Introduction
Abdominal surgical wound infections in adults are
defined as infections that occur within the maximum time of 30 days
postsurgery and contribute to increased post-surgical morbidity and
mortality. The most common complications are abscesses, wound
infections and necroses (1).
Surgical site infections (SSIs) represent 15% of all nosocomial
infections (2) and are associated
with prolonged hospitalization time and increased antibiotic
therapy costs (3). The most
frequently involved microorganisms include: Staphylococcus
aureus (S. aureus; 15–20%), Gram-negative bacilli,
coagulase-negative staphylococci, Enterococcus spp. and
Escherichia coli (E. coli) (4). From these, methicillin-resistant
S. aureus (MRSA) represents 50% of hospital-acquired
infections in the United States and Europe, and these infections
very difficult to treat due to their resistance to multiple
antibiotics (5).
The Centers for Disease Control and Prevention (CDC)
(6) classify SSIs into three major
categories: i) superficial infections, which are localized to the
skin and subcutaneous tissue, and are characterized locally by
redness, pain, warmth and swelling, and are resolved by local
incision and the discharge of the pus; ii) deep incisional
infections, affecting muscles and fascia with the presence of
abscess, which require the surgical excision of deep wound edges;
and iii) the infection of abdominal organs or anatomical spaces,
which require surgical procedures in locations other than the
initial incision site (6).
The objectives of this study were the following: i)
the identification of germs that produced surgical wound infections
in patients from the Clinical Emergency County Hospital of Craiova
(Craiova Romania) and to assess their resistance to antimicrobials;
and ii) perform a comparison of antibiotic resistance profiles
between surgical wards and the intensive care unit (ICU), in order
to determine the most effective therapeutic protocols.
Materials and methods
Patient data
This study was conducted between September 2015 and
September 2016, and included a total of 165 patients (male:female
ratio, 2.17) aged between 18 and 87 years, hospitalized at the
General Surgery departments of the Clinical Emergency County
Hospital of Craiova, and diagnosed clinically with SSIs following
various surgical interventions. We collected discharge, liquid
drainage from superficial and deep surgical wounds and pus. Each
patient included in the study provided written informed consent.
The study was conducted in accordance with the World Medical
Association Declaration of Helsinki and was approved by the
Institutional Ethics Committee of Clinical Emergency Hospital of
Craiova.
Microbiological evaluation
The strains were identified by classical
bacteriological diagnosis and the antimicrobial testing was
performed by disc diffusion according to the guidelines of the
Clinical and Laboratory Standards Institute (CLSI) (7).
Statistical analysis
The results of antimicrobial testing were stored and
analyzed using Whonet 5.6 software (World Health Organisation,
Geneva, Switzerland). Statistical analyses were performed using the
Statistical Package for the Social Sciences (SPSS, Inc., Chicago,
IL, USA) 20.0 software (IBM Corp., Armonk, NY, USA). Antibiotic
resistance was expressed as the ratio between the number of
isolates tested as resistant over the number of isolates tested to
the antibiotic.
For each isolate, we calculated the multiple
antibiotic resistance (MAR) index as the ratio between the number
of antibiotics at which the isolate was resistant over the number
of antibiotics tested for that isolate. Comparative analyses of
continuous variables were made using the Student's t-test for two
groups and one-way analysis of variance (ANOVA) for more than two
groups. A value of P<0.05 was considered to indicate a
statistically significant difference.
Hierarchical clustering was used as a method to
group the isolates based upon the antibiotic resistance profile,
knowing that strains spreading from a patient to another suffer
mutations in the mechanisms of antibiotic resistance depending on
the antibiotic treatment received by the patient. Thus, the degree
of relatedness of the antibiotic profile is an indication of the
degree of the genetic relatedness of the strains. Hierarchical
clustering was demonstrated to be a reliable method for
constructing filiation trees of genetic relatedness (8).
We performed hierarchical clustering as previously
described by Xu et al (9).
We measured the diameters of inhibition areas around each
antibiotic disk on the Petri dish and entered them into the
hierarchical clustering procedure of SPSS. For clustering, we used
Ward's minimum variance method. Each isolate was assigned to a
specific cluster based on their zone diameters.
Results
Samples were collected from 24 patients with
superficial SSIs, 42 patients with deep incisional infections and
99 patients with deep abdominal infections.
