Introduction
Patients undergoing hemodialysis are at increased
risk of acquiring GB virus C (GBV-C) and torque teno virus (TTV;
also known as transfusion transmitted virus) infection as a result
of their impaired immune system and frequent contact with blood,
blood products, equipment and surfaces contaminated with these
viruses. A high prevalence of GBV-C infection in patients
undergoing hemodialysis has previously been documented, with rates
ranging from 3.9–26.5% in Iran, Egypt, Turkey and Brazil (1–5).
Based on the results of two older studies, however, the prevalence
in Indonesian patients undergoing hemodialysis is greater.
Handajani et al (6)
reported a prevalence of 29% among patients undergoing hemodialysis
in Surabaya, and Tsuda et al (7) reported a prevalence of 55% in
Yogyakarta. In general, the prevalence of GBV-C is greater in
patients undergoing hemodialysis than in low-risk populations,
including blood donors or healthy individuals.
Multiple GBV-C genotypes (1–7) have
been identified based on the genetic diversity of full or partial
genome sequences. However, genotypes 4, 6 and 7 are highly similar
and can be classified as one group. Thus, a simpler classification
comprising of 5 groups of GBV-C genotypes has been recommended
(8). In Indonesia, genotype 4 was
reported to be predominant (55.5%) in blood donors, followed by
genotypes 3 (22.2%), 2 (11.1%) and 6 (11.1%) (6). In contrast, genotype 6 was
predominant among patients with chronic liver disease and patients
undergoing hemodialysis, being detected in 60% and 83.3% of
patients, respectively (6). These
data suggest that genotype 6 is most common in Indonesian patients
undergoing hemodialysis.
Notably, a previous study identified that the
prevalence of GBV-C infection was 88.8% in patients with human
immunodeficiency virus (HIV) in Yogyakarta, Indonesia (9). This prevalence was greater than
expected, and genotype 2 was predominant (58.3%), followed by
genotypes 6 (28.4%) and 3 (12.6%). The distribution of GBV-C
genotypes in patients with HIV differed from that observed in blood
donors, and may reflect a change in the prevalence and genotypic
distribution of GBV-C infection in Yogyakarta, Indonesia.
TTV is also common in patients undergoing
hemodialysis in certain countries, with prevalence rates ranging
from 27.8–68.8% in Iran, India, Italy and Brazil (10–14).
However, only two previous studies have examined the prevalence of
TTV infection in Indonesia. In a healthy population, TTV was
detected in 95% of individuals. The isolates were primarily
classified into genotypes 1, 2 and 3 (98%), which were prevalent
worldwide. However, genotype 22 and 23 were found to be unique in
Indonesia. Genotype 22 was more common in Indonesia than in Japan,
whereas genotype 23 was restricted to an isolated area, Kutai on
Kalimantan Island (15). Irian
Jaya, an area in the east part of Indonesia, had a different
pattern of genotype distribution from Java Island and other areas
in Indonesia (16). There are
currently no data available regarding hemodialysis patients.
A previous study of patients undergoing hemodialysis
in Yogyakarta, Indonesia, demonstrated high prevalence rates of
hepatitis B virus (HBV) and hepatitis C virus (HCV) infection,
which may have occurred via nosocomial infection (17). It is important to know whether
other blood-borne viruses circulate in patients undergoing
hemodialysis. Therefore, in the present study, the prevalence and
genotypic distribution of GBV-C and TTV were investigated in
patients undergoing hemodialysis. The possibility of nosocomial
infection was also assessed by molecular analysis.
