10−6, respectively. The remaining three missense
variants were absent from the ExAC database. Furthermore, the
combined annotation dependent depletion (CADD) pathogenicity scores
of these missense variants were high. The CADD pathogenicity score
of the p.Ala508Thr, p.Thr766Ser and p.Thr1298Ile mutations of
COL1A1 and the p.Aso37Gly, p.Arg1258His and p.Phe4Leu
mutations of COL1A2 were 13.3, 26.6, 15.8, 17.8, 23.4 and
19.8, respectively.
 | Table III.Clinical and genetic characteristics
of 44 probands with mutations in collagen type I, alpha 1. |
Table III.
Clinical and genetic characteristics
of 44 probands with mutations in collagen type I, alpha 1.
| No. | Sex | Age (years) | F/Sa | Blue sclerae | DI | Hearing loss | Height, cm (Z
value) | Weight, kg (Z
value) | Fracture
ratesb | Nucleotide
changec | Predicted amino
acid changed | Novel |
|---|
| C1 | F | 12 | F | + | – | – | 134.0 (−2.5) | 40 (0.6) | + | c.144delT |
p.His48GlnfsX26 | Yes |
| C2 | M | 2 | F | + | – | – | 87 (−0.6) | 13 (0.3) | + | c.157–158delTG |
p.Trp53GlufsX19 | Yes |
| C3 | F | 14 | F | + | – | – | 162.6 (0.7) | 68 (2.8) | + | c.268G>T | p.Glu90X | Yes |
| C4 | M | 13 | S | + | – | – | 151 (−0.3) | 51 (2.7) | + | c.433_434insC |
p.Gly145AlafsX24 | Yes |
| C5 | M | 12 | S | + | – | – | 150.8 (0.6) | 47 (1.4) | + | c.441dupC |
p.Gly148ArgfsX21 | – |
| C6 | M | 18 | F | + | + | – | 163 (−1.5) | 70 (2.1) | +++ | c.484C>T | p.Gln162X | Yes |
| C7 | M | 2.3 | S | + | – | – | 88 (−1.4) | 12 (−0.8) | + | c.569delC |
p.Pro190LeufsX75 | Yes |
| C8 | M | 8 | F | + | + | – | 133.6 (0.7) | 28 (0.5) | + |
c.573_574delCCinsG |
p.Pro193LeufsX72 | Yes |
| C9 | M | 2 | F | + | – | – | 84 (−1.5) | 12 (−0.4) | + | c579delT |
p.Gly194ValfsX71 | – |
| C10 | F | 33 | F | + | + | – | 156.1 (−0.5) | 47 (−1.0) | ++ | c.643-2A>G | Splicing
variant | – |
| C11 | F | 30 | F | + | + | + | 152.8 (−1.0) | 49 (−0.8) | + | c.757C>T | p.Arg253X | – |
| C12 | F | 28 | F | + | – | – | 147.5 (−2.1) | 44 (−1.2) | + | c.769G>A | p.Gly257Arg | – |
| C13 | M | 42 | S | + | + | – | 150 (−2.8) | 45 (−2.3) | + | c.769G>A | p.Gly257Arg | – |
| C14 | M | 11 | F | + | – | – | 150 (1.0) | 50 (2.2) | + | c.898C>T | p.Gln300X | Yes |
| C15 | M | 4 | S | + | – | – | 116 (3) | 22 (3.3) | + | c.903+1G>A | Splicing
variant | – |
| C16 | F | 6 | F | – | + | – | 115.5 (−0.4) | 17 (−0.9) | + | c.1121G>C | p.Gly374Ala | – |
| C17 | M | 2.8 | S | + | – | – | 101 (1.7) | 20 (3.7) | + | c.1128delT |
p.Gly377AlafsX164 | – |
| C18 | F | 65 | F | + | – | – | 150.2 (−0.5) | 53 (−0.5) | + | c.1155+1G>A | Splicing
variant | – |