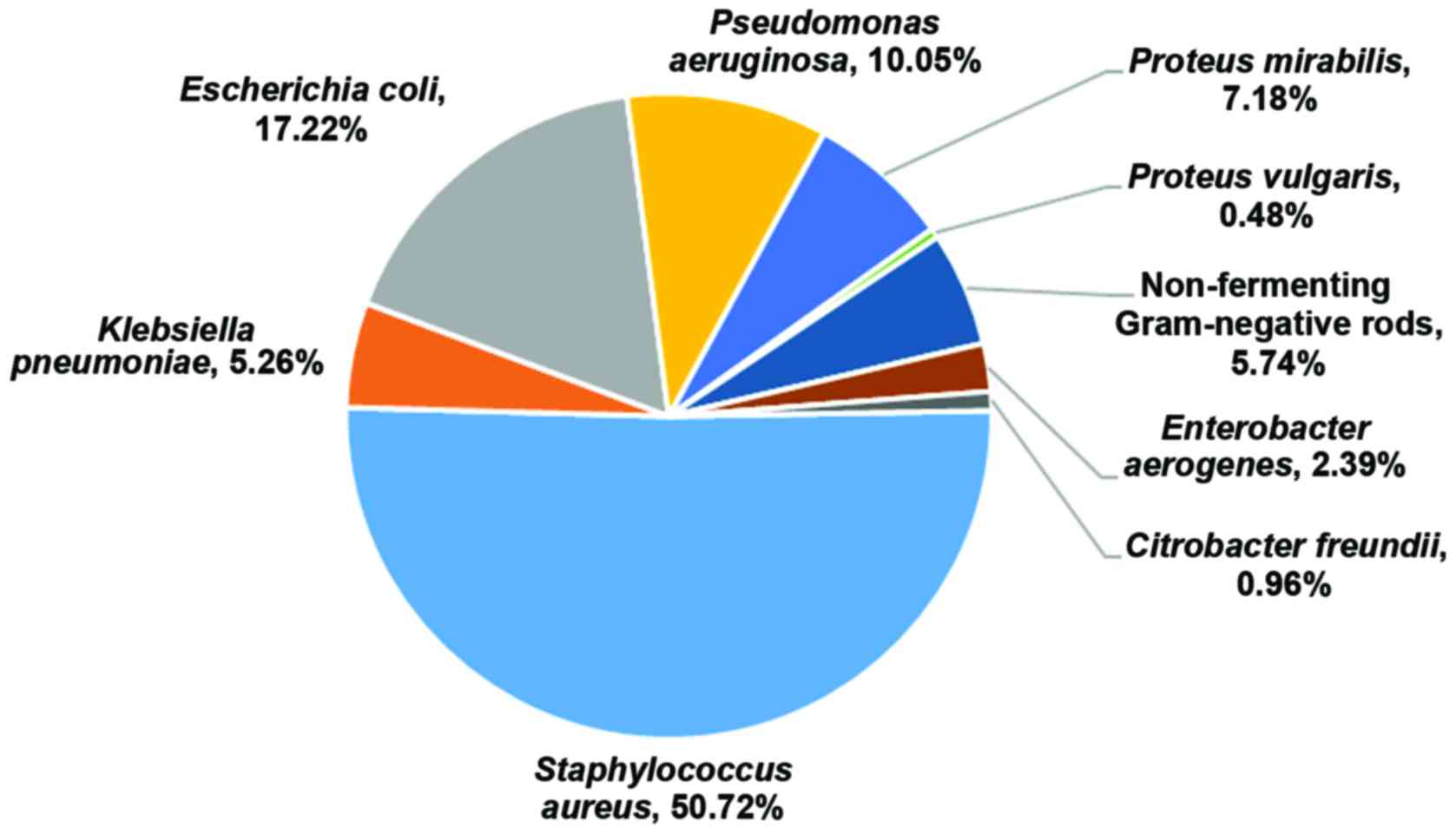
A total of 209 bacterial strains were isolated from
the patients included in this study (n=165). The most prevalent
bacterial species isolated were S. aureus (50.72%), followed
by E. coli (17.22%) and Pseudomonas aeruginosa
(10.05%). Proteus mirabilis, non-fermenting Gram-negative
rods, Klebsiella pneumoniae, Enterobacter aerogenes
and Proteus vulgaris were also isolated, although at lower
percentages (Fig. 1).
The comparison of the antibiotic resistance index by
species (Fig. 2A), revealed that
the differences between species were statistically significant
(ANOVA, P<0.001), as the non-fermenting bacteria were the most
resistant (median MAR, 0.91) and E. coli was the least
resistant (median MAR, 0.40). Pseudomonas was included in
the non-fermenting bacteria category due to the low number of
strains.
A comparison between the resistance of various
bacterial species in the ICU and surgical wards was performed
(Fig. 2B). For Enterobacteriaceae,
the resistance was only slightly higher in the ICU. As regards
Staphylococcus species, but for non-fermenting bacteria,
even if the median MAR was almost the same, the antibiotic
resistance index values were confined to the upper limit in the
ICU. For all species, the p-values from the Student's t-test
performed on the MAR index between surgical wards and the ICU were
<0.01.
The S. aureus strains were highly resistant
to ceftriaxone (100%), penicillin (91.36%), amoxicillin/clavulanate
(87.50%), amikacin (80.00%) and amoxicillin (83.33%). The
resistance to cefoxitin was 40%. Low resistance rates were
encountered to levofloxacin (37.10%), doxycycline (31.20%),
gentamycin (17.01%), tigecycline (6.90%) and teicoplanin (6.90%)
(Fig. 3A).
