Introduction
Osteonecrosis is the death of bone tissue following
microvascular injury, which results in bone resorption and,
potentially, structural collapse. The disease typically affects the
epiphyseal bone on the convex side of a joint, which may be due to
a lack of collateral circulation and most commonly affects the
femoral head, although it may also affect the humeral head, femoral
condyles, proximal tibia, vertebra, and small bones of the hand and
foot (1). Osteonecrosis of the
femoral head (ONFH) is a complex multifactorial disease that is
associated with genetic predisposition and exposure to certain
environmental factors. Various etiological factors, including the
use of corticosteroids, alcohol abuse, sickle cell anemia,
radiation and Gaucher disease, are known to be implicated in the
development of secondary osteonecrosis (2). It is possible for certain patients,
those who have received high doses of corticosteroids or consumed
excessive amounts of alcohol for a long period of time, to develop
ONFH, however, there are also rare cases of ONFH that occur
following short-duration corticosteroid treatment, thus indicating
that differences exist in the susceptibility to risk factors and
the genetic predisposition to ONFH between individuals (3,4).
Idiopathic ONFH is diagnosed when individuals are not known to have
been exposed to any of the known risk factors. The incidence or
prevalence of idiopathic ONFH reflects ethnic differences (5). Idiopathic ONFH in identical twins and
the clustering of cases in families indicate that genetic factors
may be involved in development (6,7).
Several studies have investigated genetic traits that may
predispose an individual to ONFH development, including those
involved in clotting disorders (8–10).
However, many of the results of genetic studies investigating the
association between genes involved in the clotting process and the
risk of ONFH in Caucasians have not been replicated in other ethnic
groups. For example, factor V Leiden and prothrombin G20210A
mutations have not been identified in the Korean population
(11,12). This suggests that geographic and
ethnic differences exist in the prevalence of disease-associated
genes and/or polymorphisms. In addition, Liu et al reported
that all patients with familial ONFH in the study carried collagen
type II α 1 chain (COL2A1) mutations, whereas no mutations were
identified in the COL2A1 coding region in patients with sporadic
ONFH (13). The majority of ONFH
cases are sporadic, therefore, it is essential to screen candidate
genes from a different perspective. Various genetic methods have
been developed to identify disease-associated loci or causal
variations associated with specific diseases. Genome-wide
association studies (GWAS) have been facilitated by the
availability of chip-based microarray techniques, which allow
assays of more than a million single-nucleotide polymorphisms
(SNPs) in order to identify disease-associated loci and potential
causal variants associated with diseases. GWAS has been used to map
a huge number of susceptibility genes for numerous complex
diseases, including type 1 and type 2 diabetes, inflammatory bowel
disease, prostate cancer and breast cancer (14–16).
Since GWAS was first reported in 2005, 2,041 studies have been
added to the Catalog of Published GWAS (http://www.genome.gov/26525384), however, to the best
of our knowledge, GWAS of patients with idiopathic ONFH has not
previously been performed. GWAS was performed in the present study
to identify genetic variants that influence susceptibility to and
outcomes of idiopathic ONFH in the Korean population.
Materials and methods
Study subjects
The study was initially performed at the GWAS
discovery stage to identify SNPs potentially involved in the
development of idiopathic ONFH in the Korean population. The ONFH
patients were recruited consecutively from Kyungpook National
University Hospital (Daegu, Korea) between 2002 and 2012, and
written informed consent was obtained from all study participants
prior to enrolment. This study was approved by the Institutional
Review Board of the Kyungpook National University Hospital. All
patients with ONFH were diagnosed by an orthopedist according to
the diagnostic criteria of the Association Research Circulation
Osseous classification system based on magnetic resonance imaging
and plain radiographs (17). Based
on etiological factors, patients were assigned to one of the
following groups: Idiopathic, steroid-induced and alcohol-induced
osteonecrosis. Only the patients in the idiopathic group were
included in this study. Control subjects were defined by a lack of
hip pain and the absence of any lesions with sclerotic margins or
subchondral collapse consistent with ONFH in anteroposterior and
frog-leg lateral pelvic radiographs. The GWAS included 217
idiopathic ONFH (152 males and 65 females aged 49.5±14.2 years) and
217 control (57 males and 160 females aged 54.7±12.8 years)
samples.
Genome-wide SNP genotyping
Genomic DNA was extracted from whole human blood
using the FlexiGene DNA kit (Qiagen, Inc., Valencia, CA, USA)
according to the manufacturer's protocol. For each sample,
genotyping was performed with the Axiom® 2.0 genome-wide
ASI 1 Array kit (Affymetrix, Inc., Santa Clara, CA, USA) according
to the standard protocols recommended by the manufacturer. Briefly,
for each array, ~200 ng genomic DNA was amplified and fragmented
randomly into fragments of 25–125 base pairs (bp). The initial
amplification had a reaction volume of 40 µl, which contained 20 µl
genomic DNA at a concentration of 10 ng/µl and 20 µl Denaturation
Master Mix (reagent from Module 1(P/N901711) of the Axiom 2.0
reagent kit). The initial amplification was performed for 10 min at
room temperature. Following the initial amplification, the
incubated products were amplified with 130 µl Axiom® 2.0
Neutral Solution, 225 µl Axiom® 2.0 Amp Solution, and 5
µl Axiom® 2.0 Amp Enzyme. Amplification reactions were
performed for 23±1 h at 37°C. Amplification products were then used
to amplify fragments of 200-1,100 bp. A fragmentation step reduced
the amplified products into segments of ~25–50 bp, which were then
end-labeled using reagents from Module 2-1(P/N901528) and Module
2-2(P/N901529) of the Axiom 2.0 Reagent kit. Following
hybridization, the bound target was washed under stringent
conditions to remove non-specific background and minimize any
background noise caused by random ligation events (reagents from
Module 4-1(P/N901278) and Module 4-2(P/N901276) of the Axiom 2.0
Reagent kit). Each polymorphic nucleotide was queried via a
multi-color ligation event performed on the array surface.
