Introduction
Multiple osteochondromas (MO), formerly referred to
as hereditary multiple exostoses, is an autosomal dominant
inherited disease with a recorded prevalence of 1/50,000 in
Washington, USA, in 1994 (1). MO
commonly presents as a benign bone tumor, however, 1–2% of patients
with MO progress into osteosarcoma or chondrosarcoma (2,3). MO
typically manifests as abnormalities in skeletal development,
primarily involving long bones and the knees, ankles, elbows,
wrists, shoulders and pelvis. Bony prominences of ranging size
typically emerge around the epiphysis of long bones in the limbs of
patients with MO (4,5). Osteochondromas normally emerge during
infancy and tend to present with related symptoms during
adolescence, as the number and size of tumors often increases with
age until growth terminates (6).
Owing to the expanding tumors, patients with MO often suffer from
pain, nerve and vascular compression, joint deformity and
limitation of activity among other symptoms, including a shortened
stature (7,8).
Although the molecular mechanisms underlying MO are
yet to be fully clarified, previous studies have reported that the
exotosin (EXT) family is responsible for MO (9–11).
EXT genes encode proteins that are involved in the biosynthesis of
heparan sulfate (HS), an essential molecule in the growth and
development of normal cells and dysfunction of EXT genes may lead
to MO (12,13). EXT1 (14), EXT2 (15) and EXT3 (16) are the most important members of the
EXT family (17). A common genetic
basis exists in 90% of MO cases: Mutations in EXT1 or EXT2 trigger
an early introduction of the termination codon and corresponding
partial or total EXT gene deletion, resulting in a loss of protein
function. 80% of EXT mutations are nonsense, frameshift or splicing
mutations, with large fragment deletions and insertions rarely
reported (18,19).
With the development of DNA sequencing technology,
gene mapping of diseases via exome sequencing has emerged,
exhibiting the advantage of covering all exon regions. Exome
sequencing has already been applied in the research of tumors and
genetic diseases, through the screening of novel and pathogenic
genetic variants of all exons or targeted sequences (20,21).
In the present study, exome sequencing was performed
on a patient from a three-generation Han Chinese family with
hereditary MO. Data were filtered from the 1000 Genome Project
(http://www.1000genome.org), the Single
Nucleotide Polymorphism (dbSNP) database for Chinese Han SNP (build
132; http://www.ncbi.nlm.nih.gov/project/SNP/) and other
databases. Sanger sequencing of the MO patient, an unaffected
relative and 200 unrelated healthy controls further validated the
candidate mutation. For further confirmation, four individuals from
an unrelated family with MO were examined. In addition,
immunohistochemistry and multiple sequence alignment were performed
to appraise the importance of the identified causal gene
mutation.
Materials and methods
Human subjects
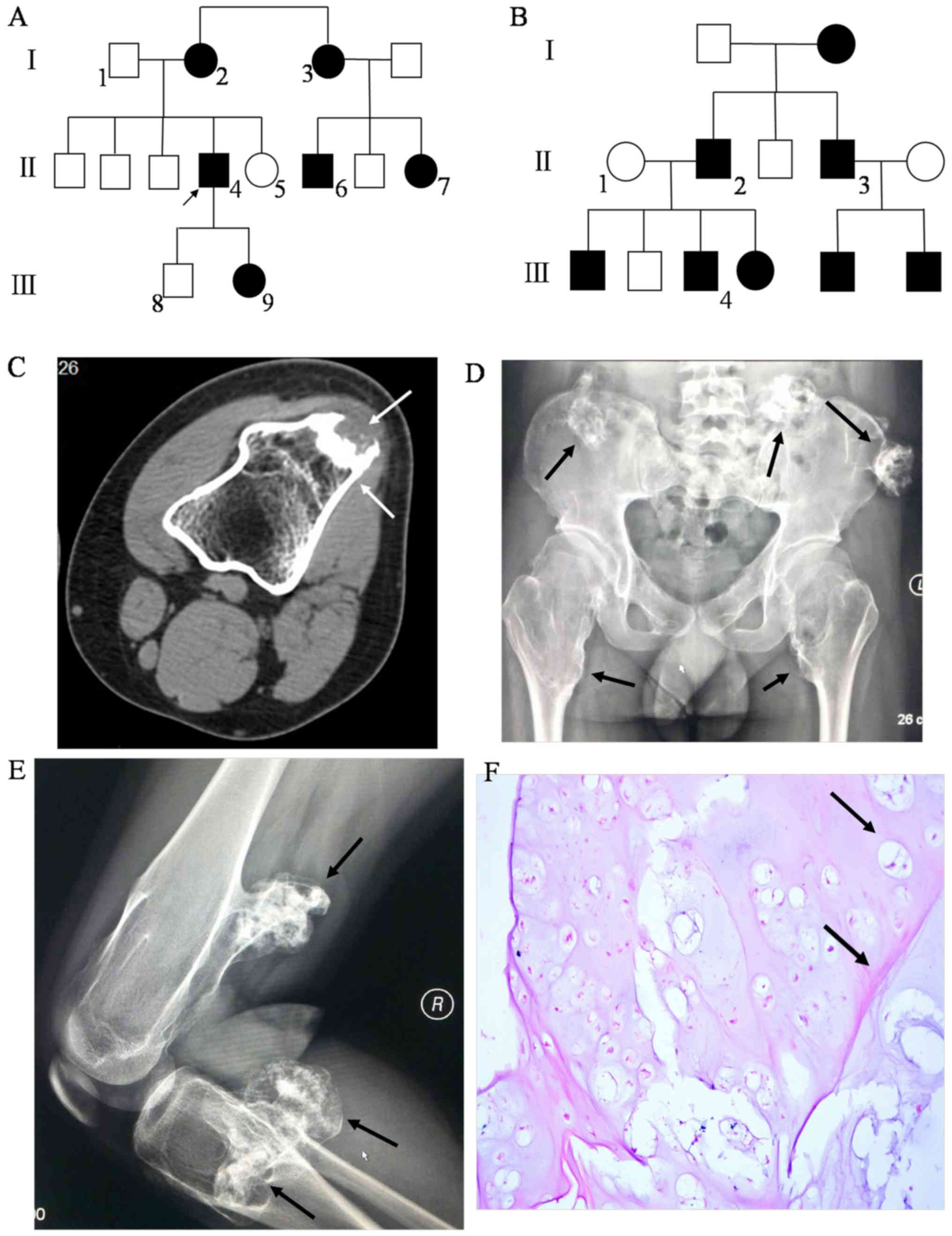
A Han Chinese family with MO from Fujian Province,
China, was included in the present study (Table I; Fig.