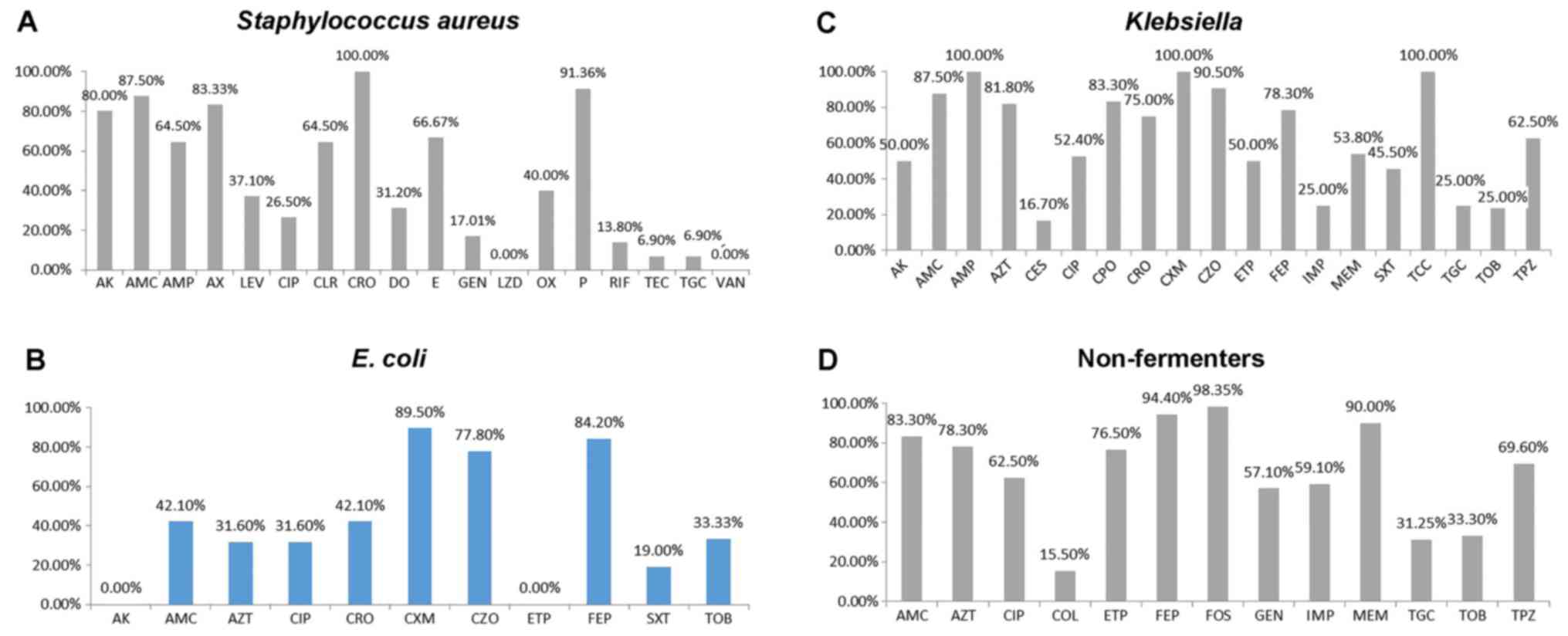 | Figure 3.Antibiotics resistance profile of (A)
Staphylococcus aureus, (B) Escherichia coli, (C)
Klebsiella pneumoniae and (D) non-fermenting Gram-negative
rods (non-fermenters). AK, amikacin; AMC, amoxicillin/clavulanate;
AMP, ampicillin; AZT, aztreonam; AX, amoxicillin; CES, cefoperasone
sulbactam; CIP, ciprofloxacin; CLR, clarithromycin; COL, colistin;
CPO, cefpirome; CRO, ceftriaxone; CXM, cefuroxime; CZO, cefazoline;
DO, doxycycline; E, erythromycin; ETP, ertapenem; FEP, cefepim;
FOS, fosfomycin; GEN, gentamycin; IMP, imipenem; LEV, levofloxacin,
LZD, linezolid; MEM, meropenem; OX, oxacillin; P, penicillin; RIF,
rifampin; SXT, trimethoprim/sulfamethoxazole; TEC, teicoplanin;
TCC, ticarcillin/clavulanate; TGC, tigecycline; TOB, tobramycin;
TPZ, piperacillin-tazobactam; VAN, vancomycin. |
After performing the antimicrobial susceptibility
testing of the isolated E. coli strains (Fig. 3B), an elevated resistance to
cefuroxime (89.50%), cefepime (84.20%) and cefazoline (77.80%) was
registered. A moderate resistance was observed to
amoxicillin/clavulanate (42.10%), ceftriaxone (42.10%), aztreonam
(31.60%) and ciprofloxacin (31.60%). The strains of E. coli
exhibited a low resistance to trimethoprim/sulfamethoxazole
(19.00%) and zero resistance to amikacin and ertapenem.
The isolated Klebsiella strains were found to
be 100% resistant (Fig. 3C) to
ampicillin, cefuroxime and ticarcillin/clavulanate, and resistant
to cefazolin (90.50%), amoxicillin/clavulanate (87,50%), cefpirome
(83.30%), aztreonam (81.80%), cefepime (78.30%), piperacillin with
tazobactam (62.50%), ciprofloxacin (52.40%), amikacin (50,00%) and
sulfametoxazole with trimethoprim (45.50%). A low level of
resistance was observed to cefoperasone-sulbactam (16.70%),
imipenem (25.00%), tigecycline (25.00%) and tobramycin
(25.00%).
The glucose non-fermenting bacteria were highly
resistant to antibiotics (Fig.
3D). For example, the resistance was very high to fosfomycin
(98.35%), cefepime (94.40%), meropenem (90.00%),
amoxicillin/clavulanate (83.30%), aztreonam (78.30%) and ertapenem
(76.50%). There was profound resistance to piperacillin-tazobactam
(69.60%), ciprofloxacin (62.50%), imipenem (59.10%) and gentamycin
(57.10%), but low resistance rates were encountered for tobramycin
(33.30%), tigecycline (31.25%) and colistin (15.50%). The
non-fermenting bacteria from patients in the ICU were generally
more resistant than those from patients in surgical wards (Fig. 4A).
