Introduction
As one of the most common types of cancer worldwide,
esophageal cancer ranks seventh in incidence and sixth in leading
cause of cancer-associated mortality (1), occurring with an incidence that
varies by ~300-fold worldwide. China and central Asia are
represented with the highest rates, particularly in certain
counties of China bordering Henan, Hebei and part of Shanxi
province; the mortality is as high as 1,100/100,000 (2–4).
Esophageal squamous cell carcinoma (ESCC) is the
dominant pathological type, affecting males more often than females
(5). Its etiology involves several
factors, including genetic, environmental, dietary and hereditary
influences (2). Early stage ESCC
clinical symptoms and signs are atypical and difficult to detect,
thus presenting challenges for diagnosis. In addition, the
progression from early to late stage ESCC may be rapid, due to the
fact that the esophagus lacks serosa and is adjacent to important
organs including the trachea and aorta. At present, the widely
recommended management of ESCC is the combined strategy of surgical
excision and concurrent chemoradiotherapy, however efficacy remains
unsatisfactory with a low 5-year survival rate and a high
prevalence of invasion and metastasis after treatment (6). Therefore, it is important for the
control and treatment of ESCC to identify and characterize
clinically applicable tumor-specific molecular biomarkers for early
detection and targeted prevention. Traditionally, the clinical
staging system and pathological standards were used to predict
clinical outcome in ESCC, however these are of limited value.
Unlike other squamous cell carcinomas, a large proportion of ESCC
progresses to metastasis and the dissemination of cancer cells,
thus novel targets for clinical intervention are required.
Chromosome 14 open reading frame 166 (C14orf166) is
a 28 kD protein whose conserved gene is located on chromosome 14 at
14q22.1, and a transcriptional regulator associated with the
repression of the centrosome architecture (7). C14orf166 was first identified as an
influenza A virus-associated protein; it can promote viral RNA
replication and transcriptional activation by positively modulating
host RNA polymerase (RNAP) II and viral RNAP activities. The
interaction between C14orf166 and the virus polymerase complex is
essential for this function (8,9).
Additionally, C14orf166 has been reported to be a host protein that
interacts with hepatitis C virus during mRNA metabolism and affects
host cellular function (10).
C14orf166 was also observed to modulate transcription and
translation by interacting with various transactivators (11,12).
It is regarded as a shuttling protein that transports RNAs between
the nucleus and cytoplasm, and serves a prominent role in RNA fate
and selective gene expression. It has been demonstrated that the
C14orf166-DDX1-HSPC117-FAM98B complex helps RNAs shuttle back and
forth (13). C14orf166 has also
been demonstrated to be a binding partner of Janus kinase (JAK) 2,
which can activate excessive signal transducer and activator of
transcription (STAT) 3 function to unbalance its tumor promotion
and anti-tumor function, hence initiating tumorigenesis (14,15).
An increasing number of studies have demonstrated that C14orf166
overexpression was identified in a variety of malignant tumor
tissues compared with their paired adjacent non-cancerous tissues,
including pancreatic cancer, brain tumor, cervical carcinoma and
nasopharyngeal carcinoma. Furthermore, C14orf166 has been regarded
as a potential serum biomarker for early diagnosis of tumors, in
addition to a predictor for poor prognosis (16–19).
However, C14orf166 expression and its role in ESCC require further
analysis.
In the present study, C14orf166 expression was
detected by immunohistochemistry, western blotting and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) on
ESCC tissues, adjacent non-cancerous tissues and several esophageal
cancer cell lines respectively. The association between C14orf166
expression and the clinicopathological characteristics of ESCC were
analyzed, highlighting the association between C14orf166
overexpression and the occurrence, development and prognosis of
ESCC.
Materials and methods
Clinical samples and cell lines
The 100 patients enrolled in the current study (83
males and 17 females) were recruited from the Department of
Thoracic Surgery (Xiangya Hospital; Central South University,
Changsha, China) between January 2010 and September 2012, and had
been diagnosed with ESCC by three pathologists. The age of the
patients ranged from 41 to 74 years and the median age was 58.7
years. They had no history of previous malignancies, and had not
previously received chemotherapy, radiotherapy or other treatments
before being sampled; the clinical and pathological data were
complete and reliable. Tumor differentiation and staging was
classified according to the 7th edition of TNM classification of
Union Internationale Contra Cancrum (20). Informed consent was acquired from
all patients before surgery and the present study was approved by
the Ethics Committee of Xiangya Hospital, Central South University.
The use of the information and specimens collected has been handled
and anonymized according to the ethical and legal standards.
ESCC tissues and paired adjacent normal tissues
(greater than 5 cm away from the tumor margin) were obtained from
those patients by esophagus resection. All specimens were
immediately snap-frozen in liquid nitrogen and stored at −80°C
until RNA and total protein extraction could be performed.
The subjects were followed-up every 3 months during
the first postoperative year and for a minimum of 6 months
afterwards for survival and recurrence inquiry until death or until
the end of the investigation.
Human ESCC cell lines (TE-1, EC109 and EC9706) were
obtained from the Cell Bank of the Chinese Academy of Sciences
(Shanghai, China). A normal human esophageal epithelial cell line
(HEEpic) was purchased from the American Type Culture Collection
(Manassas, VA, USA).