| C19 | F | 27 | S | + | – | – | 161.1 (0.2) | 51 (−0.4) | + | c.1200+1G>A | Splicing
variant | – |
| C20 | M | 7 | F | + | – | – | 134.5 (1.9) | 35 (3.0) | + | c.1243C>T | p.Arg415X | – |
| C21 | F | 4 | F | + | – | – | 105.0 (0.6) | 17 (0.7) | + | c.1299+1G>A | Splicing
variant | – |
| C22 | M | 11 | F | + | – | – | 137.2 (−0.8) | 30 (−0.5) | + | c.1299+1G>A | Splicing
variant | – |
| C23 | F | 2.5 | S | + | + | – | 80.0 (−3.3) | 10 (−2.2) | + | c.1522G>A | p.Ala508Thr | Yes |
| C24 | M | 2.5 | – | + | + | – | 76.0 (−4.9) | 9 (−3.5) | + | c.1588G>A | p.Gly530Ser | – |
| C25 | M | 29 | F | – | + | – | 157.0 (−2.3) | 58 (−0.9) | + | c.1678G>T | p.Gly560Cys | – |
| C26 | M | 8 | F | – | – | – | 112.5 (−4.2) | 20 (−1.2) | + | c.1787G>C | p.Gly596Ala | Yes |
| C27 | M | 12 | F | + | – | – | 152.0 (0.8) | 52 (2.0) | + | c.1866delT |
p.Gly623AlafsX143 | – |
| C28 | F | 11 | S | + | + | – | 160.0 (1.9) | 52 (2.6) | + | c.1930-2A>C | Splicing
variant | – |
| C29 | M | 12 | S | + | – | – | 150.0 (0.5) | 55 (2.4) | + | c.2183G>A | p.Gly728Glu | Yes |
| C30 | F | 26 | S | + | + | – | 134.5 (−4.2) | 40 (−1.7) | ++ | c.2297C>G | p.Thr766Ser | Yes |
| C31 | F | 24 | S | + | + | – | 118.0 (−6.9) | 44 (−1.0) | ++ | c.2299G>A | p.Gly767Ser | – |
| C32 | M | 8 | S | + | – | – | 120.0 (−2.5) | 20 (−1.2) | + | c.2410G>T | p.Glu804X | Yes |
| C33 | M | 28 | F | + | + | – | 173.5 (0.3) | 64 (−0.6) | + | c.2450delC |
p.Pro817LeufsX291 | – |
| C34 | F | 16 | S | – | + | – | 114.5 (−7.3) | 32 (−2.6) | +++ | c.2461G>A | p.Gly821Ser | – |
| C35 | F | 10 | F | – | – | – | 110.0 (−4.1) | 23 (−1.4) | ++ | c.2560G>A | p.Gly854Ser | – |
| C36 | M | 7 | F | + | – | – | 117.0 (−0.8) | 21 (−0.7) | + | c.2775delT |
p.Gly926ValfsX182 | – |
| C37 | F | 25 | S | + | + | – | 126.0 (−5.7) | 40 (−1.7) | ++ | c.2867G>C | p.Gly956Ala | Yes |
| C38 | M | 14 | S | + | – | – | 159.5 (−0.8) | 45 (−0.6) | + | c.3076C>T | p.Arg1026X | – |
| C39 | M | 8 | F | + | – | – | 127.4 (−0.8) | 24 (−0.4) | + | c.3328delC |
p.His1110ThrfsX129 | Yes |
| C40 | M | 13 | F | + | – | – | 154.8 (0.1) | 75 (8.0) | + | c.3559G>T | p.Gly1187Cys | Yes |
| C41 | M | 3 | S | + | – | – | 81.0 (−4.3) | 11 (−2.3) | + | c.3638delG |
p.Gly1213Alafsx26 | Yes |
| C42 | M | 4 | F | – | – | – | 106.0 (0.6) | 20 (2.1) | + | c.3655G>A | p.Asp1219Asn | – |
| C43 | M | 0.4 | S | + | – | – | 60.0 (−1.2) | 6 (−1.1) | ++ | c.3893C>T | p.Thr1298Ile | Yes |
| C44 | M | 9 | S | – | – | – | 139.0 (1.4) | 40.0 (2.0) | + | c.4363G>A | p.Gly1455Ser | – |
 | Table IV.Clinical and genetic characteristics
of 17 probands with mutations in collagen type I, alpha 2. |
Table IV.