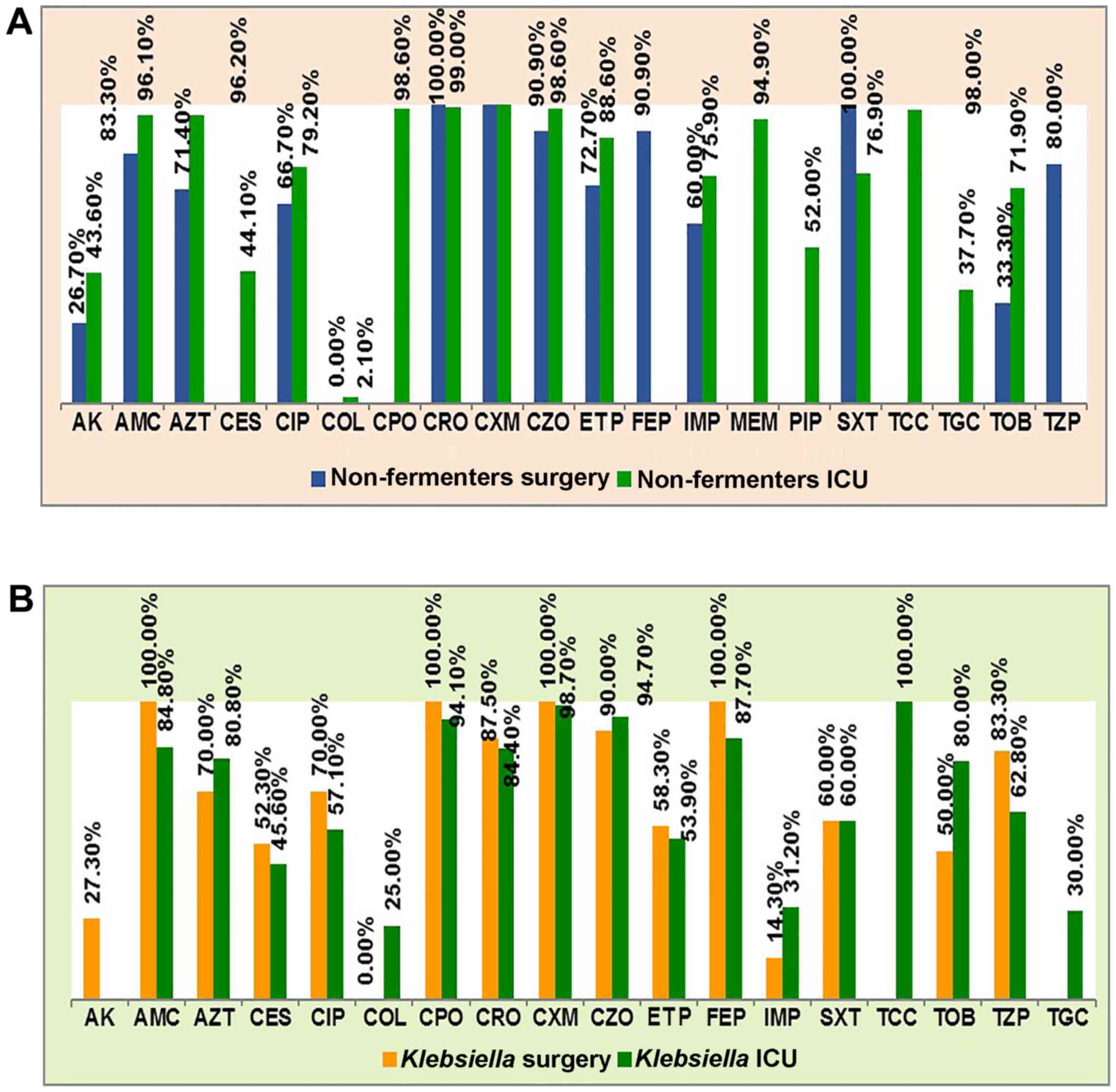 | Figure 4.Antibiotics resistance profile of (A)
non-fermenting Gram negative rods (non-fermenters) and (B)
Klebsiella pneumoniae in surgery and ICU wards. AK,
amikacin; AMC, amoxicillin/clavulanate; AZT, aztreonam; CES,
cefoperasone sulbactam; CIP, ciprofloxacin; COL, colistin; CPO,
cefpirome; CRO, ceftriaxone; CXM, cefuroxime; CZO, cefazoline; ETP,
ertapenem; FEP, cefepim; IMP, imipenem; MEM, meropenem; PIP,
piperacillin; SXT, trimethoprim/sulfamethoxazole; TCC,
ticarcillin/clavulanate; TGC, tigecycline; TOB, tobramycin; TPZ,
piperacillin-tazobactam. |
The resistance of Klebsiella strains differed
between the surgical and ICU wards for imipenem [31.20% in ICU vs.
14.30% in surgical wards, risk ratio (RR) = 2.182], piperacillin
with tazobactam (62.80 vs. 83.30%, RR = 0.754), tobramycin (80.00
vs. 50.00%, RR = 1.600) and amikacin (0 vs. 27.00%) and colistin
(25.00 vs. 0%). There were no significant differences as regards
resistance to cefuroxime, cefpirome and amoxycillin/clavulanate. We
can see that for almost all antibiotics >50% of the ICU strains
were resistant (Fig. 4B).