Following ligation, the arrays were stained and imaged using a
GeneTitan® Multi-Channel Instrument (Affymetrix, Inc.).
Images were analyzed using Genotyping Console™ software 4.14
(Affymetrix, Inc.).
Genotype quality control
Raw data in the form of CEL files were imported into
Affymetrix Power Tools (version 1.16.1; Affymetrix, Inc.) and
genotype calling was performed using Axiom GT1 algorithm
(Affymetrix, Inc.). Samples with dish quality control (QC) value of
>0.82 and call rate >0.95 were considered to have passed the
quality control assessment. Downstream analysis was performed using
PLINK (http://pngu.mgh.harvard.edu/~purcell/plink/) version
no. 1.07 (18). Markers were
considered not to meet the quality control criteria and discarded
if they were demonstrated to have any of the following: Call rate
<95%, minor allele frequency of the control <1%, and
deviations from the Hardy-Weinberg equilibrium with a P-value
<10-7. It was confirmed that the call rate per sample was
>97%.
Statistical analysis
Statistical analyses were performed using PLINK
(version no. 1.07) (18) and SAS
(version no. 9.1.3; SAS Institute, Cary, NC, USA). To test for
allelic and genotype associations between SNPs and ONFH, 2×2 and
2×3 contingency tables were constructed, which were compared using
Cochran-Armitage trend, Chi-squared, and Jonckheere-Terpstra tests.
The strength of association was estimated by the odds ratio and the
95% confidence interval, which were calculated using Cornfield
methods. Three genetic models of inheritance (recessive, dominant,
and codominant) for SNPs were assessed. Genotypes were given codes
of 0, 1 and 2; 0, 1 and 1; and 0, 0 and 1 in the codominant,
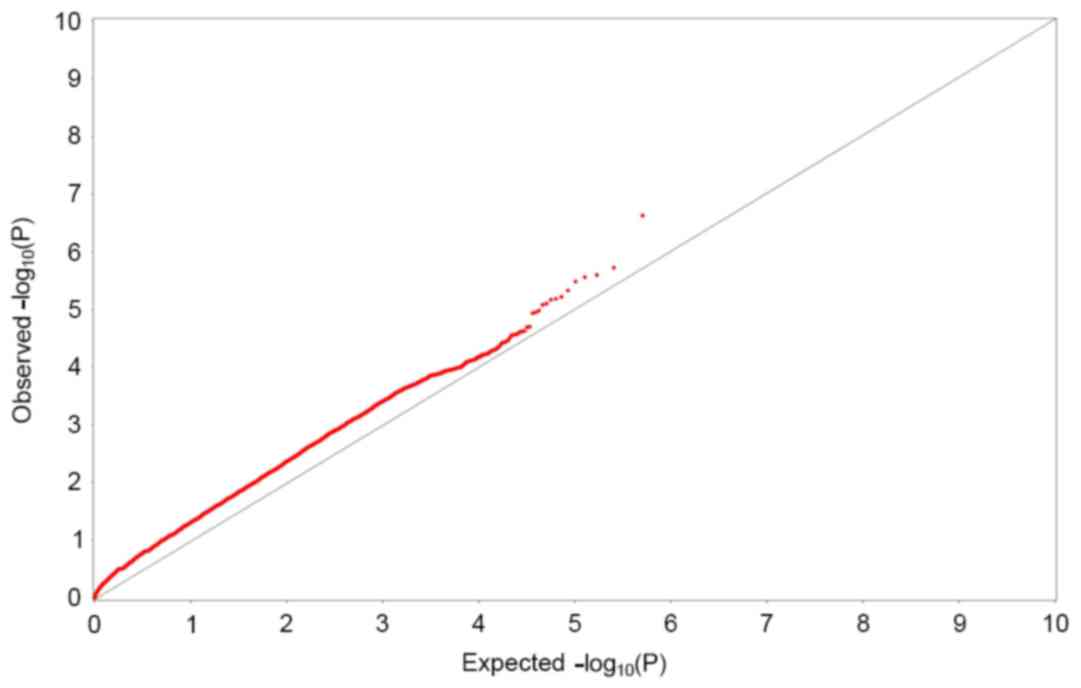
dominant, and recessive models, respectively. The quantile-quantile
(Q-Q) plot demonstrates close agreement between two distributions
if the population stratification and cryptic relatedness are
controlled adequately. Undetected population stratification or
cryptic relatedness results in deviation from the null across the
entire distribution; whereas, large-effect susceptibility loci
generate deviations at the highly significant end of the range
(19). Regional association plots
were created using LocusZoom (version no. 1.3; https://statgen.sph.umich.edu/locuszoom/genform.php?type=yourdata;
University of Michigan, Ann Arbor, MI, USA). Haploview (version no.
4.2; http://www.broad.mit.edu/mpg/haploview; Broad
Institute, Cambridge, MA, USA) was used to determine linkage
disequilibrium (LD) in the genomic region using an accelerated
expectation-maximization algorithm (20).