1A). The proband (II4) was a 27 year old man who had displayed
multiple, expanding exostoses for twenty years. Another family with
MO (affected individuals: II2, II3 and II4; unaffected individual:
II1) lacking an EXT1 gene mutation were included for further
validation (Fig. 1B). 200
unrelated healthy subjects (100 males and 100 females, aged 10–40
years) with the same geographical ancestry were included as
controls. Clinical data of all subjects was carefully recorded by
the resident, with all participants receiving a general examination
that included a computed radiography examination of long bones,
truncal and acral joints by two experienced, independent orthopedic
physicians. 5 ml venous blood was drawn from 2 family members and
the 200 healthy controls using EDTA-K2 anticoagulant vacuum blood
vessels. The present study was approved by the Ethics Committee of
The Fuzhou Second Affiliated Hospital of Xiamen University. All
subjects provided signed, informed consent prior to
participation.
 | Table I.Characteristics of family members
observed in the present study. |
Table I.
Characteristics of family members
observed in the present study.
| Subject | Age (years) | Sex | Multiple
osteochondromas | Participation in
present study |
|---|
| II4 | 27 | Male | Affected | Yes |
| II5 | 31 | Female | Unaffected | Yes |
| II6 | 28 | Male | Affected | No |
| II7 | 23 | Female | Affected | No |
| III9 | 3 | Female | Affected | No |
Exome sequencing
Genomic DNA of the subjects was extracted from
peripheral blood samples, using the QIAamp DNA Blood Midi kit
(Qiagen GmbH, Hilden, Germany) according to the manufacturer's
protocols. The extracted DNA was subsequently sent to the Huada
Gene Research Institute (Shenzhen, China) for the following
experiments, including exome sequencing. Initially, a total of 49
genes implicated in genetic tumors (Table II) were selected for exome
sequencing, which was based on the gene capture strategy from the
Huada Gene Research Institute (22). For exome sequencing, 3 µg of
extracted DNA from the patient with MO (II4) was randomly sheared
into 100–200 bp fragment libraries by the Covaris LE220
ultrasonoscope (Covaris, Inc., Woburn, MA, USA), and purified by
AMPure beads (Beckman Coulter, Inc., Brea, CA, USA). The purified
DNA fragment tails were repaired and ‘A’ adapters were ligated to
both ends of the resulting fragments which carried specific ‘T’
linkers. In this way, the cDNA library was constructed completely.
Prepared DNA was then amplified by ligation-mediated polymerase
chain reaction (LM-PCR), purified, and hybridized to the NimbleGen
Human custom array (NimbleGen; Roche Molecular Diagnostics, CA,
USA) for enrichment. Reaction conditions were as follows: 1 cycle
of 98°C for 30 sec, 15 cycles of 98°C for 10 sec, 60°C for 30 sec
and 72°C for 30 sec; 1 cycle of 72°C for 5 min. Non-hybridized
fragments were subsequently washed out. Both non-captured and
captured LM-PCR products were subjected to an Eva Green (3100;
Biotium, Inc., Hayward, CA, USA) Real-Time Fluorescence
Quantitative PCR System (StepOne; Applied Biosystems; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) to estimate the magnitude of
enrichment based on previous work (23). Each captured library was
subsequently loaded onto the Hiseq2000 platform (Illumina, Inc.,
San Diego, CA, USA), and high-throughput sequencing was performed
for each captured library independently to ensure that each sample
met the desired average fold-coverage. Raw image files were
processed by Illumina base calling software 1.7 (Illumina, Inc.)
and the sequences of each individual were generated as 90 bp
paired-end reads.
 | Table II.Genes captured in the present
study. |
Table II.
Genes captured in the present
study.