Clinical and genetic characteristics
of 17 probands with mutations in collagen type I, alpha 2.
| No. | Sex | Age (years) | F/Sa | Blue sclerae | DI | Hearing loss | Height, cm (Z
value) | Weight, kg (Z
value) | Fracture
ratesb | Nucleotide
changec | Predicted amino
acid changed | Novel |
|---|
| H1 | M | 13 | F | + | + | – | 142.3 (−1.2) | 48 (2) | + | c.12T>G | p.Phe4Leu | Yes |
| H2 | F | 9 | F | + | – | – | 122 (−1.6) | 26.5 (0.2) | + | c.110A>G | p.Asp37Gly | Yes |
| H3 | M | 7 | S | + | + | – | 113.3 (−0.8) | 23 (0.7) | + | c.812G>A | p.Gly271Asp | Yes |
| H4 | F | 8 | S | – | – | – | 120 (−1.3) | 21 (−0.8) | + | c.946G>A | p.Gly316Ser | – |
| H5 | M | 6 | S | + | – | – | 113 (−0.8) | 19 (−0.5) | + | c.1009G>A | p.Gly337Ser | – |
| H6 | F | 7 | F | + | + | – | 112.8 (−1.1) | 17.2 (−1.5) | + | c.1009G>A | p.Gly337Ser | – |
| H7 | F | 47 | F | + | – | – | 158 (−0.1) | 47.5 (−1.4) | + | c.1081C>T | P.Arg361X | Yes |
| H8 | F | 9 | F | – | – | – | 126.5 (−2.3) | 48.3 (1.5) | + | c.1648G>A | p.Gly550Ser | – |
| H9 | M | 19 | F | + | – | – | 160 (−2) | 76 (1.8) | + | c.1666G>T | p.Gly556Cys |
|
| H10 | M | 21 | S | – | – | – | 145 (−2.6) | 50 (−1.1) | + | c.2081G>A | p.Gly694Asp | Yes |
| H11 | M | 25 | S | + | + | – | 136.5 (−5.6) | 56 (−1.0) | + | c.2081G>A | p.Gly694Asp | Yes |
| H12 | M | 7 | F | + | – | – | 116.5 (−0.6) | 23 (0.5) | ++ | c.2441G>A | p.Gly814Glu | – |
| H13 | M | 13 | S | – | – | – | 150 (−0.4) | 42 (0.6) | + | c.2764G>A | p.Gly922Ser | Yes |
| H14 | F | 43 | S | – | – | – | 136.9 (−4.2) | 64 (0.8) | + | c.2918G>A | p.Gly973Asp | – |
| H15 | M | 11 | S | + | + | – | 135.0 (−1.2) | 30 (−0.5) | + | c.3197G>T | p.Gly1066Val | – |
| H16 | M | 3 | S | + | – | – | 87 (−2.6) | 11 (−2.3) | + | c.3304G>A | p.Gly1102Ser | – |
| H17 | F | 8 | F | + | – | – | 131.5 (−0.5) | 30.7 (0.9) | + | c.3773G>A | p.Arg1258His | Yes |
The number of fractures in patients with the
missense mutation close to the carboxyl-terminal end was greater
compared with patients with missense mutations close to the amino
terminal. In addition, 2 probands had a p.Gly257Arg (c.769G>A)
mutation at exon 11 of COL1A1, which is the mutation
identified in 2 unrelated patients in our previous study (14); 1 proband had a p.Gly767Ser
(c.2299G>A) mutation at exon 33, which is the mutation
identified in 4 unrelated patients in our previous study; 1 proband
had a p.Gly821Ser (c.2461G>A) mutation at exon 37, which is the
same mutation identified in 2 unrelated patients in our previous
study; and 2 probands had a p.Gly337Ser (c.1009G>A) mutation at
exon 19 of COL1A2, which is the same mutation identified in
1 unrelated patient in our previous study.