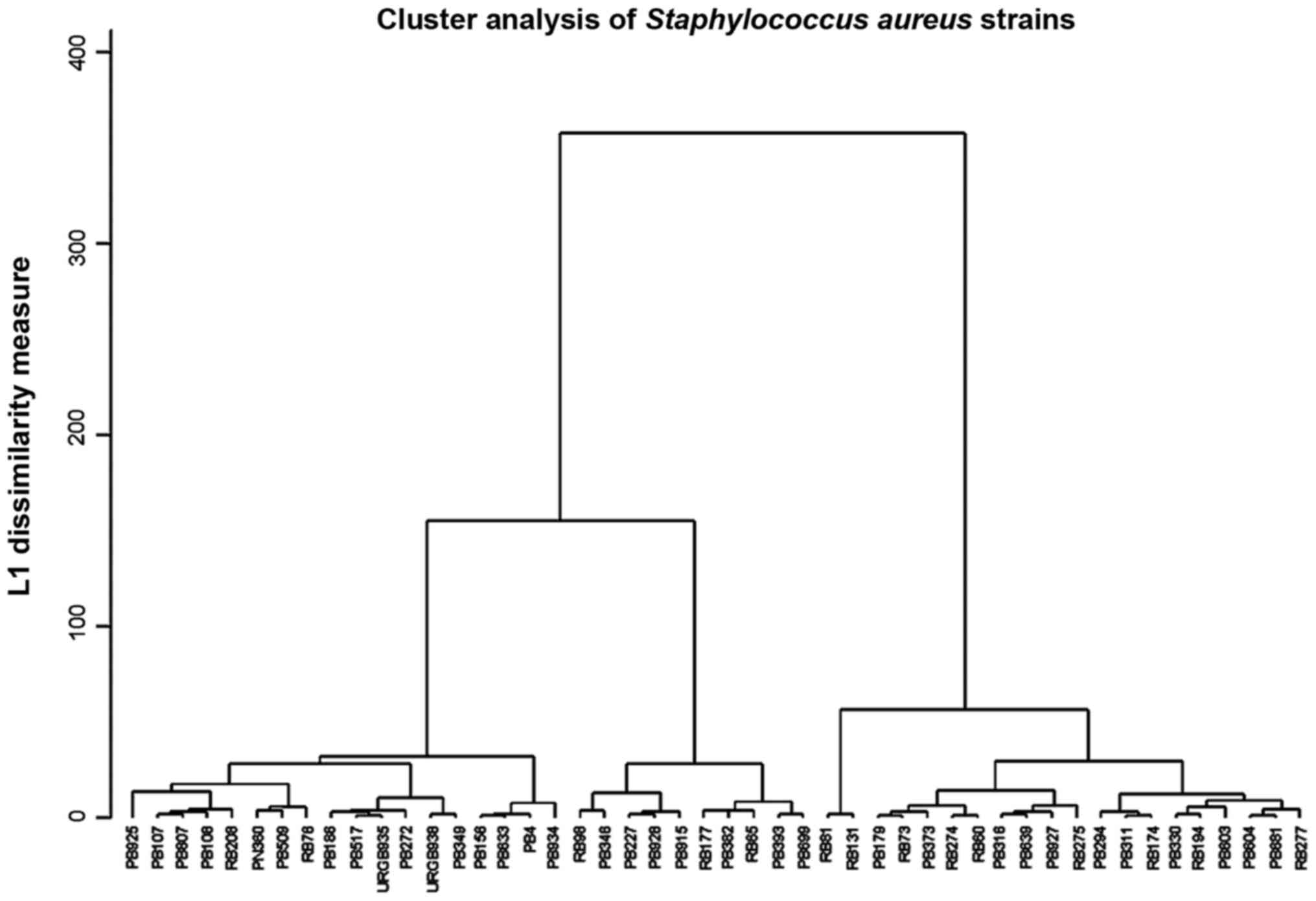
The clustering analysis revealed 12 clusters for
S. aureus; 6 in the sensitive area, probably low-resistant
S. aureus from the aeromicroflora and 6 in the high
resistance area, the MRSA (Fig.
5).
The E. coli and Klebsiella strains had
a heterogenous resistance, distributed in 11 clusters over the
whole interval, as Klebsiella can easily acquire resistance
plasmids from other enterobacteria (Fig. 6).
Discussion
The knowledge of the antibiotic resistance of
bacteria recovered from SSIs is critical in for the optimization of
the prophylactic antibiotic therapy of surgical maneuvers, in an
effort to avoid the selection of multi-resistant microorganisms.
Although the emergence of resistant strains is a natural phenomenon
which cannot be avoided, the transformation of resistant strains
into resistant populations is favored by non-rational antibiotic
therapy. It is well known that the ICU departments are the centers
of dissemination of multi-resistant strains in hospitals, a fact
that underlines the importance to compare the differences in
resistance between ICU and surgical wards.
In the case of Klebsiella, known as a
bacterium that collects easily resistance plasmids, the additional
resistance observed in surgical wards may possibly reflect the
prophylactic usage of antibiotics routinely administered prior to
surgery, that may also induce resistance to other bacteria, that
would survive in the hospital environment and would lastly transmit
the resistance genes to Klebsiella strains (10). The 52.40% resistance to
ciprofloxacin is important in treating the pleural infections, as
ciprofloxacin penetrates well in the pleural space (11).
Although increased bacterial resistance would be
expected in ICUs due to the higher usage of antibiotics, the
differences in the antibiotic resistance of bacterial strains
derived from ICU or surgical wards was minor in this study. The
differences in resistance were minor for cephalosporins. The
highest difference was observed for cefepime (100% in ICU vs.