| Chromosome | Gene symbol | Chromosome | Gene symbol |
|---|
| Chr1 | SDHB; MUTYH;
NTRK1; | Chr12 | CDKN1B; CDK4 |
|
| SDHC; CDC73;
FH |
|
|
| Chr2 | EPCAM; MSH2;
MSH6; | Chr13 | BRCA2; RB1 |
|
| TMEM127; PMS1;
BARD1 |
|
|
| Chr3 | VHL; MLH1 | Chr14 | MAX; MLH3 |
| Chr5 | APC; RAD50 | Chr16 | PALB2; CDH1 |
| Chr7 | PMS2; MET | Chr17 | TP53; FLCN; NF1;
BRCA1; |
|
|
|
| RAD51C; BRIP1;
AXIN2 |
| Chr8 | NBN; EXT1 | Chr18 | SMAD4 |
| Chr9 | CDKN2A | Chr19 | STK11 |
| Chr10 | RET; BMPR1A;
PTEN | Chr22 | CHEK2; NF2 |
| Chr11 | EXT2; SDHAF2;
MEN1; | Total | 49 genes |
|
| MRE11A; ATM;
SDHD |
|
|
Read mapping and variant analysis
Raw read sequencing quality was assessed following
completion of exome sequencing, to ensure that low quality and
contaminated reads were removed. Reads with too many N bases
(>10%) or low base quality (>50% bases with base quality
<5) were discarded. Burrows-Wheeler Aligner (BWA) software
(version hg19, build 37.1, http://bio-bwa.sourceforge.net/) was then used to
align clean reads to the (University of California Santa Cruz
(UCSC) human reference genome. Evaluation of the capture experiment
was executed at the same time. Based on the BWA alignment results,
Short Oligonucleotide Analysis Package snp software (version 1.03;
http://soap.genomics.org.cn/index.html) and SAMtools
software (version 0.9.41; http://sihua.us/samtools.htm) were used to search for
single nucleotide variant (SNV) and insertion and deletion (indel)
mutations, respectively. Once base polymorphisms of the target
region were obtained, information of interest was aligned to data
from the following databases by the Huada Gene Research Institute:
the National Center for Biotechnology Information Database of Short
Genetic Variation (dbSNP) (https://www.ncbi.nlm.nih.gov/snp/), the International
HapMap Project (ftp://ftp.ncbi.nlm.nih.gov/hapmap/), the 1000 Genomes
Project (ftp://www.1000genome.org), the Exome
Sequencing Project (ESP; esp6500siv2), the Exome Aggregation
Consortium (ExAC; version 0.3) and the Kaviar database (version no.
160,204) (24), which include
various ethnic groups. Mutations of interest were screened, marked
and analysed for gene function. Quality control, carried out before
analysis for clean data and after data processing, was present
throughout this pipeline in order to obtain clean data and
alignments.
Sanger sequencing
Confirmatory Sanger sequencing was carried out in
the proband and his sister (II5) to validate the candidate mutation
that was identified by targeted exome sequencing. Target sequences
from 200 unrelated healthy controls also underwent Sanger
sequencing to estimate population frequencies and the pathogenicity
of the candidate mutation. Sanger sequencing was performed using
the standard protocol (18).
Primers used were as follows: 5′-CAGTCCGGATCATTTCTGGCC-'3 and
5′-ACTGAGGTGACAACTGGTCTC-'3. Sequence comparisons and analyses were
performed using DNAman 8.0 (Lynnon Biosoft, San Ramon, CA, USA) and
Chromas 2.0 (Technelysium Pty, Ltd., South Brisbane, Australia),
respectively.
Histochemistry and
immunohistochemistry
Chondroma tissues from the proband and an unrelated
patient with MO who lacked the EXT1 gene mutation, were fixed with
1% paraformaldehyde for 24 h at 4°C, rinsed with phosphate buffered
saline (PBS), decalcified by immersion in 10% nitric acid and
formaldehyde overnight and embedded in paraffin. Paraffin-embedded
chondroma tissues were sectioned (5 µm thick) and placed on glass
slides. For histochemistry, the tissue slides were dewaxed in
xylene, hydrated with graded ethanol, and stained by toluidine blue
and hematoxylin-eosin, for 5 min for each at room temperature and
viewed with a fluorescence microscope (Olympus Corporation, Tokyo,
Japan) at ×40 magnification).
For immunohistochemistry, based on the standard
procedure of high sensitivity two-step detection kit (cat. no.
PV-9001; ZSGB-BIO, Beijing, China), dewaxed and hydrated chondroma
sections were treated with 3% hydrogen peroxide solution for 10
min, rinsed with PBS and incubated with rabbit polyclonal anti-EXT1
antibody (cat. no. ab177101; 1:50 working dilution; Abcam,
Cambridge, UK) at 4°C overnight. Chondroma sections were treated
with Polymer Helper (an assistant that helps the macromolecular
detection system combine with the detected primary antibody
molecule with higher sensitivity, included with the two-step
detection kit) for 15 min, rinsed with PBS and incubated with the
secondary antibody (cat. no. K152709C, polymer of anti-rabbit IgG;
OriGene Technologies, Inc.) at 37°C for 15 min and stained using
the DAB substrate kit (Vector Laboratories, Inc., Burlingame, CA,
USA) and Mayer's hematoxylin, and viewed with an fluorescence
microscope (Olympus Corporation) at ×15 magnification. Targeted
amino acid sequences of the primary antibody for EXT1 were 5–162 of
human EXT1, a region upstream of the novel mutation.
Results
Clinical imaging and pathological
examination
Specialized examination revealed that the proband
had a valgus deformity in the left knee, and prominent tumors of
ranging size in the arms, knees and pelvis. Exogenous tumors
emerged from the distal end of bilateral femurs, the proximal end
of bilateral tibias and fibulae, and around the pelvis (Fig. 1C-E), partially manifesting as
irregular cauliflowers of varying size, with calcific cartilage
caps. The tumors were connected to the wide base of the parent
bone.
Tissue structure of MO was visualized by staining
(Fig. 1F), revealing that layers
of fibrous perichondrium were present around the cartilage cap, in
which there was a typical cluster of chondrocytes growing
neatly.
Exome sequencing and validation of
Sanger sequencing
Exome sequencing of 49 genes linked to hereditary
tumors was performed for the subject. For the targeted regions, the
mean sequencing depths of >80% of the 49 genes were yielded
>100 times. The mean coverage of targeted exons for >10 reads
was 92.46% and for >20 reads 89.82%. 289 SNVs and 36 indels were
identified within the captured and flanking regions, including 21
nonsynonymous SNVs, 2 indels in coding exons and splicing mutations
in introns (Table III).