According to the Sillence classification, no
patients in the present study had OI type II. A total of 40
patients (65.6%) had OI type I, 11 patients (18.0%) had type III
and 10 patients (16.4%) had type IV. As in our previous study,
COL1A1 mutations were more frequent than COL1A2
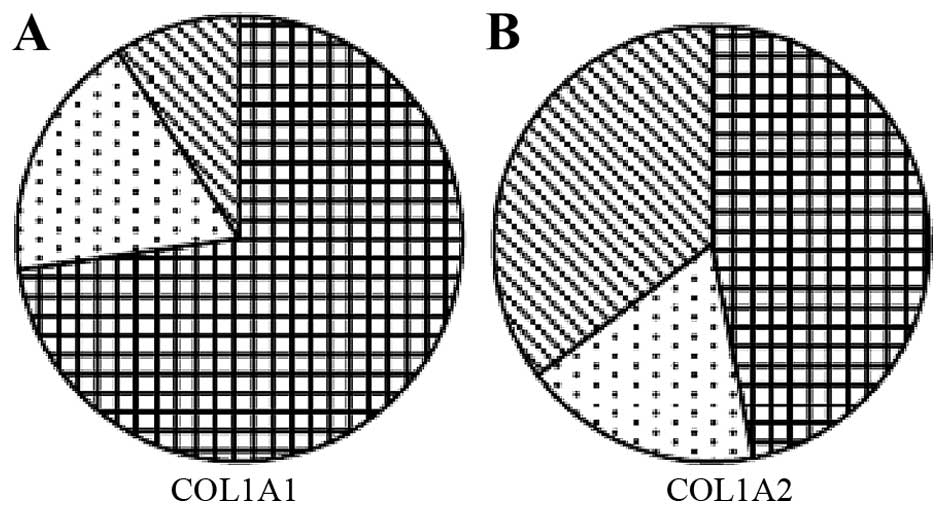
mutations in patients with OI types I and III (P<0.05; Fig. 3). In the COL1A1 mutation
group, the most common fracture site was the femur (n=21; 21% of
all fractures), followed by tibia/fibula (n=19; 19%) and
radius/ulna (n=19; 19%). In the COL1A2 mutation group, the
most common fracture site was the femur (n=12; 40% of all
fractures), followed by tibia/fibula (n=5; 17%), radius/ulna (n=3;
10%) and humerus (n=3; 10%).
There were no significant differences in the
phenotypes of patients with glycine to serine mutations of
COL1A1 and COL1A2 genes (Table V).
 | Table V.Clinical characteristics of patients
with glycine to serine substitutions. |
Table V.
Clinical characteristics of patients
with glycine to serine substitutions.
| Parameter | COL1A1
(n=5) | COL1A2
(n=6) | P-value |
|---|
| Age (years) | 12.3±18.1 | 7.7±3.3 | 0.23 |
| Height (Z
score) | −4.4±3.5 | −1.4±0.9 | 0.08 |
| Weight (Z
score) | −1.3±2.1 | −0.5±1.4 | 0.47 |
Discussion
The present study identified 61 mutations in
COL1A1 and COL1A2, 25 of them novel. Of the 25 novel
mutations, 13 missense mutations, 5 nonsense mutations and 7
frameshift mutations were identified. To date, 912 unique mutations
in COL1A1 are listed on the OI variant database (oi.gene.le.ac.uk/home.php?select_db=COL1A1), 571
unique mutations in COL1A2 (oi.gene.le.ac.uk/home.php?select_db=COL1A2), plus a
further 5 novel mutations that have recently been discovered
(18–20). Despite the number of mutations
identified, the 25 novel mutations observed in the present study
may contribute to revealing the pathogenesis of OI and improve the
disease-causing gene spectrum of OI in humans; the novel glycine
substitution and frameshift mutations are of particular interest in
this regard. In addition, the present study identified various
misssense mutations with unclear pathogenicities, including the
p.Ala508Thr, p.Thr766Ser and p.Thr1298Ile mutations of COL1A1 and
the p.Asp37Gly, p.Arg1258His and p.Phe4Leu mutations of COL1A2.
They occur at highly or relatively highly conserved positions and
their ExAC control frequencies were very low or absent.
Furthermore, the CADD pathogenicity scores of these missense
variants were high. Therefore, these 6 novel misssense mutations
may be pathogenic.
As in our previous study, almost three times the
number of mutations was observed in COL1A1 compared with
COL1A2. The COL1A1 group contains patients with stop
or frameshift mutations that lead to haploinsufficiency and OI type
I. However, these haploinsufficiency mutations are not observed in
COL1A2, as they do not result in a phenotype.
In the present study, of the COL1A1
mutations, 4 probands had a p.Gly257Arg mutation at exon 11, 5
probands had a p.Gly767Ser mutation at exon 33 and 3 probands had a
p.Gly821Ser mutation at exon 37. The Type 1 Collagen Mutation
Database lists these mutations 30, 26 and 21 times, respectively.