Results
Genotype data for 217 idiopathic ONFH cases (152
male, 65 female) and 217 control (57 male, 160 female) subjects was
analyzed. Based on stringent quality control criteria, 509,886 SNPs
were selected for association analysis for idiopathic ONFH. A
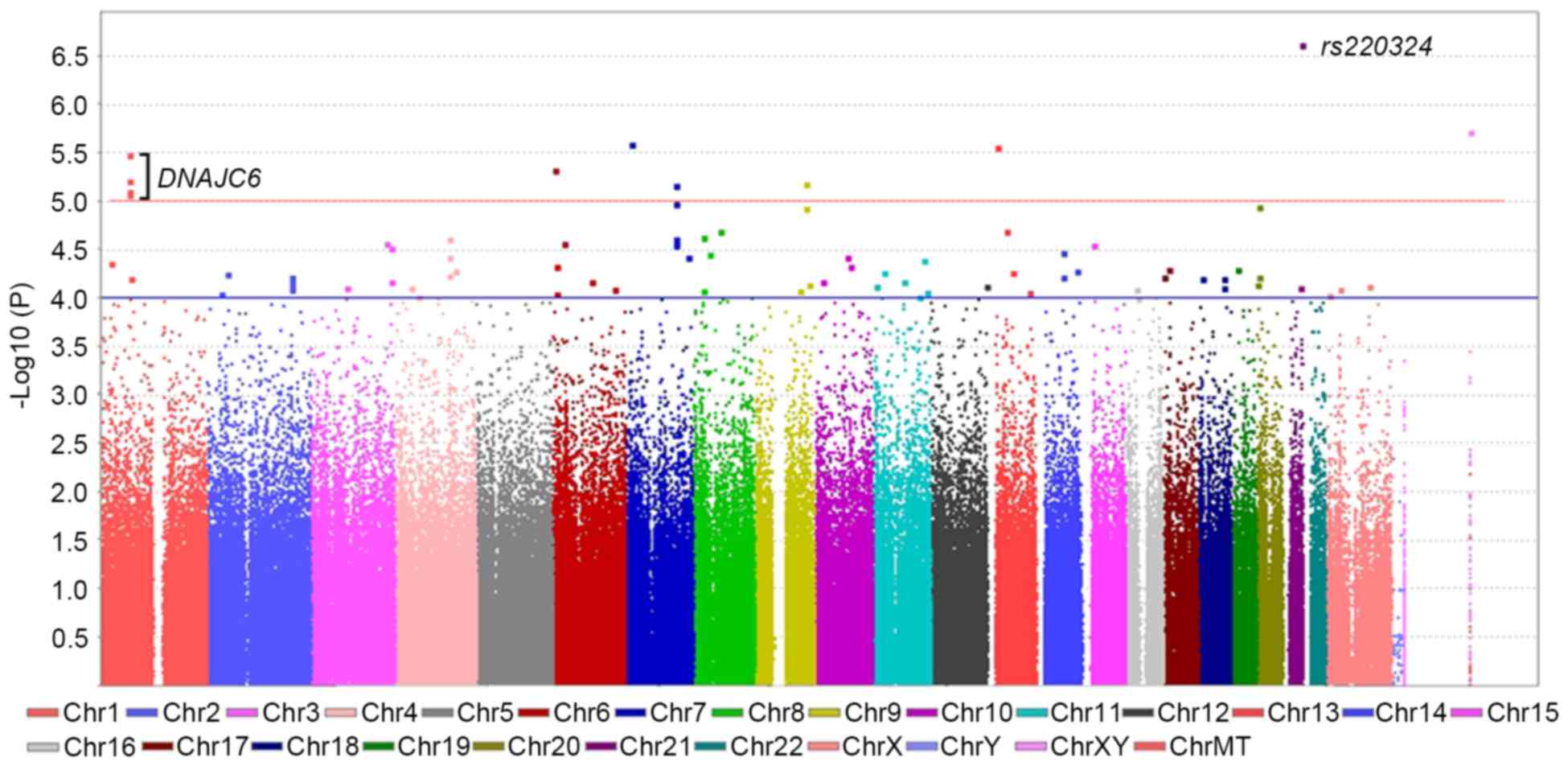
Manhattan plot of the association analyses for all SNPs was
produced using the chromosomal positions (x-axis) and negative
logarithm of P-values (y-axis) for each SNP (Fig. 1). In the Q-Q plot (Fig. 2), minimal deviation from the
expected P-value distribution was observed, and only at the upper
tail of the distribution, suggesting that population stratification
was adequately controlled. All of the association signals with
P<1×10−4 (78 SNPs) are shown in Table I. Although none of the SNPs reached
the accepted genome-wide significance level of 5×10−8,
11 SNPs on chromosomes 1, 6, 7, 9, 13 and 21 had the strongest
associations with increased susceptibility to idiopathic ONFH at a
significance level of P<10−5 (Fig. 1 and Table I). The most significant association
was detected at rs220324 (P=3.57×10−7), which is located
in an intergenic region between the uromodulin-like 1
(UMODL1) gene and the ATP-binding cassette, sub-family G,
member 1 (ABCG1) gene region on chromosome 21q22.3. Given
that certain disease-associated genes may be enriched among the top
ranked genes in GWAS, the present study then filtered the SNPs to
identify chromosomal regions that harbored clusters of associated
SNPs at a less stringent criterion (P<1×10−5). It was
observed that DnaJ heat shock protein family (Hsp40) member C6
(DNAJC6), a region between 65.37 and 65.67 Mb in the
chromosome 1p31.3 region, harbored a cluster of SNPs, which were
associated with idiopathic ONFH at a significance level of
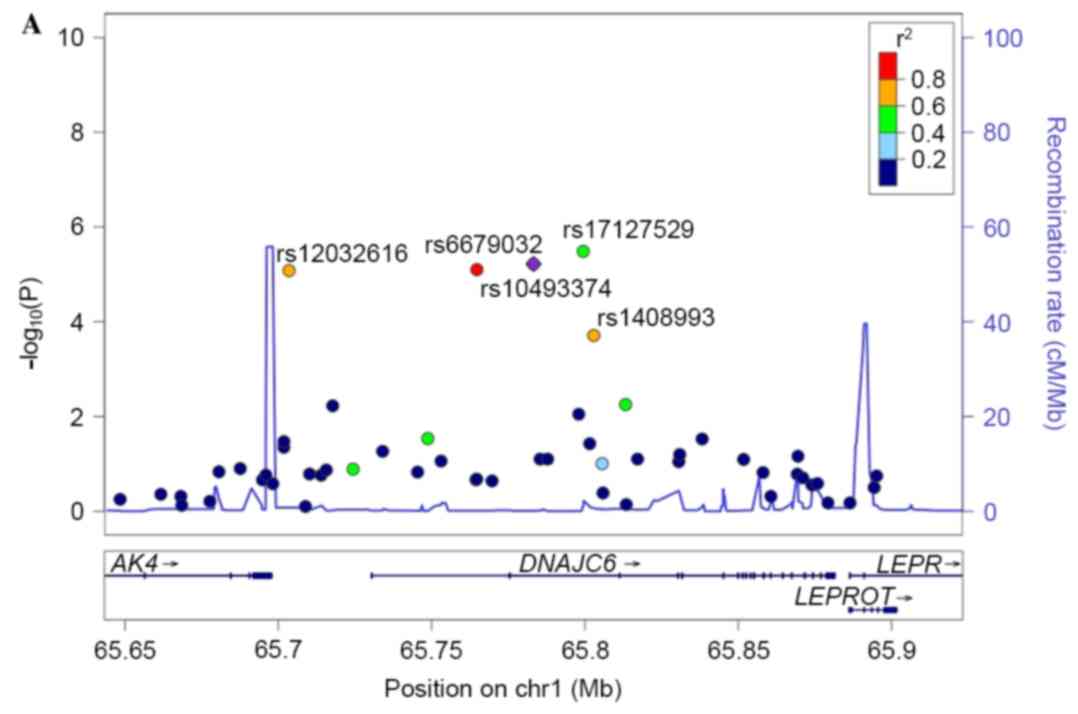
P<1×10−5. Four significant SNPs, rs10493374,
rs12032616, rs17127529 and rs6679032 in the DNAJC6 gene
region were revealed to be in strong LD with each other (Table II), and all had associations with
idiopathic ONFH at a significance level of P<1×10−5.
The regional association plot and LD plot for these loci are
presented in Fig. 3.
 | Table I.All 78 SNPs with
P<1×10−4 according to GWAS analysis of ONFH cases and
controls. |
Table I.
All 78 SNPs with
P<1×10−4 according to GWAS analysis of ONFH cases and
controls.
|
|
|
|
|
Allelec | MAF |
|
|
|
|---|
|
|
|
|
|
|
|
|
|
|---|
| rsSNP number | Chr. | Positiona | Nearest
Geneb | 1 | 2 | ONFH | Control | OR | 95% CI | P-value |
|---|
| rs220324 | 21 | 43586699 | UMODL1, ABCG1 | T | C | 0.33 | 0.484 | 0.35 | 0.23–0.52 | 2.35E-07 |
| rs306896 | X | 154979339 | SPRY3 | T | C | 0.595 | 0.444 | – | – | 1.89E-06 |