Materials and methods
Patients undergoing hemodialysis
The present study enrolled 161 patients undergoing
hemodialysis, who were tested for HBV and HCV infection by blood
chemistry, serological and molecular examination in a previous
study (17). The patients
underwent hemodialysis at a unit in Yogyakarta, Indonesia, between
January and February 2010. The age ranged from 12–79 years (mean ±
standard deviation; 48±13 years). There were 93 male patients
(57.8%) and 68 female patients (42.2%). Almost all of the patients
(97.5%) were Javanese. Blood samples (5 ml) were collected prior to
starting hemodialysis. The blood samples were allowed to clot, and
then centrifuged at 1,500 × g for 10 min at room
temperature. The sera were separated and stored at −80°C for
further use. Sociodemographic factors, risk factors, alanine
aminotransferase (ALT) and γ-glutamyl transpeptidase (GGT)
concentrations, and markers of HBV and HCV infection were obtained
as described previously (17).
The present study was reviewed and approved by the
Ethics Committees at Kobe University (Kobe, Japan) and at Gadjah
Mada University (Yogyakarta, Indonesia). All subjects provided
written informed consent.
Detection of GBV-C RNA
RNA was extracted from 140 µl sera using an RNA
extraction kit (QIAamp Viral RNA Mini kit; Qiagen GmbH, Hilden,
Germany) according to the manufacturer's protocol. The RNA was then
converted into cDNA using a SuperScript III One-Step Reverse
Transcription Polymerase Chain Reaction system (Invitrogen; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) and outer reverse
primers in E1 and 5′-untranslated region (UTR). The cDNA was used
as a DNA template for analysis by polymerase chain reaction (PCR),
with primer pairs designed to amplify the 5′-UTR and E1 region of
the GBV-C genome.
The partial 5′-UTR of the GBV-C genome was amplified
using the following outer primer sequences: Forward
5′-GCCAAAAGGTGGTGGATGGG-3′, reverse 5′-CGGAGCTGGGTGGCCCCATGC-3′;
and the following inner primer sequences: Forward
5′-TGGTAGGTCGTAAATCCCGG-3′, reverse 5′-TGGTCCTTGTCAACTCGCCG-3′ in a
nested PCR to obtain an amplicon of 262 nucleotides (nt; nt
134–395). A portion of the E1 gene was amplified using the
following outer primer sequences: Forward
5′-ATCATGGCAGTCCTTCTGCT-3′, reverse 5′-TCARTCCATCTCCAAAACTC-3′; and
the following inner primer sequences: Forward
5′-GGGCAATATTTSCTCACAAA-3′, reverse 5′-CAAAACTCACTTTCCCACTT-3′ in a
nested PCR, to obtain an amplicon of 347 nt (nt 630–976). The nt
numbers refer to the PNF2126 isolate under accession no. U44402.
The first and second round PCRs were run under the same conditions
for 35 cycles, with each cycle consisting of 1 min at 94°C, 1 min
at 45°C, and 2 min at 72°C (6,18).
Detection of TTV DNA
TTV DNA was extracted from 200 µl sera using a DNA
extraction kit (QIAamp DNA Blood Mini kit; Qiagen Sciences, Inc.,
Gaithersburg, MD, USA) according to the manufacturer's protocol.
The 5′-UTR of the TTV genome was amplified by nested PCR using the
following outer primer sequences: Forward
5′-GTAAGTGCACTTCCGAATGGCTGAG-3′, reverse
5′-GAGCCTTGCCCATRGCCCGGCCAG-3′, where R = A or G; and the following
inner primer sequences: Forward 5′-CTGAGTTTTCCACGCCCGTCCGC-3′,
mixed with an equal amount of the primer with the underlined 4 nt
replaced by ATGC, and reverse 5′-CCCATRGCCCGGCCAGTCCCGAGC-3′. The
amplicon obtained in the first round of PCR was 162 nt long (nt
91–252) while the amplicon obtained in the second round was 134 nt
long (nt 111–244). PCR comprised of 35 cycles at 95°C for 30 sec
plus 9 min in the first cycle for the first round, and 25 cycles in
the second round at 72°C for 40 sec, plus 7 min in the last cycle
(19,20).