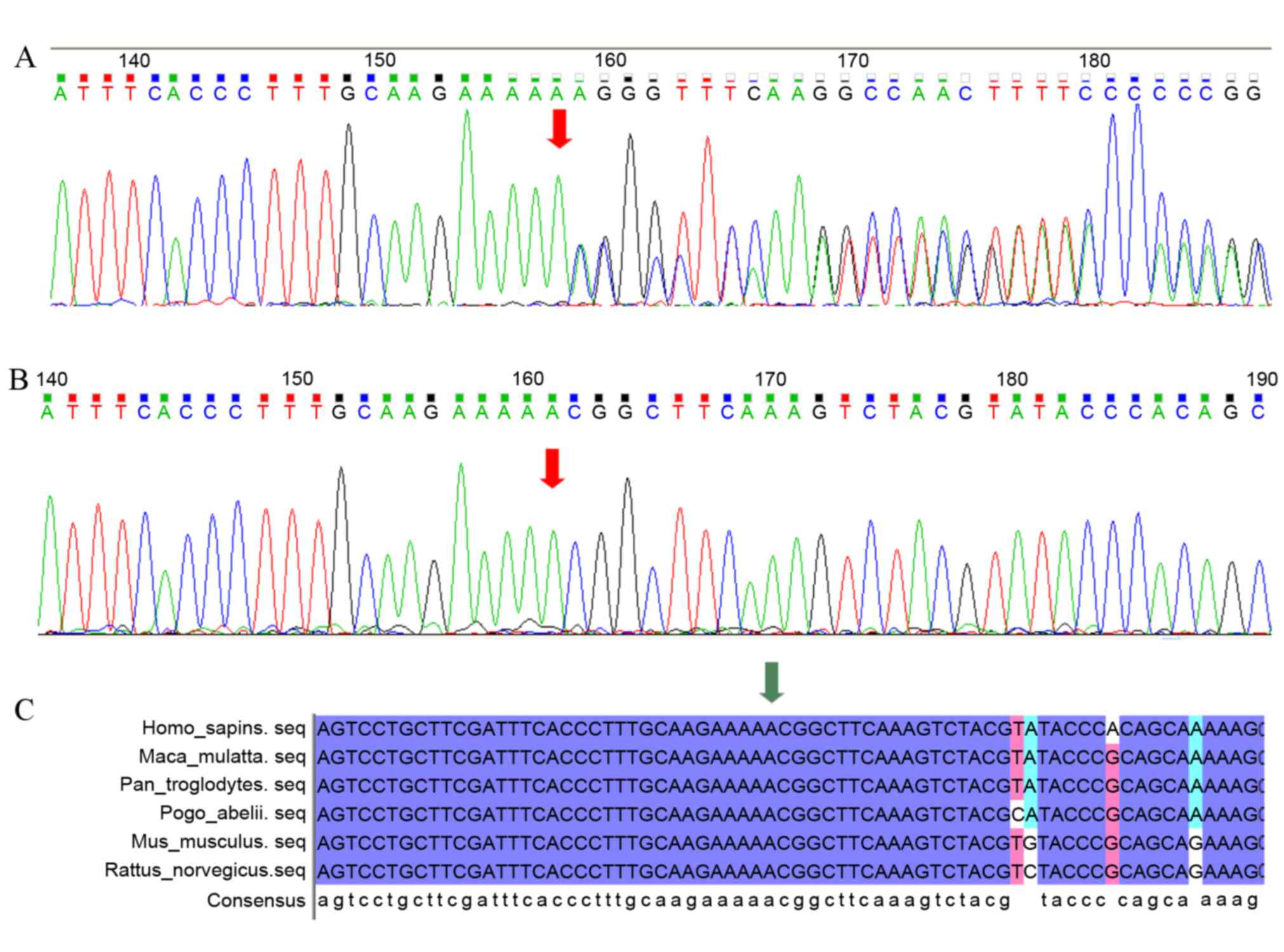
Considering the alteration of gene function caused by the mutations
and their respective population frequencies, a frameshift mutation
(c.335dupA) in EXT1 remained as candidate for causal mutations,
further validated by Sanger sequencing of the patient with MO
patient, the unaffected sister and 200 healthy controls. The
heterozygous insertion (c.335_336insA) in EXT1 was identified as a
candidate pathogenic gene mutation, as it was present in the
proband (Fig. 2A) but absent in
the unaffected individual (Fig.
2B) and the 200 healthy controls, as well as an unrelated
family with MO. Multiple sequence alignment revealed that codon 335
and codon 336 of EXT1 are highly conserved in vertebrate species
with a relatively close relationship to humans (Macaca
mulatta, Pan troglodytes and Pongo abelii) and
species with a remote relationship to humans (Mus musculus,
Rattus norvegicus) (Fig.
2C). This demonstrates its functional importance.
 | Table III.Nonsynonymous single nucleotide
variants and indels identified from targeted exome
resequencing. |
Table III.