RT-qPCR
Total RNA was extracted from tissues or cells using
TRIzol (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA,
USA) according to the manufacturer's protocol. After treatment with
the DNA-free kit (Ambion; Thermo Fisher Scientific, Inc.) to remove
the chromosomal DNA, the complementary DNA was synthesized using
the GoScript RT kit (catalog no. A5001; Promega Corporation,
Madison, WI, USA) and stored at −20°C until use. The mRNA
expression levels of C14orf166 and β-actin were determined by
RT-qPCR using the ABI PRISM 7500 sequence detector system (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The primer sequences
were sense/anti-sense: C14orf166:
5′-TGCATTGTCAGCAGTTTTTGA-3′/5′-TGACTGGCTTCTTGGTTTAGC-3′; and
β-actin: 5′-GCACCACACCTTCTACAATGAG-3′/5′-GATAGCACAGCCTGGATAGCA-3′.
The mRNA expression levels of the target genes were normalized to
the β-actin signal. All the reactions were conducted in triplicate
using 20 µl samples containing 50 ng complementary DNA. The
reaction protocol involved heating for 10 min at 95°C, followed by
40 cycles of amplification (15 sec at 95°C and 1 min at 60°C), and
a final extension step of 15 sec at 95°C and 15 sec at 60°C. The
data were analyzed using the ABI PRISM 7500 Sequence Detection
software. The expression of C14orf166 was described as
2-ΔΔCq (21).
Western blot analysis
The cells and tumor tissues were collected and lysed
on ice. Subsequent to centrifugation at 12,000 × g for 20 min, the
concentration of proteins was measured and protein samples were
denatured by boiling for 10 min, then were and loaded onto a 10%
SDS-PAGE gel for electrophoresis. The proteins were transferred
onto a PVDF membrane (EMD Millipore, Billerica, MA, USA) which was
then incubated in the blocking solution at room temperature for 2
h. Anti-C14orf166 (1:100; catalog no. 19848-1-AP; ProteinTech
Group, Inc., Chicago, IL, USA) and anti-GAPDH (1:5,000; catalog no.
MAB5718; R&D Systems, Inc., Minneapolis, MN, USA) were used for
western blotting, incubated at 4°C overnight. The membranes were
subsequently incubated with horseradish peroxidase (HRP)-labeled
goat anti-rabbit IgG (1:6,000; catalog no. NB730-H; Novus
Biologicals, LLC, Littleton, CO, USA) or HRP-labeled goat
anti-mouse IgG (1:1,000; catalog no. HAF007; Novus Biologicals,
LLC), for 1.5 h at room temperature. Protein expression was
normalized against GAPDH expression. Bands were visualized with the
BeyoECL Plus Detection System (Beyotime Institute of Biotechnology,
Haimen, China) and Bio-Rad ImageLab software version 3.0 (Bio-Rad
Laboratories, Inc., Hercules, CA, USA).
Immunohistochemistry staining
The rapid PV two-step staining method used the
following specifications: Paraffin sections of 5 µm were obtained,
which were heated at 65°C for 60 min, then dewaxed in xylene and
rehydrated via an ethanol series. Subsequently, high-temperature
antigen retrieval was conducted using a microwave in 0.1 M citrate
solution (pH 6.0) for 10 min, followed by incubation in 3%
H2O2 at room temperature for 20 min. The
sections were then incubated in goat serum at room temperature for
20 min, then incubated with anti-C14orf166 rabbit polyclonal
antibody (1:100) at 4°C overnight. Subsequently, incubation with
the secondary anti-rabbit antibody was conducted for 20 min at room
temperature, prior to staining with DAB/hematoxylin, mounting and
examination under the microscope.
Immunohistochemical (IHC) staining was scored
independently by two pathologists who had no knowledge of the
patients' history or condition and any discrepancy was solved by a
consensus. The score of immunoreactivity was obtained by
calculating the extent and intensity of the staining of the cells
in a semi-quantitative manner. As described previously (22), the standards for evaluation
included the following: Positive stain intensity (0, negative; 1,
weak positive; 2, moderate positive; 3, strong positive) and
proportion of positive areas (≤10%=1, 10–50%=2, ≥50%=3). The
staining score was the multiplication of the two previous scores. A
total of five high power fields in each specimen were selected
randomly with the final score as an average of the five scores.
Samples were classified as negative when the final scores were 0–3
and positive when >4. C14orf166 expression was considered high
when the intensity of staining in the tumor cell nuclei was
stronger than that of the non-tumorous part and detection of normal
esophageal mucosa served as the negative control.
Statistical analysis
SPSS software (version 16.0; SPSS, Inc., Chicago,
IL, USA) was used for statistical analyses. All data were presented
as the means ± standard deviation. Categorical variables were
compared by the χ2 test and continuous variables were compared
using independent two sample t-test. Multivariate analyses were
performed by the Cox proportional hazard model. Survival curves
were performed by the Kaplan-Meier method (the log-rank test). All
tests were two-tailed and P<0.05 was considered to indicate a
statistically significant difference.