87.50% in surgical wards). These data confirm those of an earlier
study that found small differences in resistance between ICU and
surgical wards to 1st and 2nd generation cephalosporins, and higher
for 4th generation cephalosporins (84.2 vs. 69.82% for cefepime)
(4). This may suggest that the
highly resistant Klebsiella strains from the ICU have
already colonized the surgical wards, due to the fact that many
surgical patients are transferred from the surgery room directly to
the ICU, where they spend a few days until they are transported
back into the surgical ward. Furthermore, the differences
identified in carbapenem resistance (31.20 vs. 14.30% for imipenem)
were probably due to the high prescription rate in the ICU
(12). It has been demonstrated
that if β-lactams are used for a prolonged period of time, the
resistance rapidly increases within several years (13,14).
In our study, non-fermenting bacteria had generally
higher resistance rates than other bacteria, particularly in 4th
generation cephalosporins, penicillins with inhibitors and
carbapenems. A notable difference between resistance to meropenem
(90.00%) and imipenem (59.10%) was detected, that can be correlated
with a greater prescription of meropenem. The aminoglicosides
resistance was also high (57.10% to gentamycin), but below that for
cephalosporins. The prophylactic use of gentamycin may be therefore
justified (15). Nevertheless, the
cephalosporins are still recommended as prophylactic antibiotherapy
in abdominal surgery (16).
Minocycline, an old antibiotic with proven safety, that has been
reported in the last few years to express improved efficacy in
preventing infections with Acinetobacter baumannii would
represent a further effective alternative (17).
The resistance differences between ICU and surgical
wards were greater for non-fermenting bacteria than for
Klebsiella. This can be at least partly explained by the
fact that in ICU, many patients may have extremely low immunity due
to their medical conditions (18).
They may therefore have an elevated risk of acquiring common
environmental pathogen infections, which would not infect patients
with normal immunity (e.g., from surgical wards) and possess high
natural resistance to antibiotics, such as Acinetobacter
species (19) or opportunistic
fungal agents, scuh as Fusarium (20). The fact that Pseudomonas is
spread in various wards of the hospital and Acinetobacter
affects mainly patients from ICU can explain the greater prevalence
of Pseudomonas (10.05%) compared with other non-fermenting
bacteria (5.74%). This difference is constantly reported by studies
on all infections, not only in SSIs (21). The high usage of antibiotics in the
ICU promotes the higher resistance of bacteria isolated from these
patients.
The isolation of MRSA from surgical wounds has been
shown to be related to lower chances of primary healing and delayed
healing (22). The current method
for testing MRSA is to test the resistance to cefoxitin (23), which is also a surrogate test for
resistance to oxacillin. Hence, we can conclude that 40% of the
S. aureus isolates were MRSA. MRSA can establish within
hospitals and becomes difficult to control. Its survival is
promoted probably by the high usage of antibiotics, that can induce
the transformation of methicillin susceptible S. aureus
(MSSA) in MRSA (24). In our
study, the prevalence of MRSA was 20.29% [MRSA prevalence = MRSA
rate (40%) × S. aureus prevalence (50.72%)]. This is similar
with other observations reporting MRSA prevalence of 28.50% in SSIs
(25).
A recent finding is that many apparently susceptible
S. aureus strains actually contain small populations of
mecA-positive cells (26).
Many patients are colonized at admission with MSSA, located mostly
in the pharynx, but also in other sites (3). For example, some of the patients in
the present study had acne, from which S. aureus was
isolated (27). These MRSA
subpopulations can be selected through exposure to cefoxitin or
oxacillin up to 48 h (28).
Probably, the real prevalence of MRSA in SSI is >20.29%
(29).
A small difference between resistance to amoxicillin
alone (83.33%) and resistance to amoxicillin/clavulanate
combination (87.50%) was documented, indicating that only a few
strains of Staphylococcus may secrete β-lactamases in the
study samples. The resistance to quinolones has been reported to
emerge following treatment with ciprofloxacin (30). In the current study, a
ciprofloxacin resistance of 26.50% was found, possibly as many
surgeons use quinolones as prophylactic therapy. The low resistance
to rifampin of 13.80% can be explained as this is a reserve
antibiotic, for cases with tuberculosis. This is in accordance with
other reports; for example, Peterson et al reported a 15.50%
resistance of S. aureus strains to rifampin (31).
Profiling the resistance to antimicrobials using the
diameters of inhibition areas obtained by disk diffusion is widely
used in studies of epidemiology and strain homology (32). The usual analysis methods of
antibiotic resistance do not take advantage of the continuous
nature of the inhibition zone diameter, as these methods analyze
the binary variable of resistance. Cluster analysis can use
continuous variables on large scale and provides homology trees
that can help us understand the relatedness of the isolates
(33).
The acquisition of resistance genes by
Klebsiella strains, a very frequent observation in a
hospital environment, can be demonstrated by cluster analysis. In
Fig. 6 we can observe two big
clusters, the strains from the 2nd cluster had a higher resistance
index. Possibly, these represent hospital strains and the next
level of clusters contains strains that acquired genes for
extended-spectrum β-lactamases (ESBLs) and carbapenemases. Strains
carrying carbapenemase genes are in the third cluster (Fig. 6). The next level of clustering,
shows three 3rd degree clusters that may correspond to different
resistance mechanisms. It is known that the ESBL production confer
resistance to many penicillin derivatives and cephalosporins as the
carbapenemases production render the bacteria resistant to most
β-lactams. Other mechanisms such as porin loss (impermeability) can
also lead to very high antibiotic resistance. The clusters of 1st,
2nd and 3rd degree may reflect the distribution of antibiotic
resistance mechanisms in the Enterobacteriaceae population in the
current study hospital rather than relatedness due to transmission
from patient to patient. Further levels of the clustering analysis
can be used for this purpose, and can be observed similarity of
resistance profiles between patients from ICU and surgical wards
could be due to ICU admission of patients shortly after the
surgery, where they may stay for several days, during which they
may acquire highly resistant strains.