The open reading frame-1 (ORF1) region of the TTV
genome was amplified by semi-nested PCR. The first round of PCR was
comprised of 35 cycles (94°C for 30 sec; 60°C for 45 sec; 72°C for
45 sec, plus 7 min in the last cycle) using the following primer
sequences: Forward 5′-ACAGACAGAGGAGAAGGCAACATG-3′, reverse
5′-CTGGCATTTTACCATTTCCAAAGTT-3′. The second round of PCR was
comprised of 25 cycles under the same conditions as the first
round, with the following primer sequences: Forward
5′-GGCAACATGYTRTGGATAGACTGG-3′, where Y=T or C; R=A or G, and
reverse as above. The amplicon obtained in the first round of PCR
was 286 nt long (nt 1900–2185), while that obtained in the second
round was 271 nt long (nt 1915–2185). The nt positions are based on
the TA278 isolate under accession number AB017610 (20–22).
Sequencing and phylogenetic
analysis
The PCR products were directly sequenced using a
BigDye Terminator v3.1. Cycle Sequencing kit and an ABI PRISM
3100-Avant Genetic Analyzer (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The sequences were manually edited and aligned
using ClustalX software (version 2.0.12; http://www.clustal.org). The GBV-C genotypes were
determined by phylogenetic analysis of the partial 5′-UTR and E1
sequences, whereas the TTV genotypes were determined using the
partial ORF1 sequence. Published sequences were retrieved from
GenBank (https://www.ncbi.nlm.nih.gov/genbank; accessed on July
18, 2013) and were used as reference sequences. Phylogenetic trees
were constructed using the neighbor-joining method based on the
Kimura two-parameter distance estimation model (23). To validate the reliability of the
tree topologies, bootstrap reconstruction was performed 1,000
times, and bootstrap values of >70% were considered
statistically significant. Analyses were conducted using Molecular
Evolutionary Genetics Analysis software version 4.0.2 (http://megasoftware.net). Sequences were also compared
in order to identify identical sequences.
Nt sequence accession numbers
The GBV-C and TTV sequences described in the present
study were submitted to the DNA Data Bank of Japan under accession
numbers LC034595-LC034680 for the 5′-UTR sequences of GBV-C,
LC034681-LC034741 for the GBV-C E1 region sequences, and
LC034742-LC034809 for the TTV ORF1 sequences.
Statistical analysis
Statistical analyses were performed using
χ2-tests or Fisher's exact tests for categorical
variables, and independent Student's t-tests or Mann-Whitney
U tests for continuous variables using PASW Statistics, version
18.0.0 (SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to
indicate a statistically significant difference. Patients were
defined as GBV-C or TTV positive if they were RNA positive in at
least in one region (5′-UTR or E1 for GBV-C; 5′-UTR or ORF1 for
TTV).
Results
Prevalence of GBV-C
GBV-C RNA was detected in 92/161 patients (57.1%).
The 5′-UTR and E1 region sequences were both amplified in samples
from 55 patients (34.2%), the 5′-UTR sequence only was detected in
31 patients (19.3%), and the E1 region sequence only was detected
in 6 patients (3.7%). These results suggest that detection of GBV-C
RNA by amplification of the 5′-UTR region is more sensitive than
amplification of the E1 region.
GBV-C positivity was not associated with age, sex,
or any of the other risk factors analyzed in this study (Table I). A total of 27 patients (29.3%)
were co-infected with GBV-C and HBV, while 79 patients (85.9%) were
co-infected with GBV-C and HCV (Table
I). Most patients from both groups had normal ALT
concentrations (≤40 IU/l), with mean ALT concentrations of
20.6±20.1 and 29.2±39.2 IU/l observed in the GBV-C positive and
GBV-C-negative groups, respectively (Table I). GBV-C-positive patients tended
to have higher GGT concentrations than GBV-C-negative patients,
however the difference was not statistically significant, with mean
GGT concentrations of 150.3±165.0 and 135.4±144.0 IU/l recorded in
GBV-C-positive and GBV-C-negative patients, respectively (Table I). The observed differences in ALT
and GGT might be due to the high frequency of co-infection with
either HBV or HCV in both groups.
 | Table I.Characteristics and possible risk
factors of GBV-C and TTV infection. |
Table I.