Results
C14orf166 was overexpressed in human
ESCC tissues and cell lines
In the present study, in order to investigate the
expression of C14orf166 in ESCC, C14orf166 protein was detected by
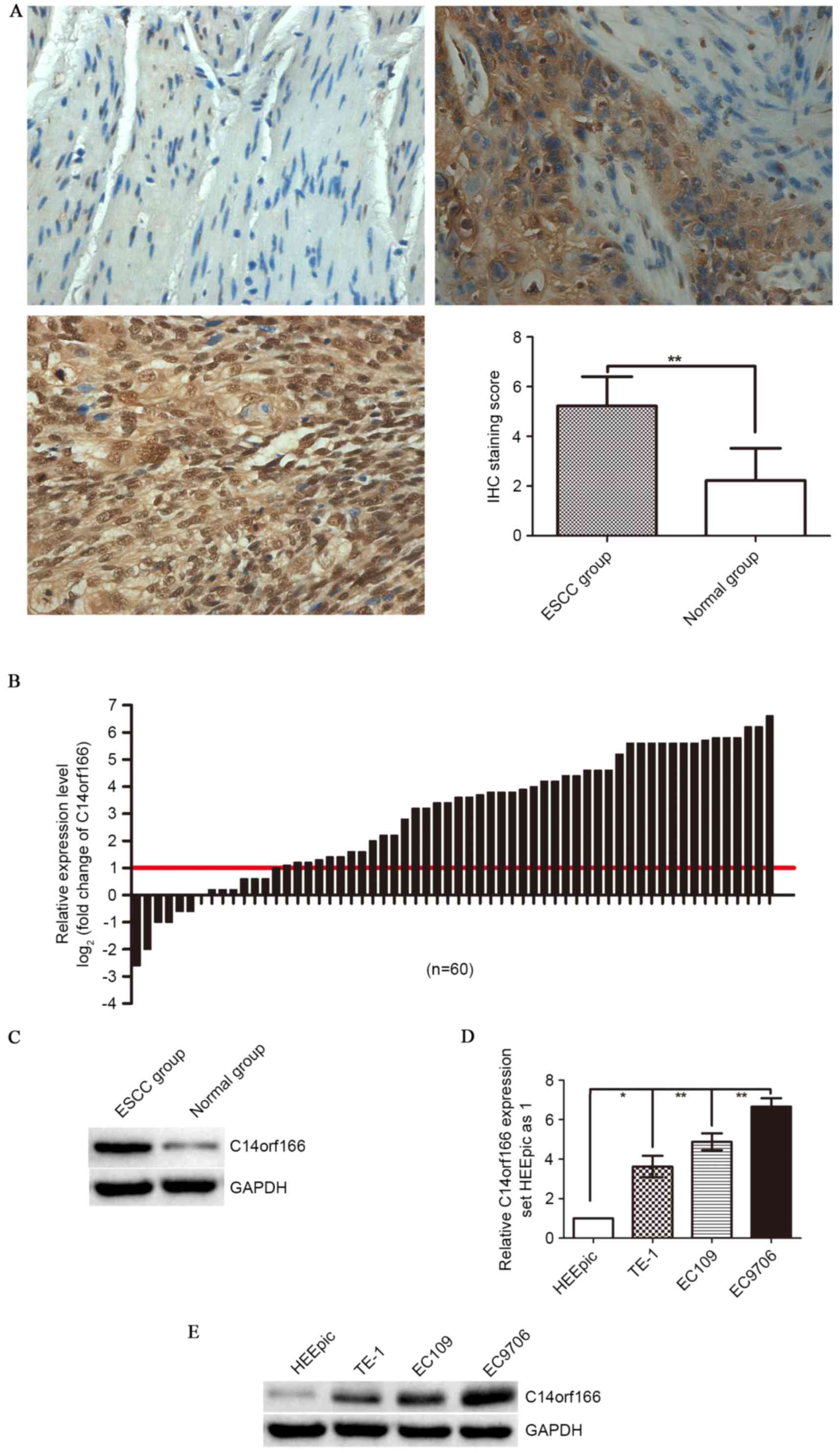
IHC staining and western blotting. According to IHC staining
results, only weak or no immunoreactivity was observed in the
adjacent normal tissues, whereas C14orf166 protein was demonstrated
to be expressed highly in ESCC tissues, predominantly localized in
the carcinoma cell nuclei with cytoplasmic reaction occasionally
displayed (Fig. 1A). According to
the aforementioned standards, the positive expression rate of
C14orf166 protein was 90% (90/100) in the ESCC group, while it was
14% (14/100) in the paired normal tissues group, a difference that
is statistically significant (P<0.001; Fig 1A), this tendency was verified by
western blot analysis (P<0.00; Fig.
1D). Furthermore, when the C14orf166 mRNA expression in the 60
cases of ESCCs and the paired normal tissues was analyzed by
RT-qPCR, there was a similar trend of C14orf166 expression in the
mRNA level observed, compared with the paired adjacent normal
tissues; a 2.3-fold increase for C14orf166 expression was noted in
ESCC tissues (Fig. 1B).
Comparative analysis of ESCCs with paired adjacent normal tissues
further demonstrated that increased C14orf166 expression [more than
2-fold (namely log2 (fold change)>1)] was observed in
76.66% cases (46/60), suggesting that the overexpression of
C14orf166 was a frequent event in human ESCC at mRNA and protein
level.
Subsequently, C14orf166 expression in ESCC cell
lines and normal esophagus epithelial cell line was detected by
RT-qPCR and western blotting. As Fig.
1D and E demonstrate, compared with the expression in normal
cells (HEEpic), C14orf166 mRNA and protein expression levels were
elevated significantly in human ESCC cell lines (TE-1, EC109 and
EC9706).