Analysis of the Staphylococcus resistance
cluster dendrogram, reveals that the MRSA clearly differentiate
from the sensitive staphylococci. There is a hierarchy in
resistance of MRSA which suggests acquisition of supplementary
resistance genes (Fig. 5). These
could be erm genes that produce of the ribosomal methylation.
In conclusion, up to 40% of surgeries are associated
to postoperative infectious complications.
The most common bacterial pathogen of SSIs is S.
aureus, followed by gram-negative organisms. This high
incidence can be partially be explained by the high load of S.
aureus in air flora, the colonization of the patients at
admission and probably to other unidentified perioperative
factors.
The bacteria isolated from ICU wards have a higher
resistance to certain antimicrobials, especially carbapenems,
compared with those from surgical wards. The use of these agents
should be avoided in ICU wards to prevent the development of the
resistance.
Beyond the detection of antibiotic resistance, the
molecular mechanisms that underlie resistance have been elucidated
by specific phenotypic and molecular methods, such as PCR and
mass-spectroscopy.
The drug sensitivity profile expressed as diameters
of inhibition areas of antibiotics in the diffusimetric method can
be used for phenotypic typing and epidemiological tracing of
hospital strains. This can contribute to antibiotic stewardship
measures, in hospitals that cannot afford expensive analyzers that
can type the strains as mass spectrometers or DNA sequencers.
The anti-infectious therapy optimized based upon
antibiotic resistance of the bacterial strains will improve the
quality of medical care, by discarding the antibiotics which loosed
their efficacy.
The data gathered from this study can help the
infection control team to establish effective guidelines for
antibiotic therapies in various surgical procedures, with the aim
to minimize the risk of developing SSIs and to the correct and
efficient use of the anti-infectious armamentarium.
Glossary
Abbreviations
Abbreviations:
|
SSIs
|
surgical site infections
|
|
ICU
|
intensive care unit
|
References
|
1
|
Staicus C, Calina D, Rosu L, Rosu AF and
Zlatian O: Involvement of microbial flora in aetiology of surgical
site infections. Eur J Hosp Pharm Sci Pract. 22:A592015.
|
|
2
|
Watanabe A, Kohnoe S, Shimabukuro R,
Yamanaka T, Iso Y, Baba H, Higashi H, Orita H, Emi Y, Takahashi I,
et al: Risk factors associated with surgical site infection in
upper and lower gastrointestinal surgery. Surg Today. 38:404–412.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Owens CD and Stoessel K: Surgical site
infections: Epidemiology, microbiology and prevention. J Hosp
Infect. 70 Suppl 2:3–10. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cristea OM, Zlatian OM, Dinescu SN,
Avramescu CS, Balasoiu M, Niculescu M and Calina DC: A comparative
study on antibiotic resistance of Klebsiella strains from surgical
and intensive care wards. Curr Heal Sci. 42:169–179. 2016.
|
|
5
|
Dulon M, Haamann F, Peters C, Schablon A
and Nienhaus A: MRSA prevalence in European healthcare settings: a
review. BMC Infect Dis. 11:1382011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
House of Commons Public Accounts
Committee, . Reducing Healthcare associated infection in hospitals
in England. Fifty-second Report of Session 2008–09. http://www.publications.parliament.uk/pa/cm200809/cmselect/cmpubacc/812/812.pdfNovember
10–2009.
|
|
7
|
Kassim A, Omuse G, Premji Z and Revathi G:
Comparison of Clinical Laboratory Standards Institute and European
Committee on Antimicrobial Susceptibility Testing guidelines for
the interpretation of antibiotic susceptibility at a University
teaching hospital in Nairobi, Kenya: a cross-sectional study. Ann
Clin Microbiol Antimicrob. 15:212016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li JY, Ma Y, Sun Z, Yao L, Zhang L, Hu C
and Jin S: Cluster analysis to detect the homogeneous strains of E.
coli isolated from clinical specimens. Chin J Microbiol Immunol.
23:384–387. 2003.
|
|
9
|
Xu J, Shi C, Song M, Xu X, Yang P, Paoli G
and Shi X: Phenotypic and genotypic antimicrobial resistance traits
of foodborne Staphylococcus aureus isolates from Shanghai. J Food
Sci. 79:M635–M642. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dzidic S and Bedeković V: Horizontal gene
transfer-emerging multidrug resistance in hospital bacteria. Acta
Pharmacol Sin. 24:519–526. 2003.PubMed/NCBI
|
|
11
|
Liapakis IE, Kottakis I, Tzatzarakis MN,
Tsatsakis AM, Pitiakoudis MS, Ypsilantis P, Light RW, Simopoulos CE
and Bouros DE: Penetration of newer quinolones in the empyema
fluid. Eur Respir J. 24:466–470. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kumar M, Dutta R, Saxena S and Singhal S:
Risk Factor Analysis in Clinical Isolates of ESBL and MBL
(Including NDM-1) Producing Escherichia coli and Klebsiella Species
in a Tertiary Care Hospital. J Clin Diagn Res. 9:DC08–DC13.