| rs1432486 | 7 | 13481626 |
Intergenic/Unknown | C | T | 0.491 | 0.333 | 1.93 | 1.47–2.54 | 2.52E-06 |
| rs9285223 | 13 | 23124968 |
Intergenic/Unknown | C | T | 0.204 | 0.288 | – | – | 2.77E-06 |
| rs17127529 | 1 | 65799440 |
DNAJC6 | T | C | 0.311 | 0.463 | 0.52 | 0.40–0.69 | 3.27E-06 |
| rs17548629 | 6 | 3114457 | RIPK1 | C | T | 0.183 | 0.318 | 0.48 | 0.35–0.66 | 4.69E-06 |
| rs10493374 | 1 | 65783231 |
DNAJC6 | G | T | 0.15 | 0.276 | 0.46 | 0.33–0.65 | 6.05E-06 |
| rs12376144 | 9 | 119488641 | ASTN2 | C | A | 0.394 | 0.254 | – | – | 6.53E-06 |
| rs7794364 | 7 | 117508506 | CTTNBP2 | C | T | 0.201 | 0.095 | – | – | 6.82E-06 |
| rs6679032 | 1 | 65764742 |
DNAJC6 | A | G | 0.152 | 0.276 | 0.47 | 0.34–0.66 | 7.94E-06 |
| rs12032616 | 1 | 65703484 |
DNAJC6, AK4 | A | G | 0.16 | 0.286 | 0.47 | 0.34–0.66 | 8.32E-06 |
| rs1012084 | 7 | 117529421 |
Intergenic/Unknown | G | A | 0.2 | 0.097 | – | – | 1.05E-05 |
| rs6037866 | 20 | 4521824 |
Intergenic/Unknown | T | C | 0.229 | 0.365 | – | – | 1.12E-05 |
| rs756118 | 9 | 119487330 | ASTN2 | T | C | 0.392 | 0.256 | – | – | 1.16E-05 |
| rs3794431 | 13 | 44544511 | DGKZP1 | A | G | 0.171 | 0.293 | 0.5 | 0.36–0.69 | 2.00E-05 |
| rs16927830 | 8 | 62658110 | ASPH, NKAIN3,
MIR4470 | T | C | 0.585 | 0.47 | 2.76 | 1.71–4.44 | 2.04E-05 |
| rs2466224 | 8 | 22785361 | PEBP4 | A | C | 0.277 | 0.411 | – | – | 2.37E-05 |
| rs28407041 | 4 | 127437492 |
Intergenic/Unknown | C | T | 0.449 | 0.32 | 2.33 | 1.57–3.45 | 2.40E-05 |
| rs4730811 | 7 | 117582806 |
Intergenic/Unknown | A | G | 0.21 | 0.108 | – | – | 2.45E-05 |
| rs7775145 | 6 | 25419880 | LRRC16A | G | A | 0.47 | 0.353 | 2.39 | 1.58–3.60 | 2.66E-05 |
| rs10513755 | 3 | 177115806 |
Intergenic/Unknown | C | G | 0.558 | 0.482 | 1.36 | 1.04–1.77 | 2.71E-05 |
| rs11631923 | 15 | 25764385 | LOC105370737 | T | C | 0.32 | 0.449 | – | – | 2.75E-05 |
| rs6466637 | 7 | 117576675 |
Intergenic/Unknown | A | G | 0.21 | 0.109 | – | – | 2.79E-05 |
| rs9882205 | 3 | 186570398 | ADIPOQ | A | G | 0.347 | 0.472 | 0.42 | 0.28–0.64 | 3.05E-05 |
| rs16887454 | 8 | 38413475 | LOC105379383 | G | A | 0.217 | 0.309 | 0.45 | 0.30–0.66 | 3.52E-05 |
| rs10999727 | 10 | 72960515 |
Intergenic/Unknown | G | A | 0.468 | 0.354 | 2.33 | 1.55–3.49 | 3.71E-05 |
| rs11098882 | 4 | 127447572 |
Intergenic/Unknown | A | C | 0.38 | 0.25 | 1.84 | 1.38–2.46 | 3.74E-05 |
| rs17170575 | 7 | 147218996 | CNTNAP2,
MIR548I4 | C | T | 0.243 | 0.204 | 20.7 | 2.75–156 | 3.76E-05 |
| rs1886461 | 14 | 61944882 | PRKCH | A | G | 0.498 | 0.388 | 2.43 | 1.59–3.72 | 3.93E-05 |
| rs11216126 | 11 | 116617240 |
Intergenic/Unknown | A | C | 0.24 | 0.131 | 2.08 | 1.46–2.97 | 4.06E-05 |
| rs7514144 | 1 | 20735869 | LOC339505 | T | C | 0.124 | 0.23 | 0.48 | 0.33–0.68 | 4.37E-05 |