Elevated C14orf166 expression was
associated with clinicopathological characteristics in ESCC
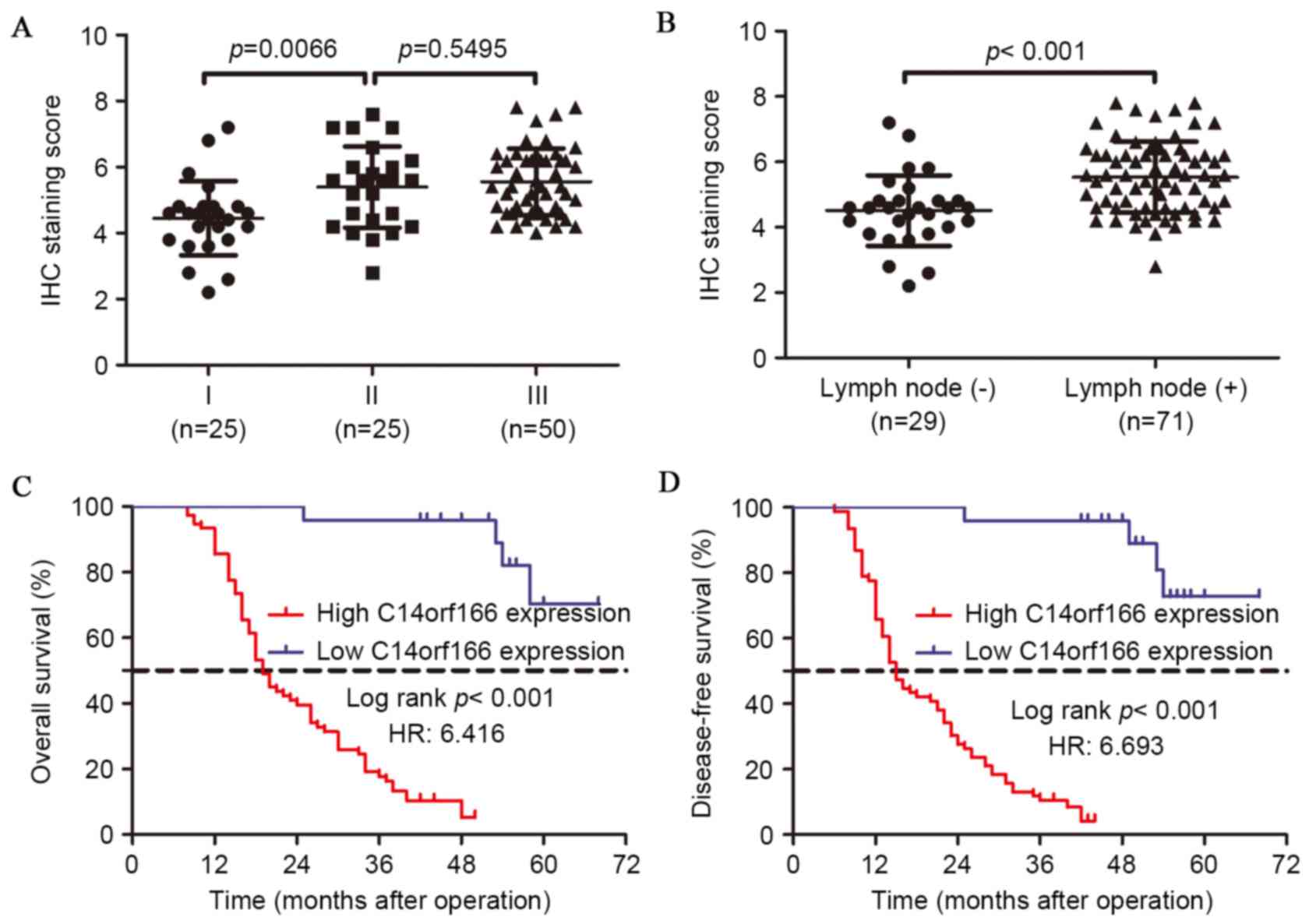
The association between C14orf166 expression and the
clinicopathological features of ESCC was explored by the χ2 test.
As summarized in Table I, the high
expression of C14orf166 was significantly associated with T staging
(P<0.001), lymph node metastasis (P=0.000) and TNM stage
(P=0.000) respectively. However, no significant association was
observed between C14orf166 expression and variables including age
(P=0.058), gender (P=0.239), drinking history (P=0.954), tumor size
(P=0.093) or tumor differentiation (P=0.518). Subgroup analysis
indicated that overexpressed C14orf166 was associated with advanced
stage and lymph node metastasis (Fig.
2A and B). As presented in Table
I, no statistical difference was identified between C14orf166
overexpression and the success of subsequent adjuvant therapy
following the operation, which was likely affected by multiple
factors, including disease degree and family income. In addition,
in the 44 patients in the study who received chemotherapy or
radiotherapy, no association between the increased C14orf166
expression and therapeutic effect was identified (P=0.227).
 | Table I.C14orf166 expression levels and
clinicopathological features in 100 cases of esophageal squamous
cell carcinoma. |
Table I.