Nonsynonymous single nucleotide
variants and indels identified from targeted exome
resequencing.
| Gene | Mut_type | Intron/Exon | Mutation | Function |
Freq_1000g2015aug | esp6500siv2 |
|---|
| MUTYH | Het | Exon12 | C>G | nonsynonymous | 0.313498 | 0.2541 |
| EPCAM | Hom | Exon3 | T>C | nonsynonymous | 0.666134 | 0.5667 |
| MSH6 | Het | Exon1 | G>A | nonsynonymous | 0.200879 | 0.1796 |
| MSH6 | Het | Exon2/4 | C>Ta | nonsynonymous | – | 0.000077 |
| APC | Hom | Exon14/16/17 | T>A | nonsynonymous | 0.865415 | 0.8263 |
| PMS2 | Het | Exon11 | T>C | nonsynonymous | 0.883187 | 0.8704 |
| PMS2 | Het | Exon11 | G>T | nonsynonymous | 0.112021 | 0.0335 |
| MET | Het | Exon2 | A>G | nonsynonymous | 0.0329473 | 0.0146 |
| NBN | Het | Exon5/6 | C>G | nonsynonymous | 0.357029 | 0.2866 |
| BMPR1A | Het | Exon3 | C>A | nonsynonymous | 0.4998 | 0.3895 |
| MEN1 | Het | Exon10/11 | T>C | nonsynonymous | 0.834465 | 0.9084 |
| ATM | Hom | Exon40 | A>G | nonsynonymous | 1 | – |
| BRCA2 | Het | Exon10 | A>C | nonsynonymous | 0.249401 | 0.2332 |
| BRCA2 | Hom | Exon14 | T>C | nonsynonymous | 0.975839 | 0.9777 |
| MLH3 | Het | Exon5 | C>T | nonsynonymous | 0.00459265 | 0.000077 |
| MLH3 | Hom | Exon2 | T>C | nonsynonymous | 0.990415 | 0.9888 |
| TP53 | Hom | Exon4 | G>C | nonsynonymous | 0.542931 | 0.63 |
| FLCN | Hom | Exon8 | C>T | nonsynonymous | 0.0996406 | 0.0775 |
| FLCN | Het | Exon8 | C>Tb | nonsynonymous | – | – |
| AXIN2 | Het | Exon6 | G>A | nonsynonymous | 0.00359425 | – |
| AXIN2 | Het | Exon2 | G>A | nonsynonymous | 0.33766 | 0.3948 |
| EXT1 | Het | Exon1 | insAc | Indel | – | – |
| PTEN | Hom | Exon1/2 | delT | Indel
(splicing) | 1 | – |
Histochemical staining and
immunohistochemistry
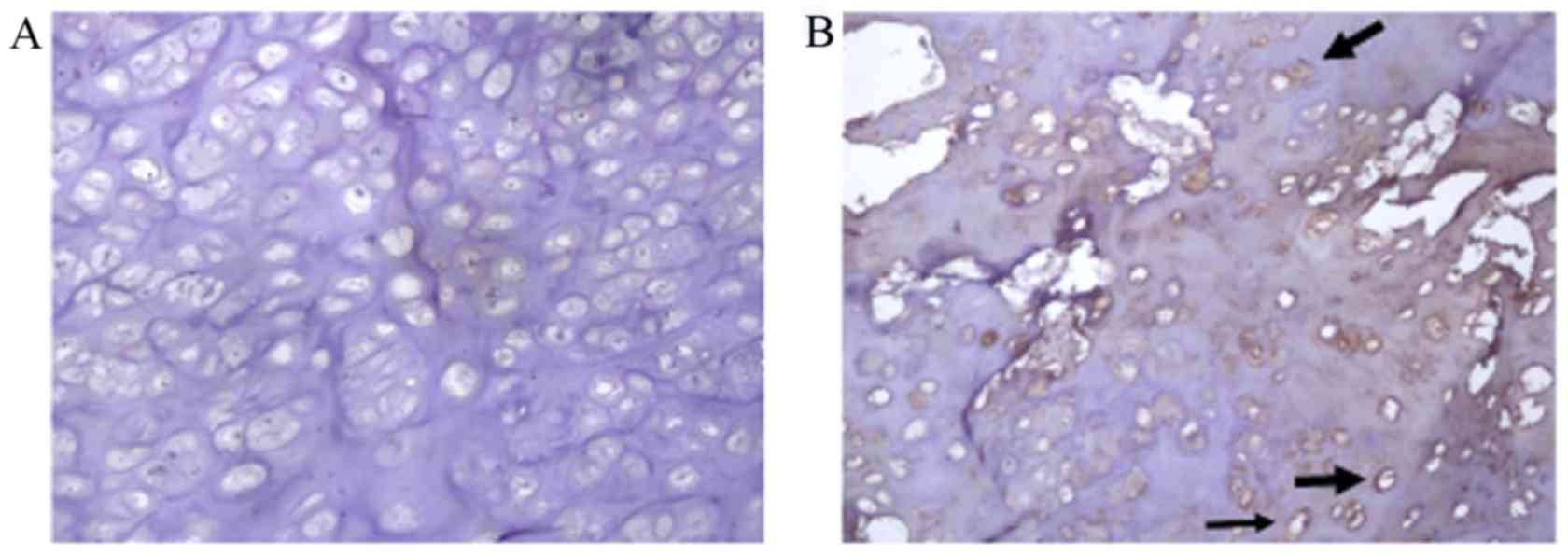
To evaluate the functional impact of c.335_336insA
on the EXT1 protein, protein expression levels of EXT1 in two
unrelated families with MO were detected using a supersensitive
two-step immunohistochemistry method. EXT1 protein expression
levels in the proband patient with MO and the c.335_336insA
mutation (Fig. 3A) were markedly
decreased compared with those in a patient with MO that lacked any
EXT1 mutation (Fig. 3B).