2015.PubMed/NCBI
|
|
13
|
Neonakis IK, Baritaki S, Georgiladakis A
and Spandidos DA: Analysis of the beta-lactam resistance phenotypes
of Escherichia coli. An 8-year survey conducted in Greece. Roum
Arch Microbiol Immunol. 67:10–13. 2008.PubMed/NCBI
|
|
14
|
Neonakis IK, Samonis G, Messaritakis H,
Baritaki S, Georgiladakis A, Maraki S and Spandidos DA: Resistance
status and evolution trends of Klebsiella pneumoniae isolates in a
university hospital in Greece: Ineffectiveness of carbapenems and
increasing resistance to colistin. Chemotherapy. 56:448–452. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sifakis S, Angelakis E, Makrigiannakis A,
Orfanoudaki I, Christakis-Hampsas M, Katonis P, Tsatsakis A and
Koumantakis E: Chemoprophylactic and bactericidal efficacy of 80 mg
gentamicin in a single and once-daily dosing. Arch Gynecol Obstet.
272:201–206. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Petrakis I, Gonianakis C, Vrachassotakis
N, Tsatsakis A, Vallilakis IS and Chalkiadakis G: Prospective study
of preincisional intraparietal single-dose ceftriaxone in reducing
postoperative wound infection in type I and II diabetic patients.
Acta Diabetol. 36:159–162. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Neonakis IK, Spandidos DA and Petinaki E:
Is minocycline a solution for multidrug-resistant Acinetobacter
baumannii? Future Microbiol. 9:299–305. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Youinou P, Garré M, Menez JF, Morin JF and
Masse R: Protein malnutrition and deficient cell-mediated immunity
in intensive care (author's transl). Nouv Presse Med. 10:3835–3837.
1981.(In French). PubMed/NCBI
|
|
19
|
Neonakis IK, Spandidos DA and Petinaki E:
Confronting multidrug-resistant Acinetobacter baumannii: A review.
Int J Antimicrob Agents. 37:102–109. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tanase A, Colita A, Ianosi G, Neagoe D,
Branisteanu DE, Calina D, Docea AO, Tsatsakis A and Ianosi SL: Rare
case of disseminated fusariosis in a young patient with graft vs.
host disease following an allogeneic transplant. Exp Ther Med.
12:2078–2082. 2016.PubMed/NCBI
|
|
21
|
Malini A, Deepa E, Gokul B and Prasad S:
Nonfermenting gram-negative bacilli infections in a tertiary care
hospital in kolar, karnataka. J Lab Physicians. 1:62–66. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Solomkin JS: Antibiotic resistance in
postoperative infections. Crit Care Med. 29 Suppl 4:N97–N99. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cockerill FR and Patel JB: M100-S25
Performance Standards for Antimicrobial Susceptibility Testing;
Twenty-Fifth Informational Supplement. Clin Lab Stand Inst.
35:44–49. 2015.
|
|
24
|
Schentag JJ, Hyatt JM, Carr JR, Paladino
JA, Birmingham MC, Zimmer GS and Cumbo TJ: Genesis of
methicillin-resistant Staphylococcus aureus (MRSA), how treatment
of MRSA infections has selected for vancomycin-resistant
Enterococcus faecium, and the importance of antibiotic management
and infection control. Clin Infect Dis. 26:1204–1214. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Manian FA, Meyer PL, Setzer J and Senkel
D: Surgical site infections associated with methicillin-resistant
Staphylococcus aureus: Do postoperative factors play a role? Clin
Infect Dis. 36:863–868. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Taiwo SS: Methicillin resistance in
Staphylococcus aureus: A review of the molecular epidemiology,
clinical significance and laboratory detection methods. West Afr J
Med. 28:281–290. 2009.PubMed/NCBI
|
|
27
|
Ianoşi S, Ianoşi G, Neagoe D, Ionescu O,
Zlatian O, Docea AO, Badiu C, Sifaki M, Tsoukalas D, Tsatsakis AM,
et al: Age-dependent endocrine disorders involved in the
pathogenesis of refractory acne in women. Mol Med Rep.
14:5501–5506. 2016.PubMed/NCBI
|
|
28
|
Thornsberry C and McDougal LK: Successful
use of broth microdilution in susceptibility tests for
methicillin-resistant (heteroresistant) staphylococci. J Clin
Microbiol. 18:1084–1091. 1983.PubMed/NCBI
|
|
29
|
Hiramatsu K, Kihara H and Yokota T:
Analysis of borderline-resistant strains of methicillin-resistant
Staphylococcus aureus using polymerase chain reaction. Microbiol
Immunol. 36:445–453. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Høiby N, Jarløv JO, Kemp M, Tvede M,
Bangsborg JM, Kjerulf A, Pers C and Hansen H: Excretion of
ciprofloxacin in sweat and multiresistant Staphylococcus
epidermidis. Lancet. 349:167–169. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Peterson LR, Quick JN, Jensen B, Homann S,
Johnson S, Tenquist J, Shanholtzer C, Petzel RA, Sinn L and Gerding
DN: Emergence of ciprofloxacin resistance in nosocomial
methicillin-resistant Staphylococcus aureus isolates. Resistance
during ciprofloxacin plus rifampin therapy for
methicillin-resistant S aureus colonization. Arch Intern Med.
150:2151–2155. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tarale P, Gawande S and Jambhulkar V:
Antibiotic susceptibility profile of bacilli isolated from the skin
of healthy humans. Braz J Microbiol. 46:1111–1118. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Berge AC, Atwill ER and Sischo WM:
Assessing antibiotic resistance in fecal Escherichia coli in young
calves using cluster analysis techniques. Prev Vet Med. 61:91–102.
2003. View Article : Google Scholar : PubMed/NCBI
|