In addition, 3 probands had a p.Gly337Ser mutation at exon 19 of
COL1A2. The Type 1 Collagen Mutation Database lists this
mutation 23 times. Therefore, these 3 mutations in COL1A1
and 1 mutation in COL1A2 may occur at a mutation hotspot for
human OI.
Type I procollagen chains form a heterotrimer
containing two copies of the α1 (I) chain and one copy of the α2
(I) chain; it has therefore been suggested that phenotypes
resulting from mutations in COL1A1 were more severe compared
with those from COL1A2 (21). However, the present study observed
no significant differences between COL1A1 and COL1A2
glycine to serine mutation groups, in contrast to a previous study
by Rauch et al (22). The
number of patients examined in the present study was small, which
may limit the statistical power. As mutations toward the
carboxyl-terminus are more disruptive to helix formation, it has
been suggested that mutations closest to the carboxyl-terminal end
would be more severe compared with those closer to the
amino-terminal end of the α(I) chains (23,24).
However, Rauch et al (22)
demonstrated this ‘gradient model’ of disease severity for a2(I)
mutations but not for mutations affecting a1(I). In the present
study, probands with the missense mutation at the amino-terminal
end had a milder phenotype compared with those closer to the
carboxyl-terminal end. It has previously been demonstrated that the
most common mutations associated with OI are substitutions for
glycine by another amino acid in the triple helical domain of
COL1A1 or COL1A2 (22). In the present study, 33 missense
mutations were identified from 61 probands. Substitutions of
glycine by serine were the most common in COL1A1 and
COL1A2, in accordance with our previous study and Rauch
et al (14,22).
In contrast to our previous study, disease in the 61
patients of the present study was 54% familial and 44% sporadic
disease. This was in accordance with a previous study on Korean OI
patients (51% familial and 49% sporadic OI cases) (25). In the present study, OI type I
(65.6%) was the most common type. A total of 80.3% probands
presented with blue sclerae and all patients with mutations in the
amino-terminal end of the a1(I) chain had blue sclera, as described
by Rauch et al (22), who
additionally observed that dentinogenesis imperfecta was absent in
patients with mutations in the amino-terminal end of the a1(I) or
a2(I) triple helical domain. In the present study, patients were
identified with COL1A1 mutations at the amino-terminal and
carboxyl-terminal ends without dentinogenesis imperfecta. Hearing
loss is a common secondary feature of OI in adults, often with
mixed conductive and sensorineural deficiency (4). Previous studies have reported that in
a Scottish population ~50% of patients have subjective hearing loss
(26), and >60% of Finnish OI
adult patients had hearing loss (27); however, only ~5% of OI pediatric
patients have been reported to suffer hearing loss (28). In the present study, only 1.6% of
probands had hearing loss; this may due to the fact that the
majority of the patients were children. The most common fracture
site was the femur, followed by tibia/fibula, radius/ulna in the
patients of the COL1A1 and COL1A2 mutation groups;
this is in accordance with the study by Ben Amor et al
(29).
The present study is the first, to the best of our
knowledge, to observe Chinese OI patients in such large numbers.
However, the present study had certain limitations. The study was
limited to a single center and was cross-sectional. No patients had
lethal type II OI as the patients were recruited from the
outpatient center and newborns with the lethal type were not yet
diagnosed.
In conclusion, the present study identified 61
mutations in COL1A1 and COL1A2, 25 of which were
novel. Serine substitutions were the most frequently encountered
mutation type. The mutations p.Gly257Arg, p.Gly767Ser and
p.Gly821Ser in COL1A1, and p.Gly337Ser in COL1A2 may
be located at a mutation hotspot for human OI. Family history was
positive for OI in 33 probands (54%). All probands had suffered
fractures and the most common fracture site was the femur. A total
of 49 probands presented with blue sclerae (80.3%), 20 probands
suffered from dentinogenesis imperfecta (32.8%) and 1 patient had
hearing loss (1.6%). These findings may improve understanding of
the pathogenic gene mutation spectrum and clinical manifestations
of mutations in COL1A1 and COL1A2 genes of Chinese
patients with OI, and may be useful for future clinical diagnosis
and genetic counseling.