C14orf166 expression levels and
clinicopathological features in 100 cases of esophageal squamous
cell carcinoma.
|
|
| C14orf166
expression |
|
|---|
|
|
|
|
|---|
| Clinicopathological
variable | n | Low | High | P-value |
|---|
| Age (years) |
|
|
|
|
| ≤60 | 54 | 17 | 37 |
|
|
>60 | 46 | 7 | 39 | 0.058 |
| Gender |
|
|
|
|
|
Female | 16 | 2 | 14 |
|
| Male | 84 | 22 | 62 | 0.239 |
| Drinking history |
|
|
|
|
|
Yes | 37 | 9 | 28 |
|
| No | 63 | 15 | 48 | 0.954 |
| Tumor size
(cm) |
|
|
|
|
| ≤4 | 56 | 17 | 39 |
|
|
>4 | 44 | 7 | 37 | 0.093 |
|
Differentiation |
|
|
|
|
|
Well | 27 | 6 | 21 |
|
|
Moderate | 49 | 14 | 35 |
|
|
Poor | 24 | 4 | 20 | 0.518 |
| T Stage |
|
|
|
|
| T1 | 35 | 15 | 20 |
|
| T2 | 14 | 7 | 7 |
|
| T3 | 39 | 1 | 38 |
|
| T4 | 12 | 1 | 11 | <0.001 |
| N Stage |
|
|
|
|
| N0 | 28 | 18 | 10 |
|
| N1 | 42 | 3 | 39 |
|
| N2 | 21 | 2 | 19 |
|
| N3 | 9 | 1 | 8 | <0.0001 |
| TNM Stage |
| I | 25 | 21 | 4 |
|
| II | 25 | 2 | 23 |
|
|
III | 50 | 1 | 49 | <0.0001 |
| Adjuvant therapy
following operation |
|
|
|
|
|
Yes | 44 | 8 | 36 |
|
| No | 56 | 16 | 40 | 0.227 |
Overexpression of C14orf166 was
associated with the poor prognosis of ESCC
To assess the role of C14orf166 expression in ESCC
prognosis, the 100 patients were followed up subsequent to the
operations. Kaplan-Meier survival analysis and the log-rank test
were performed and demonstrated that a high level of C14orf166
expression is closely associated with a shorter overall survival
(OS) time (P<0.001) and disease-free survival (DFS) time
(P<0.001); the patients with low C14orf166 expression had an
approximately 3-fold longer OS time and 4-fold longer DFS time than
those with overexpressed C14orf166.
To validate the feasibility of C14orf166 expression
in ESCC prognosis, the Cox proportional hazards regression model
was introduced. Multivariate survival analysis on all
characteristics demonstrated that survival time was significantly
dependent on lymph node involvement, TNM stage and C14orf166
expression level (Table II), they
were independent prognostic factors for OS and DFS, indicating that
C14orf166 may be a potential clinical prognostic predicator for
patients with ESCC.
 | Table II.Cox regression multivariate analysis
of overall and disease-free survival in 100 patients with
esophageal squamous cell carcinoma. |
Table II.
Cox regression multivariate analysis
of overall and disease-free survival in 100 patients with
esophageal squamous cell carcinoma.
|
|
| Overall
survival | Disease-free
survival |
|---|
|
|
|
|
|
|---|
| Variable | n | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age (years) |
|
≤60 | 54 | 1 |
| 1 |
|
|
>60 | 46 | 0.79
(0.42–1.51) | 0.483 | 0.73
(0.39–1.35) | 0.313 |
| Gender |
|
|
|
|
|
|
Female | 16 | 1 |
| 1 |
|
|
Male | 84 | 0.75
(0.36–1.58) | 0.449 | 0.60
(0.28–1.30) | 0.198 |
| Drinking
history |
|
|
|
|
|
|
Yes | 37 | 1 |
| 1 |
|
| No | 63 | 1.14
(0.61–2.13) | 0.683 | 1.36
(0.74–2.52) | 0.320 |
| Tumor size
(cm) |
|
|
|
|
|
| ≤4 | 56 | 1 |
| 1 |
|
|
>4 | 44 | 1.43
(0.81–2.53) | 0.218 | 1.64
(0.95–2.83) | 0.074 |
|
Differentiation |
|
| 0.930 |
| 0.809 |
|
Well | 27 | 1 |
| 1 |
|
|
Moderate | 49 | 0.97
(0.48–1.99) | 0.942 | 1.24
(0.63–2.45) | 0.532 |
|
Poor | 24 | 0.87
(0.39–1.95) | 0.737 | 1.11
(0.51–2.41) | 0.794 |
| T stage |
|
| 0.620 |
| 0.254 |
| T1 | 35 | 1 |
| 1 |
|
| T2 | 14 | 1.88
(0.67–5.28) | 0.233 | 2.53
(0.91–7.00) | 0.075 |
| T3 | 39 | 1.56
(0.45–5.39) | 0.482 | 1.77
(0.55–5.75) | 0.339 |
| T4 | 12 | 2.42
(0.32–18.08) | 0.391 | 3.18
(0.43–23.71) | 0.260 |
| N stage |
|
| 0.023 |
| 0.032 |
| N0 | 28 | 1 |
| 1 |
|
| N1 | 42 | 2.41
(0.42–13.67) | 0.322 | 2.16
(0.44–10.62) | 0.345 |
| N2 | 21 | 3.50
(0.45–27.28) | 0.231 | 3.81
(0.56–25.94) | 0.172 |
| N3 | 9 | 46.19
(3.13–681.5) | 0.005 | 20.19
(1.86–218.74) | 0.013 |
| TNM stage |
|
| 0.004 |
| 0.006 |
| I | 25 | 1 |
| 1 |
|
| II | 25 | 1.93
(0.24–15.64) | 0.537 | 2.66
(0.35–20.39) | 0.348 |
|
III | 50 | 36.85
(1.51–900.7) | 0.027 | 42.89
(1.83–1003.01) | 0.019 |
| C14orf166
expression |
|
Low | 24 | 1 |
| 1 |
|
|
High | 76 | 16.96
(1.89–152.41) | 0.012 | 20.33
(2.25–183.34) | 0.007 |
Discussion
As a constituent of nuclear and cytoplasmic protein
complexes, C14orf166 is located in the nuclear and cytoplasm and,
partly as a result of this location, it has been identified to be a
protein serving a vital role in cellular RNA expression regulation,
including RNA transcription, maturation and translation. C14orf166