Characteristics and possible risk
factors of GBV-C and TTV infection.
|
| GBV-C RNA |
| TTV DNA |
|
|---|
|
|
|
|
|
|
|---|
| Variable | Positive
(n=92) | Negative
(n=69) | P-value | ORF1-positive
(n=68) | ORF1-negative
(n=93) | P-value |
|---|
| Age, mean ±
standard deviation | 48.4±12.3 | 47.6±14.2 | 0.7 | 48.8±12.6 | 47.6±13.4 | 0.5 |
| Male/female
ratio | 50/42 | 43/26 | 0.3 | 43/25 | 50/43 | 0.2 |
| Hemodialysis
duration ≥1 year, n (%) | 56 (60.8) | 47 (68.1) | 0.3 | 43 (63.2) | 60 (64.5) | 0.9 |
| History of blood
transfusion, n (%) | 87 (94.6) | 66 (95.6) | 1.0 | 66 (97.1) | 87 (93.5) | 0.5 |
| Number of blood
transfusions, >5 times, n (%) | 37 (40.2) | 29 (42.0) | 0.8 | 32 (47.0) | 34 (36.5) | 0.2 |
| History of kidney
transplantation, n (%) | 0 (0) | 1 (1.4) | 0.3 | 1 (1.5) | 0 (0) | 0.4 |
| History of multiple
sexual partners, n (%) | 0 (0) | 2 (2.9) | 0.1 | 1 (1.5) | 1 (1.1) | 1.0 |
| History of
suffering sexually transmitted disease, n (%) | 0 (0) | 0 (0) | – | 0 (0) | 0 (0) | – |
| History of
injecting drug use, n (%) | 0 (0) | 1 (1.4) | 0.2 | 0 (0) | 1 (1.1) | 1.0 |
| Elevated ALT level,
n (%) | 10 (10.9) | 13 (18.8) | 0.1 | 10 (14.7) | 13 (14.0) | 0.9 |
| Elevated GGT level,
n (%) | 61 (66.3) | 41 (59.4) | 0.4 | 45 (66.2) | 57 (61.3) | 0.5 |
| HBV positive, n
(%) | 27 (29.3) | 12 (17.4) | 0.08 | 14 (20.6) | 25 (26.8) | 0.4 |
| HCV positive, n
(%) | 79 (85.9) | 55 (79.7) | 0.3 | 55 (80.9) | 79 (84.9) | 0.5 |
Genotypic distribution of GBV-C
To investigate the genotypic distribution of GBV-C
and the possibility of nosocomial infection phylogenetic analysis
of the 86-nt 5′-UTR sequence and the 61-nt E1 region sequences was
conducted. Phylogenetic trees were constructed for both sequences.