Discussion
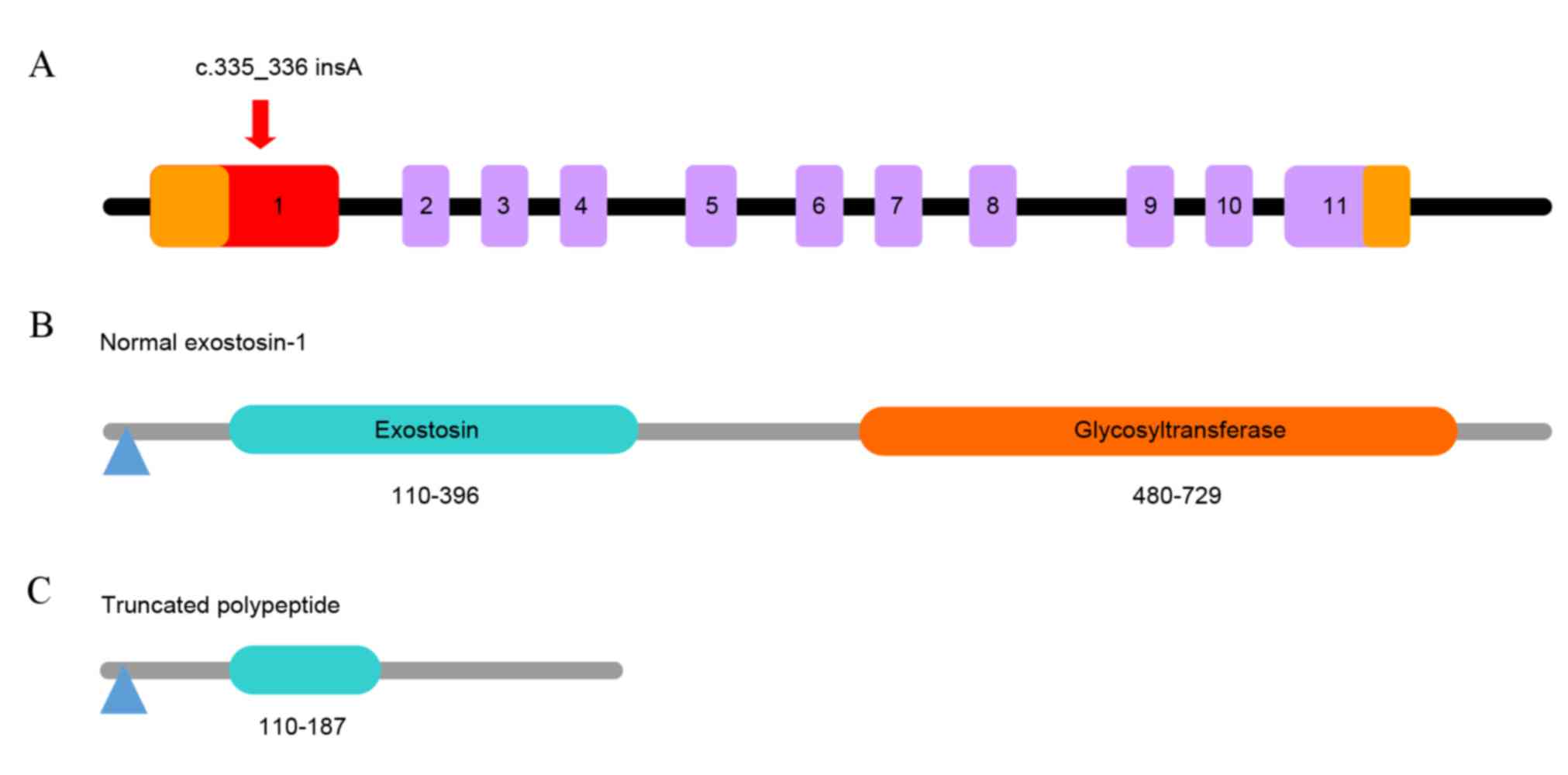
To date, 196 different mutations distributed in the
exon/intron 1 of the EXT1 gene have been identified from more than
200 families with MO, according to the Multiple Osteochondromas
Mutation Database (version 2.0, http://medgen.ua.ac.be/LOVDv.2.0/home.php). EXT1 is
comprised of 11 constructive exons (Fig. 4A) encoding 746 amino acids and
possesses two domains: exostosin and glycosyltransferase (Fig. 4B). The mutation identified in the
present study, c.335_336insA, appeared between codon 335 and codon
336 in exon 1 of EXT1 (Fig. 4A),
resulting in a frameshift mutation in the first domain of the EXT1
gene and creating a premature stop codon at amino acid 187
(Fig. 4C). This resulted in the
loss of part of the exostosin domain and the entire
glycosyltransferase domain, producing a truncated polypeptide
(Fig. 4C).
As a putative tumor-suppressor in the EXT gene
family, EXT1 mutations have been suggested to be a major cause of
MO, in addition, patients with MO who have EXT1 gene mutations tend
to display more severe symptoms and suffer a greater risk for
malignant transformation than patients with EXT2 gene mutations
(25–27). EXT1 and EXT2 proteins form a stable
complex as a glucuronic acid sugar-based transfer enzyme and
N-acetylglucosamine glycosyltransferase, catalyzing the
polymerization of HS chains in the endoplasmic reticulum and Golgi
apparatus (28). HS is
ubiquitously expressed on the cell surface, and as a component of
extracellular matrix glycoprotein, it is involved in the mediation
of cell adhesion, signal transduction and the receptor ligand
binding process (29). Previous
studies have indicated that HS is an essential molecule for the
growth and differentiation of normal chondrocytes and its
dysfunction may be associated with MO (30–33).