participates in RNAP II-directed RNA transcription by interacting
with the RNAP II directly, aiding to shuttle important elements. It
has also been reported to transport specific mRNAs from the cell
body to the dendrites in the developing brain, which additionally
implies a significant role in brain development (23). Functions of C14orf166 account for
normal RNA transcription, mRNA expression, signal transduction, and
important cellular phenotypes including proliferation, apoptosis
and differentiation. Blocking C14orf166 will reduce mRNA
translation by 50%, severely impairing cellular activity (12). However, there are also genes
associated with oncogenesis and tumor invasion behavior under the
regulation of C14orf166, and overexpression of C14orf166 was
identified in multiple tumor tissues relative to its adjacent
normal tissues (16–19).
To the best of our knowledge, there have been no
investigations into a potential role for C14orf166 in ESCC at
present and this is the first study on the possible association
between C14orf166 and ESCC. The present study investigated the
protein and mRNA expression of C14orf166 in a series of paired ESCC
specimens with intact follow-up data. C14orf166 was first
identified to be consistently upregulated in ESCC tissues and cell
lines. Consistent with the expression in other carcinoma tissues,
IHC staining demonstrated that C14orf166 protein expression was
clearly elevated in ESCC tissues, as demonstrated by western
blotting. RT-qPCR analysis additionally displayed a high frequency
of C14orf166 mRNA overexpression in the majority of ESCC cases,
which demonstrated C14orf166 was transcriptionally and
translationally upregulated in ESCC, indicating that C14orf166 may
have a significant role in ESCC initiation.
From the clinical data, C14orf166 expression was
correlated with respect to various clinicopathological
characteristics in 100 patients with ESCC. As predicted, the high
expression of C14orf166 had no correlation with age, gender,
drinking history, tumor size or even tumor differentiation, however
it demonstrated a strong association with the T staging, lymph node
involvement and TNM stage. This indicated that the higher
expression of C14orf166 leads to the more malignant biological
characteristics, despite the tumor differentiation; this is
consistent with a previous study in pancreatic cancer (18). Additionally, the data produced by
the current study demonstrated that elevated C14orf166 expression
was a negative prognostic predicator for patients with ESCC. As an
adjuvant therapy, concurrent radiochemotherapy is used clinically
to improve the prognosis of patients with ESCC, while patients
undergoing adjuvant therapy experience a higher frequency of severe
side effects including oral mucous ulceration and vomiting. Thus,
C14orf166 may represent a potential biomarker to guide the
selection of therapeutic strategy and improve survival, although
those who received adjuvant therapy in the present study could not
demonstrate this advantage, which was limited by the small size of
the study. Furthermore, C14orf166 has been reported to be detected
in the serum of pancreatic carcinoma (13), which indicates a potential novel
biomarker for early diagnosis and an aid in therapy design.
Therefore, further investigations are required to fully elucidate
this potential biomarker.
In addition to previous studies, the results of the
present study indicate that C14orf166 may participate in
oncogenesis and tumor progression, which may be partly explained by
the fact that, in addition to regulation of mRNA expression,
C14orf166 serves a vital role in signal transduction during cell
division; protein correlation profiling analysis in diverging cells
demonstrated that C14orf166 interplayed with ninein in the
centrosomes (24), which was
confirmed by Howng et al (25) whose research demonstrated that
C14orf166 could bind to the ninein, thus inhibiting phosphorylation
by glycogen synthase kinase 3 β (GSK-3β). GSK-3β can phosphorylate
a variety of substrates participating in signal transduction and
cell proliferation, in addition to organ development. Dysfunction
of GSK-3β is prevalent in tumor formation, promoting progression
and contributing to drug resistance by interfering with signaling
pathways, including phosphatidylinositol-3-kinase (PI3K)-protein
kinase B (AKT)-mammalian target of rapamycin (mTOR), Wnt/β-catenin
and JAK2/STAT3 signaling pathways (26–28).