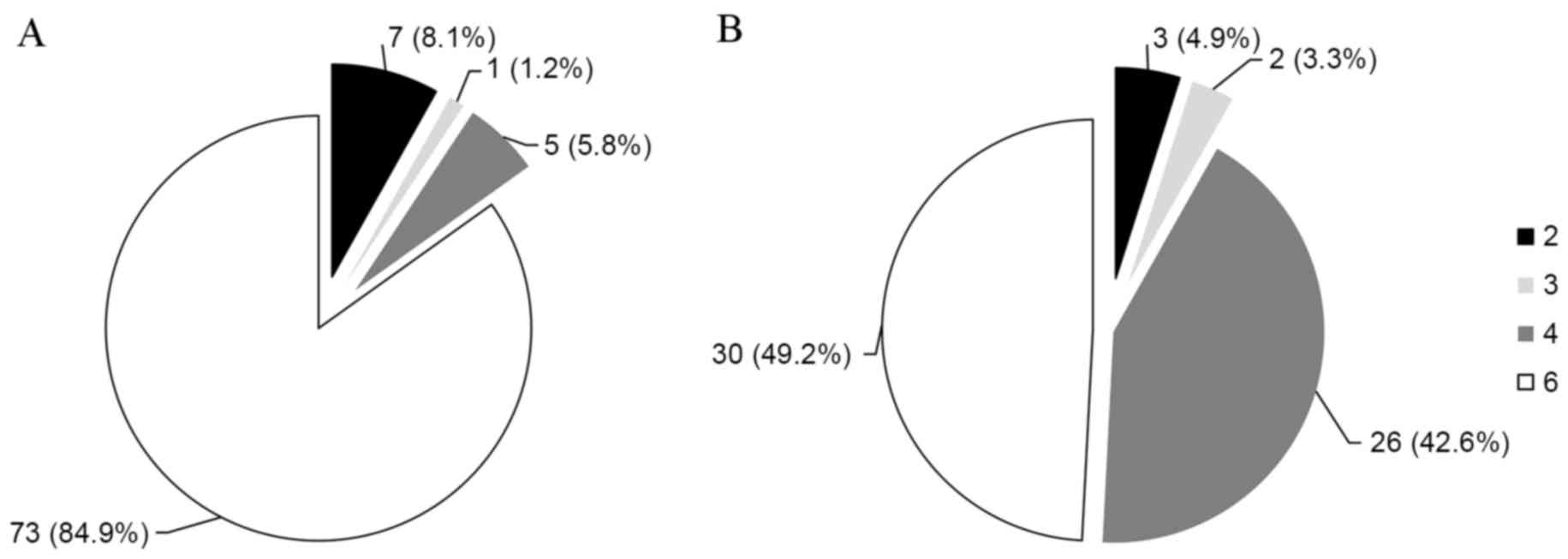
Analysis of the 5′-UTR revealed that genotype 6 was the most common
genotype (73 patients; 84.9%; Fig.
1A), followed by genotypes 2 (7 patients; 8.1%; Fig. 1A), 4 (5 patients; 5.8%; Fig. 1A), and 3 (1 patients; 1.2%;
Fig. 1A). Analysis of the E1
region revealed that genotype 6 was the most common genotype (30
patients; 49.2%; Fig. 1B),
followed by genotypes 4 (26 patients; 42.6%; Fig. 1B), 2 (3 patients; 4.9%; Fig. 1B), and 3 (2 patients; 3.3%;
Fig. 1B). The difference in