In the present study a novel frameshift mutation in
exon 1 of EXT1 gene, c.335_336insA, was identified in one affected
individual of a family with MO and was not identified in either an
unaffected family member or in any of 200 unrelated healthy
controls. It was also not present in individuals from an unrelated
family with MO. Multiple sequence alignment and
immunohistochemistry both confirmed the pathogenicity of the
c.335_336insA mutation. Furthermore, according to the number and
distribution of the affected patients in the family with MO
(Fig. 1A), it is possible to infer
that the c.335_336insA mutation originated from the mother of the
proband and was transmitted to the daughter of the proband in an
autosomal dominant manner.
The c.335_336insA mutation is located within the
exostosin domain of the EXT1 gene, and results in a frameshift at
codon 335 and a change of an asparagine to a lysine. The occurrence
of a premature stop codon and truncated EXT1 usually triggers
nonsense-mediated mRNA decay and reduces the level of functional
EXT1 (34,35). Immunohistochemistry also confirmed
that the EXT1 expression level was decreased. This result is
consistent with previous studies, which demonstrated that truncated
EXT1 fails to fold correctly with exostosin-2 and rapidly degrades,
resulting in decreased EXT1 levels in the chondrocytes of patients
with MO (21,36).
Multiple alignment of the sequences spanning the
mutation site suggested that this point of interest is
extraordinarily ancient and conserved, with scarcely any
differences in mammalian species either closely or distantly
related to humans. Referring to evidence from previous studies, it
is possible to infer that MO in the proband of the present study
was caused by the c.335_336insA mutation, which directly resulted
in the functional loss of the two important domains of EXT1. This
hindered the effective folding of EXT1 and efficient biosynthesis
of HS, leading to disruption of signal transduction. Ultimately,
the proliferation and differentiation of chondrocytes escaped
regulation and initiated the development of MO. Further studies are
required to illuminate the molecular mechanisms underlying the
actions of the two EXT1 domains, in particular the
glycosyltransferase domain.
To conclude, a novel frameshift mutation
c.335_336insA in EXT1 gene was discovered through exome sequencing
and validated by Sanger sequencing. The pathogenicity of the
mutation was further confirmed via multiple alignment and
immunohistochemistry. The present study also emphasized the
dysfunction of the EXT genes in the development of MO. This finding
will aid early diagnosis and prenatal genetic screening of the
remaining members in the studied family with MO.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (81371902), the Fujian Medical
Innovation Project (2012-CX-30) and the Innovative Team Training
Project in Fuzhou (2013- S-wt2).
References
|
1
|
Schmale GA, Conrad EU III and Raskind WH:
The natural history of hereditary multiple exostoses. J Bone Joint
Surg Am. 76:986–992. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jennes I, Pedrini E, Zuntini M, Mordenti
M, Balkassmi S, Asteggiano CG, Casey B, Bakker B, Sangiorgi L and
Wuyts W: Multiple osteochondromas: Mutation update and description
of the multiple osteochondromas mutation database (MOdb). Hum
Mutat. 30:1620–1627. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tian C, Yan R, Wen S, Li X, Li T, Cai Z,
Du H and Chen H: A splice mutation and mRNA decay of EXT2 provoke
hereditary multiple exostoses. PLoS One. 9:e948482014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Stieber JR and Dormans JP: Manifestations
of hereditary multiple exostoses. J Am Acad Orthop Surg. 13:110–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jamsheer A, Socha M, Sowińska-Seidler A,
Telega K, Trzeciak T and Latos-Bieleńska A: Mutational screening of
EXT1 and EXT2 genes in Polish patients with hereditary multiple
exostoses. J Appl Genet. 55:183–188. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Vanita V, Sperling K, Sandhu HS, Sandhu PS
and Singh JR: Novel EXT1 and EXT2 mutations in hereditary multiple
exostoses families of Indian origin. Genet Test Mol Biomarkers.
13:43–49. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wu Y, Xing X, Xu S, Ma H, Cao L, Wang S
and Luo Y: Novel and recurrent mutations in the EXT1 and EXT2 genes
in Chinese kindreds with multiple osteochondromas. J Orthop Res.
31:1492–1499. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jones KB, Datar M, Ravichandran S, Jin H,
Jurrus E, Whitaker R and Capecchi MR: Toward an understanding of
the short bone phenotype associated with multiple osteochondromas.
J Orthop Res. 31:651–657. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wuyts W and Van Hul W: Molecular basis of
multiple exostoses: Mutations in the EXT1 and EXT2 genes. Hum
Mutat. 15:220–227. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wuyts W, Radersma R, Storm K and Vits L:
An optimized DHPLC protocol for molecular testing of the EXT1 and
EXT2 genes in hereditary multiple osteochondromas. Clin Genet.
68:542–547. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lonie L, Porter DE, Fraser M, Cole T, Wise
C, Yates L, Wakeling E, Blair E, Morava E, Monaco AP and Ragoussis
J: Determination of the mutation spectrum of the EXT1/EXT2 genes in
British Caucasian patients with multiple osteochondromas, and
exclusion of six candidate genes in EXT negative cases. Hum Mutat.
27:11602006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
McCormick C, Leduc Y, Martindale D,
Mattison K, Esford LE, Dyer AP and Tufaro F: The putative tumour
suppressor EXT1 alters the expression of cell-surface heparan
sulfate. Nat Genet. 19:158–161. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zak BM, Crawford BE and Esko JD:
Hereditary multiple exostoses and heparan sulfate polymerization.