| rs3804483 | 6 | 6637677 | LY86 | C | T | 0.267 | 0.391 | 0.45 | 0.30–0.66 | 4.62E-05 |
| rs1515416 | 10 | 79364940 | KCNMA1 | A | C | 0.385 | 0.279 | 4.64 | 2.09–10.3 | 4.64E-05 |
| rs2969183 | 17 | 11344773 | SHISA6 | G | A | 0.067 | 0.014 | 5.43 | 2.21–13.4 | 5.00E-05 |
| rs714919 | 19 | 15151667 |
Intergenic/Unknown | G | A | 0.272 | 0.384 | 0.45 | 0.31–0.67 | 5.02E-05 |
| rs34299628 | 4 | 142506694 |
Intergenic/Unknown | G | A | 0.214 | 0.326 | 0.45 | 0.31–0.67 | 5.09E-05 |
| rs11629059 | 14 | 96093508 |
Intergenic/Unknown | G | A | 0.565 | 0.482 | 2.56 | 1.61–4.08 | 5.16E-05 |
| rs9569891 | 13 | 59150370 |
Intergenic/Unknown | C | T | 0.623 | 0.486 | 1.74 | 1.33–2.29 | 5.32E-05 |
| rs731365 | 11 | 24334057 | LOC105376595 | G | T | 0.317 | 0.451 | 0.57 | 0.43–0.75 | 5.47E-05 |
| rs7561623 | 2 | 45694417 | SRBD1 | G | A | 0.249 | 0.141 | 2.03 | 1.43–2.87 | 5.61E-05 |
| rs28558216 | 4 | 127437795 |
Intergenic/Unknown | C | T | 0.376 | 0.249 | 1.82 | 1.36–2.43 | 5.86E-05 |
| rs1902249 | 2 | 198521356 | RFTN2 | T | C | 0.604 | 0.467 | 1.74 | 1.33–2.27 | 5.94E-05 |
| rs3783786 | 14 | 61937224 | PRKCH | G | A | 0.599 | 0.468 | – | – | 5.94E-05 |
| rs297745 | 20 | 4473609 |
Intergenic/Unknown | C | T | 0.224 | 0.346 | – | – | 6.00E-05 |
| rs9789059 | 17 | 129225 | RPH3AL | C | T | 0.349 | 0.418 | 0.32 | 0.18–0.57 | 6.10E-05 |
| rs547810 | 18 | 58202247 |
Intergenic/Unknown | G | C | 0.376 | 0.264 | 2.19 | 1.49–3.23 | 6.14E-05 |
| rs1965881 | 18 | 9659377 |
Intergenic/Unknown | C | T | 0.528 | 0.393 | – | – | 6.17E-05 |
| rs17130890 | 1 | 69683475 | LOC105378786 | T | C | 0.178 | 0.294 | 0.52 | 0.38–0.72 | 6.19E-05 |
| rs9444695 | 6 | 90253629 | ANKRD6 | G | A | 0.588 | 0.456 | 1.7 | 1.30–2.23 | 6.63E-05 |
| rs6719832 | 2 | 198533369 | RFTN2 | A | G | 0.602 | 0.468 | 2.38 | 1.55–3.66 | 6.69E-05 |
| rs11253891 | 10 | 16332723 | LOC102724039 | G | A | 0.157 | 0.267 | 0.51 | 0.36–0.71 | 6.72E-05 |
| rs3774262 | 3 | 186571814 | ADIPOQ | G | A | 0.357 | 0.235 | 1.85 | 1.26–2.70 | 6.73E-05 |
| rs1660849 | 11 | 71100302 | FLJ42102,
SHANK2 | C | T | 0.521 | 0.405 | 2.59 | 1.61–4.18 | 6.78E-05 |
| rs6136898 | 20 | 2047818 |
Intergenic/Unknown | C | T | 0.126 | 0.051 | – | – | 7.24E-05 |
| rs7869304 | 9 | 124946021 | MORN5 | A | G | 0.336 | 0.461 | 0.45 | 0.30–0.67 | 7.25E-05 |
| rs7974644 | 12 | 130593579 |
Intergenic/Unknown | G | A | 0.618 | 0.484 | – | – | 7.43E-05 |
| rs916220 | X | 102254602 |
Intergenic/Unknown | G | A | 0.346 | 0.198 | 3.31 | 1.81–6.07 | 7.44E-05 |
| rs11820502 | 11 | 5688024 | TRIM5 | C | G | 0.336 | 0.465 | 0.34 | 0.19–0.59 | 7.51E-05 |
| rs6708239 | 2 | 198525084 | RFTN2 | A | G | 0.604 | 0.47 | 1.72 | 1.31–2.25 | 7.57E-05 |