Abnormal expression of PI3K-AKT-mTOR is frequent in melanoma and
indicates a poor prognosis, and thus inhibition of the mTOR pathway
will benefit numerous patients in the clinic (29). Ge et al (30) demonstrated that through abnormal
activation of Wnt/β-catenin pathway, miR-942 aided to maintain
cancer stem cell-like traits in ESCC, inducing poor prognosis and
unsatisfactory drug effects. In addition, GSK-3β can promote
esophagus carcinoma cells metastasis and spread by degrading
β-catenin (31,32). The dysregulated intracellular
JAK2/STAT3 signaling pathway is common in numerous types of
carcinoma and is associated with malignancy and poor prognosis, an
antagonist of the pathway attenuates tumor bearing and improves
drug efficacy (33,34). Zhang et al (19) identified that C14orf166
overexpression was associated with lymph node involvement and
shorter OS and DFS in cervical cancer, which was associated with
abnormal JAK2/STAT3 pathway activity. Chen et al (35) reported that C14orf166, together
with acylglycerol kinase, were identified as JH2-interacting
proteins, which consecutively activated the JAK2/STAT3 signaling
pathway to promote esophageal squamous cell generation and enhanced
the cancer stem cell population. A JAK2 inhibitor will block the
growth of the ESCC through the JAK/STAT3 pathway (36). The current study hypothesized that
influenced by this signaling pathway, C14orf166 modifies the
downstream signal transduction via GSK-3β, affecting the
constitution and stability of centrosomes, and excessive C14orf166
expression promotes gene translation, resulting in tumor
origination and progression.
Taken together, these data in the present study
suggest that C14orf166 may serve a vital role in the formation and
progression of ESCC. C14orf166 may be a potential biomarker for
lymph node metastasis and poor prognosis in ESCC, which may aid in
the identification of patients at high risk and offer a rationale
for selecting appropriate treatment. Although the present study
shows preliminary data for the potential association between
C14orf166 overexpression and ESCC, further investigation is
required to fully elucidate the underlying mechanism of C14orf166
in regulating the oncogenesis, progression, metastasis and
prognosis of ESCC.
Acknowledgements
The current study was supported by the National
Natural Scientific Foundation of China (grant no. 81372515) and the
Science Funds for Young Scholar of Xiangya Hospital (grant no.
2013Q02).
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Guohong Z, Min S, Duenmei W, Songnian H,
Min L, Jinsong L, Hongbin L, Feng Z, Dongping T, Heling Y, et al:
Genetic heterogeneity of oesophageal cancer in high-incidence areas
of southern and northern China. PLoS One. 5:e96682010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jemal A, Murray T, Samuels A, Ghafoor A,
Ward E and Thun MJ: Cancer statistics, 2003. CA Cancer J Clin.
53:5–26. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tran GD, Sun XD, Abnet CC, Fan JH, Dawsey
SM, Dong ZW, Mark SD, Qiao YL and Taylor PR: Prospective study of
risk factors for esophageal and gastric cancers in the Linxian
general population trial cohort in China. Int J Cancer.
113:456–463. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Enzinger PC and Mayer RJ: Esophageal
cancer. N Engl J Med. 349:2241–2252. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Siegel R, DeSantis C, Virgo K, Stein K,
Mariotto A, Smith T, Cooper D, Gansler T, Lerro C, Fedewa S, et al:
Cancer treatment and survivorship statistics, 2012. CA Cancer J
Clin. 62:220–241. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lupi I, Broman KW, Tzou SC, Gutenberg A,
Martino E and Caturegli P: Novel autoantigens in autoimmune
hypophysitis. Clin Endocrinol (Oxf). 69:269–278. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rodriguez A, Pèrez-González A and Nieto A:
Cellular human CLE/C14orf166 protein interacts with influenza virus
polymerase and is required for viral replication. J Virol.
85:12062–12066. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Naili MN Natasya, Hasnita CH, Shamim AK,
Hasnan J, Fauziah MI, Narazah MY, James A, Zulkiflee S, Nidzam MM
and Zilfalil BA: Chromosomal alterations in Malaysian patients with
nasopharyngeal carcinoma analyzed by comparative genomic
hybridization. Cancer Genet Cytogenet. 203:309–312. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lee JW, Liao PC, Young KC, Chang CL, Chen
SS, Chang TT, Lai MD and Wang SW: Identification of hnRNPH1, NF45l,
and C14orf166 as novel host interacting partners of the mature
hepatitis C virus core protein. J Proteome Res. 10:4522–4534. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Schamel K, Staeheli P and Hausmann J:
Identification of the immunodominant H-2K(k)-restricted cytotoxic
T-cell epitope in the Borna disease virus nucleoprotein. J Virol.
75:8579–8588. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Huarte M, Sanz-Ezquerro JJ, Roncal F,
Ortin J and Nieto A: PA subunit from influenza virus polymerase
complex interacts with a cellular protein with homology to a family
of transcriptional activators. J Virol. 75:8597–8604. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pèrez-González A, Pazo A, Navajas R,
Ciordia S, Rodriguez-Frandsen A and Nieto A: hCLE/C14orf166
associates with DDX1-HSPC117-FAM98B in a novel
transcription-dependent shuttling RNA-transporting complex. PLoS
One. 9:e909572014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yu H, Pardoll D and Jove R: STATs in
cancer inflammation and immunity: A leading role for STAT3. Nat Rev
Cancer. 9:798–809. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ernst M and Putoczki TL: Stat3: Linking
inflammation to (gastrointestinal) tumourigenesis. Clin Exp
Pharmacol Physiol. 39:711–718. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang J, Gu Y, Wang L, Hang X, Gao Y, Wang
H and Zhang C: HUPO BPP pilot study: A proteomics analysis of the
mouse brain of different developmental stages. Proteomics.