genotypic distribution was because 20 strains classified as
genotype 6 based on the 5′-UTR were reclassified as genotype 4 and
1 strain classified as genotype 6 was reclassified as genotype 3
based on the E1 region.
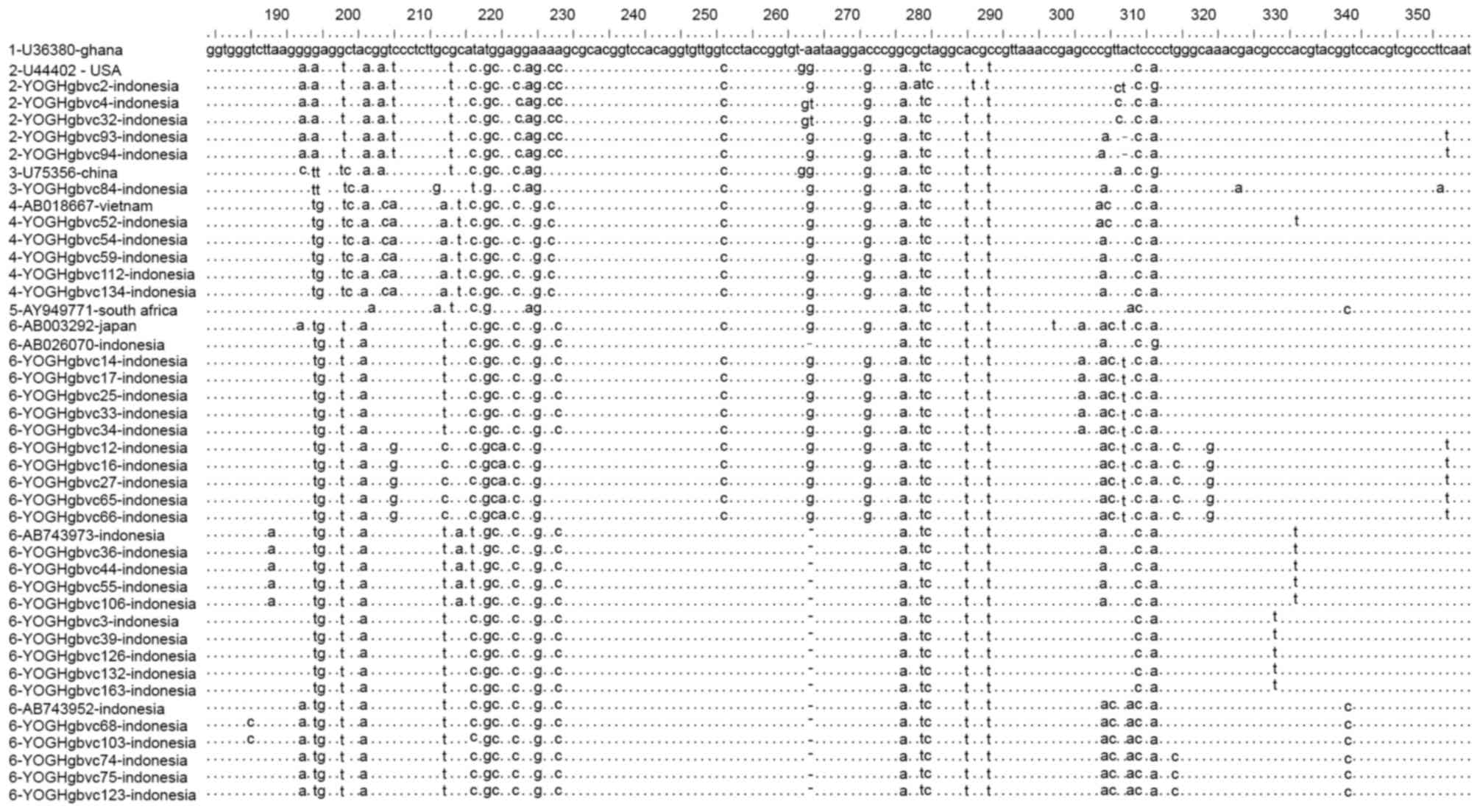
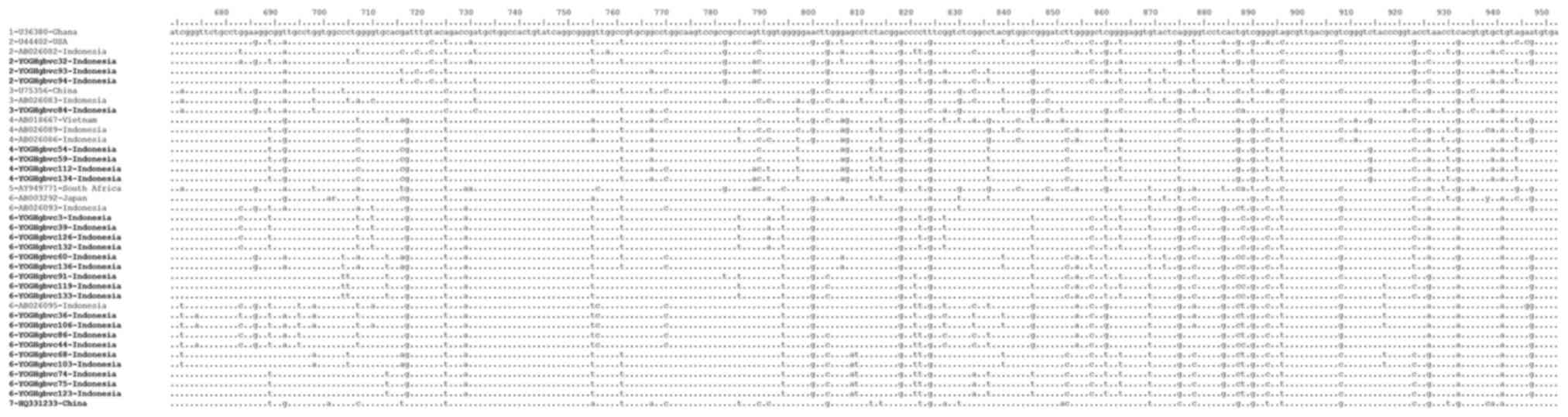
Alignment of representative 5′-UTR and E1 region
sequences demonstrated that these sequences were identical for some
of the isolates, with isolates from Indonesia containing some
unique substitutions and deletions which differed from those of
isolates from other countries (Figs.