| rs6778265 | 3 | 81985745 | LOC105377179 | C | T | 0.12 | 0.221 | 0.48 | 0.33–0.70 | 7.65E-05 |
| rs10009706 | 4 | 38079475 | TBC1D1 | G | A | 0.332 | 0.446 | 0.31 | 0.17–0.57 | 7.67E-05 |
| rs17657655 | 18 | 58229575 |
Intergenic/Unknown | A | C | 0.346 | 0.228 | – | – | 7.77E-05 |
| rs2835984 | 21 | 39188787 | KCNJ6, TMPRSS3 | A | T | 0.302 | 0.41 | 0.3 | 0.16–0.56 | 7.82E-05 |
| rs9967823 | 2 | 198531529 | RFTN2 | A | G | 0.604 | 0.47 | 1.72 | 1.31–2.25 | 7.88E-05 |
| rs7192193 | 16 | 26171345 |
Intergenic/Unknown | G | A | 0.519 | 0.424 | 2.63 | 1.61–4.29 | 7.94E-05 |
| rs1091272 | X | 32622348 | DMD | G | T | 0.476 | 0.356 | 5.24 | 2.16–12.7 | 7.98E-05 |
| rs9496856 | 6 | 144411489 |
Intergenic/Unknown | A | C | 0.101 | 0.164 | – | – | 8.09E-05 |
| rs7032668 | 9 | 105027288 |
Intergenic/Unknown | C | T | 0.168 | 0.265 | 0.45 | 0.31–0.67 | 8.23E-05 |
| rs17088580 | 8 | 22578660 | PEBP4,
LOC100507139 | G | A | 0.154 | 0.263 | – | – | 8.24E-05 |
| rs12574001 | 11 | 124112644 | OR8G5 | C | T | 0.134 | 0.06 | 2.74 | 1.64–4.58 | 8.71E-05 |
| rs7326256 | 13 | 99788540 |
Intergenic/Unknown | G | A | 0.062 | 0.141 | 0.37 | 0.22–0.61 | 8.71E-05 |
| rs1971866 | 2 | 30262542 | LOC101929418 | G | C | 0.507 | 0.384 | 2.28 | 1.51–3.46 | 8.81E-05 |
| rs3804486 | 6 | 6637995 | LY86 | G | A | 0.267 | 0.387 | 0.47 | 0.32–0.68 | 8.98E-05 |
| rs5979043 | X | 9164003 | FAM9B | C | G | 0.339 | 0.214 | 7.89 | 2.41–25.8 | 9.37E-05 |
| rs564569 | 11 | 107706991 | SLC35F2 | G | A | 0.502 | 0.371 | 1.71 | 1.31–2.24 | 9.59E-05 |
| rs1459821 | 4 | 53340238 |
Intergenic/Unknown | T | G | 0.074 | 0.154 | 0.38 | 0.23–0.63 | 9.62E-05 |
| rs566238 | 11 | 107706832 | SLC35F2 | C | T | 0.502 | 0.371 | 1.71 | 1.31–2.24 | 9.75E-05 |
| rs12933766 | 16 | 29922011 | KCTD13, ASPHD1 | C | T | 0.375 | 0.281 | 3.61 | 1.83–7.13 | 9.89E-05 |
 | Table II.Linkage disequilibrium relationships
(|D| and r2) between the four SNPs that had strong
associations with the DNAJC6 gene. |
Table II.
Linkage disequilibrium relationships
(|D| and r2) between the four SNPs that had strong
associations with the DNAJC6 gene.
|
| SNP |D'| |
|---|
|
|
|
|---|
| SNP
r2 | rs12032616 | rs6679032 | rs10493374 | rs17127529 |
|---|
| rs12032616 |
| 0.860 | 0.860 | 0.749 |
| rs6679032 | 0.687 |
| 1 | 1 |
| rs10493374 | 0.685 | 1 |
| 1 |
| rs17127529 | 0.256 | 0.428 | 0.426 |
Discussion
ONFH usually affects young adults and, without
treatment, progresses to the collapse of femoral head leading to
osteoarthritis and destruction of the hip joint (21). ONFH is one of the most common
diseases of the hip joint in Korea, where its incidence is
relatively high compared with other countries (5,22).