7:4008–4015. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Guo J, Wang W, Liao P, Lou W, Ji Y, Zhang
C, Wu J and Zhang S: Identification of serum biomarkers for
pancreatic adenocarcinoma by proteomic analysis. Cancer Sci.
100:2292–2301. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cui Y, Wu J, Zong M, Song G, Jia Q, Jiang
J and Han J: Proteomic profiling in pancreatic cancer with and
without lymph node metastasis. Int J Cancer. 124:1614–1621. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang W, Ou J, Lei F, Hou T, Wu S, Niu C,
Xu L and Zhang Y: C14ORF166 overexpression is associated with
pelvic lymph node metastasis and poor prognosis in uterine cervical
cancer. Tumour Biol. 37:369–379. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sobin LH, Gospodarowicz MK and Wittekind
C: TNM classification of malignant tumors. 7th edition. Oxford:
Wiley-Blackwell; 2010
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C (T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang S, Tang W, Weng S, Liu X, Rao B, Gu
J, Chen S, Wang Q, Shen X, Xue R and Dong L: Apollon modulates
chemosensitivity in human esophageal squamous cell carcinoma.
Oncotarget. 5:7183–7197. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Elvira G, Wasiak S, Blandford V, Tong XK,
Serrano A, Fan X, del Rayo Sánchez-Carbente M, Servant F, Bell AW,
Boismenu D, et al: Characterization of an RNA granule from
developing brain. Mol Cell Proteomics. 5:635–651. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Andersen JS, Wilkinson CJ, Mayor T,
Mortensen P, Nigg EA and Mann M: Proteomic characterization of the
human centrosome by protein correlation profiling. Nature.
426:570–574. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Howng SL, Hsu HC, Cheng TS, Lee YL, Chang
LK, Lu PJ and Hong YR: A novel ninein-interaction protein, CGI-99,
blocks ninein phosphorylation by GSK3beta and is highly expressed
in brain tumors. FEBS Lett. 566:162–168. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhao P, Li Q, Shi Z, Li C, Wang L, Liu X,
Jiang C, Qian X, You Y, Liu N, et al: GSK-3β regulates tumor growth
and angiogenesis in human glioma cells. Oncotarget. 6:31901–31915.
2015.PubMed/NCBI
|
|
27
|
Gao Y, Liu Z, Zhang X, He J, Pan Y, Hao F,
Xie L, Li Q, Qiu X and Wang E: Inhibition of cytoplasmic GSK-3β
increases cisplatin resistance through activation of Wnt/β-catenin
signaling in A549/DDP cells. Cancer Lett. 336:231–239. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Haraguchi K, Ohsugi M, Abe Y, Semba K,
Akiyama T and Yamamoto T: Ajuba negatively regulates the Wnt
signaling pathway by promoting GSK-3beta-mediated phosphorylation
of beta-catenin. Oncogene. 27:274–284. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kong Y, Si L, Li Y, Wu X, Xu X, Dai J,
Tang H, Ma M, Chi Z, Sheng X, et al: Analysis of mTOR gene
aberrations in melanoma patients and evaluation of their
sensitivity to PI3K-AKT-mTOR pathway inhibitors. Clin Cancer Res.
22:1018–1027. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ge C, Wu S, Wang W, Liu Z, Zhang J, Wang
Z, Li R, Zhang Z, Li Z, Dong S, et al: miR-942 promotes cancer stem
cell-like traits in esophageal squamous cell carcinoma through
activation of Wnt/β-catenin signalling pathway. Oncotarget.
6:10964–10977. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
He H, Ding F, Li Y, Luo A, Chen H, Wu C
and Liu Z: Migfilin regulates esophageal cancer cell motility
through promoting GSK-3β-mediated degradation of β-catenin. Mol
Cancer Res. 10:273–281. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang W, Yan S, Liu M, Zhang G, Yang S, He
S, Bai J, Quan L, Zhu H, Dong Y and Xu N: Beta-Catenin/TCF pathway
plays a vital role in selenium induced-growth inhibition and
apoptosis in esophageal squamous cell carcinoma (ESCC) cells.
Cancer Lett. 296:113–122. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mukthavaram R, Ouyang X, Saklecha R, Jiang
P, Nomura N, Pingle SC, Guo F, Makale M and Kesari S: Effect of the
JAK2/STAT3 inhibitor SAR317461 on human glioblastoma tumorspheres.
J Transl Med. 13:2692015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhao H, Guo Y, Li S, Han R, Ying J, Zhu H,
Wang Y, Yin L, Han Y, Sun L, et al: A novel anti-cancer agent
Icaritin suppresses hepatocellular carcinoma initiation and
malignant growth through the IL-6/Jak2/Stat3 pathway. Oncotarget.
6:31927–31943. 2015.PubMed/NCBI
|
|
35
|
Chen X, Ying Z, Lin X, Lin H, Wu J, Li M
and Song L: Acylglycerol kinase augments JAK2/STAT3 signaling in
esophageal squamous cells. J Clin Invest. 123:2576–2589. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Fang J, Chu L, Li C, Chen Y, Hu F, Zhang
X, Zhao H, Liu Z and Xu Q: JAK2 inhibitor blocks the inflammation
and growth of esophageal squamous cell carcinoma in vitro through
the JAK/STAT3 pathway. Oncol Rep. 33:494–502. 2015.PubMed/NCBI
|