Acknowledgements
The authors thank the patients and their family
members for their invaluable cooperation, and the Center for
Genetic & Genomic Analysis, Genesky Biotechnologies Inc.
(Shanghai, China) for their assistance with gene identification.
The present study was supported by the National Basic Research
Program of China (grant no. 2014CB942903), the Science and
Technology Commission of Chongqing Municipality (grant no.
CSTC2013jcyjC00009), the Science and Technology Commission of
Shanghai Municipality (grant nos. 14JC1405000 and 14ZR1431900), the
National Natural Science Foundation of China (grant nos. 81170803,
81370978 and 30800387), the Frontier Technology Joint Research
Program of the Shanghai Municipal Hospitals (grant no.
SHDC12013115) and the Shanghai Leading Talent Plan (grant no.
051).
Glossary
Abbreviations
Abbreviations:
|
OI
|
osteogenesis imperfecta
|
|
COL1A1
|
collagen type I, alpha 1
|
|
COL1A2
|
collagen type I, alpha 2
|
|
BMD
|
bone mineral density
|
|
DXA
|
dual-energy X-ray absorptiometry
|
References
|
1
|
Basel D and Steiner RD: Osteogenesis
imperfecta: Recent findings shed new light on this once
well-understood condition. Genet Med. 11:375–385. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sillence DO and Rimoin DL: Classification
of osteogenesis imperfect. Lancet. 1:1041–1042. 1978. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rauch F and Glorieux FH: Osteogenesis
imperfecta. Lancet. 363:1377–1385. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Forlino A, Cabral WA, Barnes AM and Marini
JC: New perspectives on osteogenesis imperfecta. Nat Rev
Endocrinol. 7:540–557. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Van Dijk FS and Sillence DO: Osteogenesis
imperfecta: Clinical diagnosis, nomenclature and severity
assessment. Am J Med Genet A. 164A:1470–1481. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rauch F, Fahiminiya S, Majewski J,
Carrot-Zhang J, Boudko S, Glorieux F, Mort JS, Bächinger HP and
Moffatt P: Cole-Carpenter syndrome is caused by a heterozygous
missense mutation in P4HB. Am J Hum Genet. 96:425–431. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Garbes L, Kim K, Rieß A, Hoyer-Kuhn H,
Beleggia F, Bevot A, Kim MJ, Huh YH, Kweon HS, Savarirayan R, et
al: Mutations in SEC24D, encoding a component of the COPII
machinery, cause a syndromic form of osteogenesis imperfecta. Am J
Hum Genet. 96:432–439. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mendoza-Londono R, Fahiminiya S, Majewski
J; Care4Rare Canada Consortium, ; Tétreault M, Nadaf J, Kannu P,
Sochett E, Howard A, Stimec J, et al: Recessive osteogenesis
imperfecta caused by missense mutations in SPARC. Am J Hum Genet.
96:979–985. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cheung MS and Glorieux FH: Osteogenesis
imperfecta: Update on presentation and management. Rev Endocr Metab
Disord. 9:153–160. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu W, Gu F, Ji J, Lu D, Li X and Ma X: A
novel COL1A1 nonsense mutation causing osteogenesis imperfecta in a
Chinese family. Mol Vis. 13:360–365. 2007.PubMed/NCBI
|
|
11
|
Qin W, He JX, Shi J, Xing QH, Gao JJ and
He L, Qian XQ, Liu ZJ, Shu AL and He L: Mutation detection of
COL1A1 gene in a pedigree with osteogenesis imperfecta. Yi Chuan
Xue Bao. 32:248–252. 2005.(In Chinese). PubMed/NCBI
|
|
12
|
Xia XY, Cui YX, Huang YF, Pan LJ, Yang B,
Wang HY, Li XJ, Shi YC, Lu HY and Zhou YC: A novel RNA-splicing
mutation in COL1A1 gene causing osteogenesis imperfecta type I in a
Chinese family. Clin Chim Acta. 398:148–151. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lu Y, Ren X, Wang Y, Li T, Li F, Wang S,
Xu C, Wu G, Li H, Li G, et al: Mutational and structural
characteristics of four novel heterozygous C-propeptide mutations
in the proα1(I) collagen gene in Chinese osteogenesis imperfecta
patients. Clin Endocrinol (Oxf). 80:524–531. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang ZL, Zhang H, Ke YH, Yue H, Xiao WJ,
Yu JB, Gu JM, Hu WW, Wang C, He JW and Fu WZ: The identification of
novel mutations in COL1A1, COL1A2, and LEPRE1 genes in Chinese
patients with osteogenesis imperfecta. J Bone Miner Metab.