Approximately 50–60% of total primary hip replacements are thought
to be the result of ONFH in Korea (5). In Korea, non-traumatic ONFH is
associated with idiopathic factors in 36%, alcoholism in 35%,
steroids in 14% and other factors in 15% of cases (23). In a previous study, Kang et
al (22) reported the
prevalence of ONFH in Korea using medical claims data obtained from
the Korean National Health Insurance Service (Wonju, Korea). The
estimated average number of annual cases was 14,103, thus,
indicating an average prevalence of 28.91 per 100,000 over a 5-year
period (2002–2006). It was demonstrated that 32.4% had a history of
alcohol abuse and 14.6% of cases were associated with steroid
usage. In recent years, the clinical significance of ONFH for hip
diseases has received attention, however, the details of its
pathogenesis and epidemiology are not well understood; although it
is generally assumed that venous thrombosis resulting in blood flow
obstruction to the femoral head, mediated by thrombophilia and/or
hypofibrinolysis, is important in the development of ONFH (24,25).
Several studies have investigated the genetic traits that may
predispose subjects to ONFH development. However, contrary to
expectations, the results of various studies have demonstrated that
differences are observed between reports with different study
designs and/or ethnic groups involved in the analyses. The present
study conducted a GWAS in the Korean population to identify new
susceptibility loci associated with idiopathic ONFH. To the best of
our knowledge, it is the first high-density GWA scan of ONFH in a
Korean population, and the lowest P-value, and most significant
association, this study identified was obtained for the association
of idiopathic ONFH with rs220324 (P=3.57×10−7), which is
located in an intergenic region between the UMODL1 gene and
ABCG1 gene region on chromosome 21q22.3. Notably, the
cholesterol transporter ABCG1 is involved in cholesterol
homeostasis, where it promotes cholesterol efflux from cells and
regulates intracellular cholesterol homeostasis (26). However, none of the SNPs identified
by the present study satisfied the recognized GWA criteria
(P<1×10−8). Given that certain disease-associated
genes may be enriched among the top ranked genes in GWAS, the
present study searched for chromosomal regions containing two or
more significant SNPs within 100 kb of the significant marker SNPs
using a less stringent P-value, and the LD structure of the region
was analyzed using GWAS data. Subsequently, a chromosomal block
containing genes that clustered with SNPs thought to be associated
with ONFH (P<1×10−5) was identified. The four
variants, rs10493374, rs12032616, rs17127529 and rs6679032, with
modest associations were located in and around the DNAJC6
locus. A previous GWAS of the anticoagulant pathway also detected
significant associations between certain SNPs in the DNAJC6
gene and free protein S plasma levels (27). The protein C anticoagulant pathway
regulates blood coagulation by preventing the formation of thrombi.
This pathway has two main plasma components, protein C and protein
S. Deficiencies in antithrombin, protein C and protein S, or an
impaired anticoagulant pathway, increase the incidence of
thromboembolic disorders (27).
Notably, a number of researchers have suggested that intravascular
coagulation and microcirculatory thrombotic occlusion may provide a
common pathway to non-traumatic osteonecrosis. It is assumed that
venous thrombosis with blood flow obstruction to the femoral head,
mediated by thrombophilia and/or hypofibrinolysis, leads to
increased intraosseous pressure, reduced arterial flow and hypoxia,
which appear to be important in the development of ONFH (24,25).
The exact function of DNAJC6 remains unknown, however, the
protein encoded by DNAJC6 resembles a tyrosine-protein
phosphatase auxilin, which is an enzyme that promotes the uncoating
of clathrin-coated vesicles, thus, it potentially has a role in
endocytosis. Endocytosis itself, which is followed by partial
proteolysis, is involved in coagulation via molecular modification
of factor V and factor VIII (27,28).
Thus, a similar mechanism involving DNAJC6 and free protein
S plasma levels may be associated with ONFH, although validation of
this hypothesis requires further investigation. When a less
stringent P-value (P<1×10−4) was applied to select
candidate regions for further investigation, in phase 2 of the
present study, several chromosomal regions that clustered with SNPs
thought to be associated with idiopathic ONFH were discovered (data
not shown). The present study performed one of the first GWAS of
idiopathic ONFH in Koreans and aimed to identify genetic variants
that influence susceptibility to idiopathic ONFH. The results
indicated that several SNPs in the DNAJC6 gene are
potentially associated with idiopathic ONFH. Further
investigations, including replicate studies, are required to
confirm whether DNAJC6 is an ONFH susceptibility gene, but
the preliminary findings of the present study provide novel insight
into the genetic factors associated with the risk of ONFH.
Acknowledgements
This research was supported by Biomedical Research
Institute grant, Kyungpook National University Hospital (grant no.
15-04; 2015), and partially supported by a by a grant of the Korea
Health Technology R&D Project through the Korea Health Industry
Development Institute, funded by the Ministry of Health &
Welfare, Republic of Korea (grant no. HI15C0001).
References
|
1
|
Nishimura T, Matsumoto T, Nishino M and
Tomita K: Histopathologic study of veins in steroid treated
rabbits. Clin Orthop Relat Res. 334:37–42. 1997. View Article : Google Scholar
|
|
2
|
Assouline-Dayan Y, Chang C, Greenspan A,
Shoenfeld Y and Gershwin ME: Pathogenesis and natural history of
osteonecrosis. Semin Arthritis Rheum. 32:94–124. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Orlić D, Jovanović S, Anticević D and
Zecević J: Frequency of idiopathic aseptic necrosis in medically
treated alcoholics. Int Orthop. 14:383–386. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jones LC and Hungerford DS: The
pathogenesis of osteonecrosis. Instr Course Lect. 56:179–196.
2007.PubMed/NCBI
|
|
5
|
Kim SY and Rubash HE: Avascular necrosis
of the femoral head: the Korean experienceThe adult hip. 2. 2nd
edition. Lippincott Williams & Wilkins; Philadelphia, PA: pp.