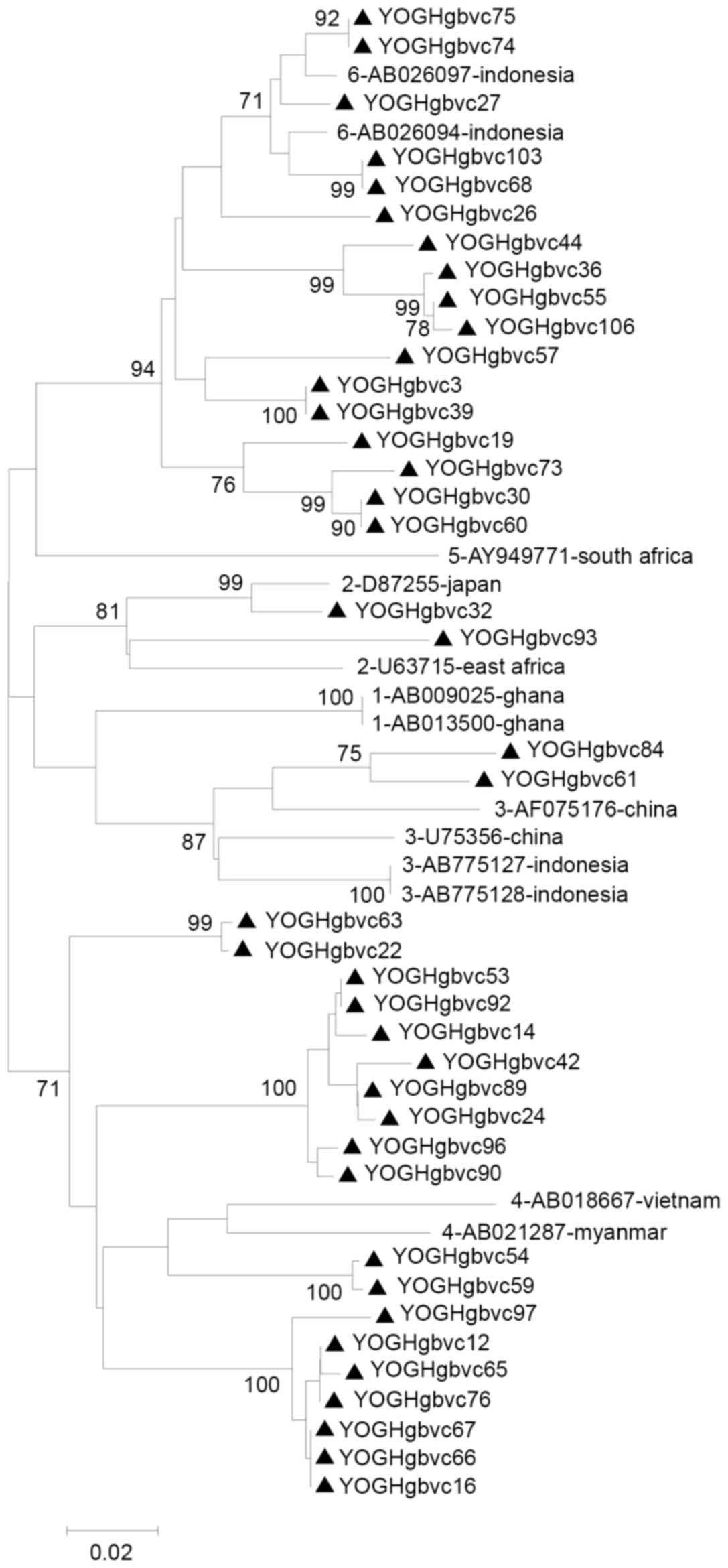
2 and 3). Similar and
identical strains were clustered together in the phylogenetic tree
with high bootstrap values (Figs.