30:69–77. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang Z, Li M, He JW, Fu WZ, Zhang CQ and
Zhang ZL: Phenotype and genotype analysis of Chinese patients with
osteogenesis imperfecta type V. PLoS One. 8:e723372013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang H, He JW, Gao G, Yue H, Yu JB, Hu
WW, Gu JM, Hu YQ, Li M, Fu WZ, et al: Polymorphisms in the HOXD4
gene are not associated with peak bone mineral density in Chinese
nuclear families. Acta Pharmacol Sin. 31:977–983. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Maynard LM, Guo SS, Chumlea WC, Roche AF,
Wisemandle WA, Zeller CM, Towne B and Siervogel RM: Total-body and
regional bone mineral content and areal bone mineral density in
children aged 8–18 y: The Fels Longitudinal Study. Am J Clin Nutr.
68:1111–1117. 1998.PubMed/NCBI
|
|
18
|
Wang Y, Cui Y, Zhou X and Han J:
Development of a high-throughput resequencing array for the
detection of pathogenic mutations in osteogenesis imperfecta. PLoS
One. 10:e01195532015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hoefele J, Mayer K, Marschall C, Alberer
M, Klein HG and Kirschstein M: Rare co-occurrence of osteogenesis
imperfecta type I and autosomal dominant polycystic kidney disease.
World J Pediatr. April 8–2016.(Epub ahead of print). doi:
10.1007/s12519-016-0014-1. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lin HY, Chuang CK, Su YN, Chen MR, Chiu
HC, Niu DM and Lin SP: Genotype and phenotype analysis of Taiwanese
patients with osteogenesis imperfecta. Orphanet J Rare Dis.
10:1522015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Prockop DJ, Constantinou CD, Dombrowski
KE, Hojima Y, Kadler KE, Kuivaniemi H, Tromp G and Vogel BE: Type I
procollagen: The gene-protein system that harbors most of the
mutations causing osteogenesis imperfecta and probably more common
heritable disorders of connective tissue. Am J Med Genet. 34:60–67.
1989. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Rauch F, Lalic L, Roughley P and Glorieux
FH: Genotype-phenotype correlations in nonlethal osteogenesis
imperfecta caused by mutations in the helical domain of collagen
type I. Eur J Hum Genet. 18:642–647. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Byers PH: Brittle bones-fragile molecules:
Disorders of collagen gene structure and expression. Trends Genet.
6:293–300. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Marini JC, Forlino A, Cabral WA, Barnes
AM, San Antonio JD, Milgrom S, Hyland JC, Körkkö J, Prockop DJ, De
Paepe A, et al: Consortium for osteogenesis imperfecta mutations in
the helical domain of type I collagen: Regions rich in lethal
mutations align with collagen binding sites for integrins and
proteoglycans. Hum Mutat. 28:209–221. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lee KS, Song HR, Cho TJ, Kim HJ, Lee TM,
Jin HS, Park HY, Kang S, Jung SC and Koo SK: Mutational spectrum of
type I collagen genes in Korean patients with osteogenesis
imperfecta. Hum Mutat. 27:5992006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Paterson CR, Monk EA and McAllion SJ: How
common is hearing impairment in osteogenesis imperfecta? J Laryngol
Otol. 115:280–282. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kuurila K, Kaitila I, Johansson R and
Grénman R: Hearing loss in Finnish adults with osteogenesis
imperfecta: A nationwide survey. Ann Otol Rhinol Laryngol.
111:939–946. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kuurila K, Grenman R, Johansson R and
Kaitila I: Hearing loss in children with osteogenesis imperfecta.
Eur J Pediatr. 159:515–519. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ben Amor IM, Roughley P, Glorieux FH and
Rauch F: Skeletal clinical characteristics of osteogenesis
imperfecta caused by haploinsufficiency mutations in COL1A1. J Bone
Miner Res. 28:2001–2007. 2013. View Article : Google Scholar : PubMed/NCBI
|