1078–1086. 2006
|
|
6
|
Glueck CJ, Glueck HI, Welch M, Freiberg R,
Tracy T, Hamer T and Stroop D: Familial idiopathic osteonecrosis
mediated by familial hypofibrinolysis with high levels of
plasminogen activator inhibitor. Thromb Haemost. 71:195–198.
1994.PubMed/NCBI
|
|
7
|
Nobillot R, Le Parc JM, Benoit J and
Paolaggi JB: Idiopathic osteonecrosis of the hip in twins. Ann
Rheum Dis. 53:7021994. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ferrari P, Schroeder V, Anderson S,
Kocovic L, Vogt B, Schiesser D, Marti HP, Ganz R, Frey FJ and
Kohler HP: Association of plasminogen activator inhibitor-1
genotype with avascular osteonecrosis in steroid-treated renal
allograft recipients. Transplantation. 74:1147–1152. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zalavras CG, Vartholomatos G, Dokou E and
Malizos KN: Factor V Leiden and prothrombin gene mutations in
femoral head osteonecrosis. Thromb Haemost. 87:1079–1080.
2002.PubMed/NCBI
|
|
10
|
Zalavras CG, Malizos KN, Dokou E and
Vartholomatos G: The 677C->T mutation of the
methylene-tetrahydrofolate reductase gene in the pathogenesis of
osteonecrosis of the femoral head. Haematologica. 87:111–112.
2002.PubMed/NCBI
|
|
11
|
Kim SY, Suh JS, Park EK, Jung WB, Kim JW,
Koo KH and Kim CY: Factor V Leiden gene mutation in femoral head
osteonecrosis. J Korean Ortho Res Soc. 6:259–264. 2003.
|
|
12
|
Chang JD, Hur M, Lee SS, Yoo JH and Lee
KM: Genetic background of nontraumatic osteonecrosis of the femoral
head in the Korean population. Clin Orthop Relat Res.
466:1041–1046. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu YF, Chen WM, Lin YF, Yang RC, Lin MW,
Li LH, Chang YH, Jou YS, Lin PY, Su JS, et al: Type II collagen
gene variants and inherited osteonecrosis of the femoral head. N
Engl J Med. 352:2294–2301. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gordon H, Moller F Trier, Andersen V and
Harbord M: Heritability in inflammatory bowel disease: From the
first twin study to genome-wide association studies. Inflamm Bowel
Dis. 21:1428–1434. 2015.PubMed/NCBI
|
|
15
|
Veron A, Blein S and Cox DG: Genome-wide
association studies and the clinic: A focus on breast cancer.
Biomark Med. 8:287–296. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Grarup N, Sandholt CH, Hansen T and
Pedersen O: Genetic susceptibility to type 2 diabetes and obesity:
from genome-wide association studies to rare variants and beyond.
Diabetologia. 57:1528–1541. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gardeniers JWM: The Arco Perspective for
Reaching One Uniform Staging System of OsteonecrosisBone
Circulation and Vascularization in Normal and Pathological
Conditions. Scoutens A, Arlet J, Gardeniers JWM and Hughes SPF:
247. Plenum Press; New York, NY: pp. 375–380. 1993, View Article : Google Scholar
|
|
18
|
Purcell S, Neale B, Todd-Brown K, Thomas
L, Ferreira MA, Bender D, Maller J, Sklar P, De Bakker PI, Daly MJ
and Sham PC: PLINK: A tool set for whole-genome association and
population-based linkage analyses. Am J Hum Genet. 81:559–575.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Clayton DG, Walker NM, Smyth DJ, Pask R,
Cooper JD, Maier LM, Smink LJ, Lam AC, Ovington NR, Stevens HE, et
al: Population structure, differential bias and genomic control in
a large-scale, case-control association study. Nat Genet.
37:1243–1246. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Barrett JC, Fry B, Maller J and Daly MJ:
Haploview: Analysis and visualization of LD and haplotype maps.
Bioinformatics. 21:263–265. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Glimcher MJ and Kenzora JE: The biology of
osteonecrosis of the human femoral head and its clinical
implications. III. Discussion of the etiology and genesis of the
pathological sequelae; commments on treatment. Clin Orthop Relat
Res. 140:273–312. 1979.
|
|
22
|
Kang JS, Park S, Song JH, Jung YY, Cho MR
and Rhyu KH: Prevalence of osteonecrosis of the femoral head: A
nationwide epidemiologic analysis in Korea. J Arthroplasty.
24:1178–1183. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Koo KH, Jeong ST and Jones JP Jr:
Borderline necrosis of the femoral head. Clin Orthop Relat Res.
358:158–165. 1999. View Article : Google Scholar
|
|
24
|
Jones LC, Mont MA, Le TB, Petri M,
Hungerford DS, Wang P and Glueck CJ: Procoagulants and
osteonecrosis. J Rheumatol. 30:783–791. 2003.PubMed/NCBI
|
|
25
|
Kerachian MA, Harvey EJ, Cournoyer D, Chow
TY and Séguin C: Avascular necrosis of the femoral head: Vascular
hypotheses. Endothelium. 13:237–244. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sag D, Cekic C, Wu R, Linden J and Hedrick
CC: The cholesterol transporter ABCG1 links cholesterol homeostasis
and tumour immunity. Nat Commun. 6:63542015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Athanasiadis G, Buil A, Souto JC, Borrell
M, López S, Martinez-Perez A, Lathrop M, Fontcuberta J, Almasy L
and Soria JM: A genome-wide association study of the Protein C
anticoagulant pathway. PLoS One. 6:e291682011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Camire RM and Bos MH: The molecular basis
of factor V and VIII procofactor activation. J Thromb Haemost.
7:1951–1961. 2009. View Article : Google Scholar : PubMed/NCBI
|