Biochim Biophys Acta. 1573:346–355. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ahn J, Lüdecke HJ, Lindow S, Horton WA,
Lee B, Wagner MJ, Horsthemke B and Wells DE: Cloning of the
putative tumour suppressor gene for hereditary multiple exostoses
(EXT1). Nat Genet. 11:137–143. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Stickens D, Clines G, Burbee D, Ramos P,
Thomas S, Hogue D, Hecht JT, Lovett M and Evans GA: The EXT2
multiple exostoses gene defines a family of putative tumour
suppressor genes. Nat Genet. 14:25–32. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Le Merrer M, Legeai-Mallet L, Jeannin PM,
Horsthemke B, Schinzel A, Plauchu H, Toutain A, Achard F, Munnich A
and Maroteaux P: A gene for hereditary multiple exostoses maps to
chromosome 19p. Hum Mol Genet. 3:717–722. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Trebicz-Geffen M, Robinson D, Evron Z,
Glaser T, Fridkin M, Kollander Y, Vlodavsky I, Ilan N, Law KF,
Cheah KS, et al: The molecular and cellular basis of exostosis
formation in hereditary multiple exostoses. Int J Exp Pathol.
89:321–331. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pedrini E, De Luca A, Valente EM, Maini V,
Capponcelli S, Mordenti M, Mingarelli R, Sangiorgi L and
Dallapiccola B: Novel EXT1 and EXT2 mutations identified by DHPLC
in Italian patients with multiple osteochondromas. Hum Mutat.
26:2802005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sarrión P, Sangorrin A, Urreizti R,
Delgado A, Artuch R, Martorell L, Armstrong J, Anton J, Torner F,
Vilaseca MA, et al: Mutations in the EXT1 and EXT2 genes in Spanish
patients with multiple osteochondromas. Sci Rep. 3:13462013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Huang XF, Xiang P, Chen J, Xing DJ, Huang
N, Min Q, Gu F, Tong Y, Pang CP, Qu J and Jin ZB: Targeted exome
sequencing identified novel USH2A mutations in Usher syndrome
families. PLoS One. 8:e638322013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu H, Wu S, Duan L, Zhu W, Zhang S, Hu X,
Jia W, Yang G, Liu C, Li W, et al: Identification of a novel EXT1
mutation in patients with hereditary multiple exostosis by exome
sequencing. Oncol Rep. 33:547–552. 2015.PubMed/NCBI
|
|
22
|
Wu J, Matthaei H, Maitra A, Dal Molin M,
Wood LD, Eshleman JR, Goggins M, Canto MI, Schulick RD, Edil BH, et
al: Recurrent GNAS mutations define an unexpected pathway for
pancreatic cyst development. Sci Transl Med. 3:92ra662011.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Glusman G, Caballero J, Mauldin DE, Hood L
and Roach JC: Kaviar: An accessible system for testing SNV novelty.
Bioinformatics. 27:3216–3217. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Porter DE, Lonie L, Fraser M, Dobson-Stone
C, Porter JR, Monaco AP and Simpson AH: Severity of disease and
risk of malignant change in hereditary multiple exostoses. A
genotype-phenotype study. J Bone Joint Surg Br. 86:1041–1046. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Francannet C, Cohen-Tanugi A, Le Merrer M,
Munnich A, Bonaventure J and Legeai-Mallet L: Genotype-phenotype
correlation in hereditary multiple exostoses. J Med Genet.
38:430–434. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Alvarez CM, De Vera MA, Heslip TR and
Casey B: Evaluation of the anatomic burden of patients with
hereditary multiple exostoses. Clin Orthop Relat Res. 462:73–79.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
McCormick C, Duncan G, Goutsos KT and
Tufaro F: The putative tumor suppressors EXT1 and EXT2 form a
stable complex that accumulates in the Golgi apparatus and
catalyzes the synthesis of heparan sulfate. Proc Natl Acad Sci USA.
97:668–4073. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Esko JD and Lindahl U: Molecular diversity
of heparan sulfate. J Clin Invest. 108:169–173. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jones KB, Pacifici M and Hilton MJ:
Multiple hereditary exostoses (MHE): Elucidating the pathogenesis
of a rare skeletal disorder through interdisciplinary research.
Connect Tissue Res. 55:80–88. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Koziel L, Kunath M, Kelly OG and Vortkamp
A: Ext1-dependent heparan sulfate regulates the range of Ihh
signaling during endochondral ossification. Dev Cell. 6:801–813.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Anower-E-Khuda MF, Matsumoto K, Habuchi H,
Morita H, Yokochi T, Shimizu K and Kimata K: Glycosaminoglycans in
the blood of hereditary multiple exostoses patients: Half reduction
of heparan sulfate to chondroitin sulfate ratio and the possible
diagnostic application. Glycobiology. 23:865–876. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jochmann K, Bachvarova V and Vortkamp A:
Reprint of: Heparan sulfate as a regulator of endochondral
ossification and osteochondroma development. Matrix Biol.
35:239–247. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gerards M, van den Bosch B, Calis C,
Schoonderwoerd K, van Engelen K, Tijssen M, De Coo R, van der Kooi
A and Smeets H: Nonsense mutations in CABC1/ADCK3 cause progressive
cerebellar ataxia and atrophy. Mitochondrion. 10:510–515. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Inoue K, Khajavi M, Ohyama T, Hirabayashi
S, Wilson J, Reggin JD, Mancias P, Butler IJ, Wilkinson MF, Wegner
M and Lupski JR: Molecular mechanism for distinct neurological
phenotypes conveyed by allelic truncating mutations. Nat Genet.
36:361–369. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang F, Liang J, Guo X, Zhang Y, Wen Y,
Li Q, Zhang Z, Ma W, Dai L, Liu X, et al: Exome sequencing and
functional analysis identifies a novel mutation in EXT1 gene that
causes multiple osteochondromas. PLoS One. 8:e723162013. View Article : Google Scholar : PubMed/NCBI
|