4 and 5). Multiple strains
were completely identical, particularly in the 5′-UTR sequences
(YOGHgbvc3, 39, 126, 132 and 163; YOGHgbvc9, 14, 17, 25, 33, 35,
37, 42, 43, 45, 49, 61, 86, 92, 96, 97, 98, 114, 116, 127, 138,
144, 159 and 165; YOGHgbvc12, 16 and 65; YOGHgbvc27 and 66;
YOGHgbvc36, 44, 55 and 106; YOGHgbvc46 and 93; YOGHgbvc54, 59, 112
and 134; YOGHgbvc67 and 76; YOGHgbvc68 and 103; YOGHgbvc69 and 73;
YOGHgbvc74, 75 123; YOGH91 and 133; YOGHgbvc149 and 162; Fig. 4). Regarding the E1 region, there
were fewer identical sequences identified (YOGH3, 39, 126 and 132;
YOGHgbvc12 and 76; YOGH16, 66, 67 and 139; YOGHgbvc42 and 89;
YOGHgbvc53, 92 and 127; YOGH60 and 136; YOGHgbvc68 and 103;
YOGHgbvc74, 75 and 123; YOGHgbvc112 and 134; YOGHgbvc119 and 133;
Fig. 5). Overall, 13 isolates were
identical in terms of both the 5′-UTR and E1 region sequences
(YOGHgbvc3, 39, 126 and 132; YOGHgbvc68 and 103; YOGHgbvc74, 75 and
123; YOGHgbvc92 and 127; YOGH112 and 134; Figs. 4 and 5). A total of 8 of these isolates (61.5%)
were obtained from patients who had been on hemodialysis for ≥1
year.
Genetic diversity was greater for the E1 region
sequences than for the 5′-UTR sequences with an overall mean
distance of 0.14 and 0.06, respectively (data not shown). The
diversity observed in the E1 region probably occurred prior to the
start of hemodialysis in these patients.
Prevalence of TTV
TTV DNA was detected in all patients. The 5′-UTR
sequence was amplified in 160 (99.4%) samples, whereas the ORF1
sequence was amplified in just 68 samples (42.2%). This suggests
that detection of GBV-C by amplification of the 5′-UTR is more
sensitive than amplification of the ORF1 sequence.
As TTV infected all of the patients, the
associations between ORF1 positivity and demographics, liver enzyme
concentrations and possible risk factors were analyzed. ORF1
positivity was not associated with age, sex or any of the risk
factor analyzed in the current study (Table I). A total of 39 patients (24.2%)
were co-infected with TTV and HBV, while 134 patients (83.2%)
patients were co-infected with TTV and HCV (Table I). The majority of patients in the
two groups had normal ALT concentrations, with a mean ± standard
deviation of 27.2±33.0 and 22.2±27.5 IU/l recorded in ORF1-positive
and ORF1-negative patients, respectively (Table I). ORF1-positive patients tended to
have higher GGT concentrations than ORF1-negative patients with a
mean ± standard deviation of 138.4±143.8 and 147.9±165.1 IU/l,
respectively, although the difference was not statistically
significant (Table I). The
observed differences in ALT and GGT concentrations might be due to
the high frequency of co-infection with HBV and HCV.
Genotypic distribution of TTV
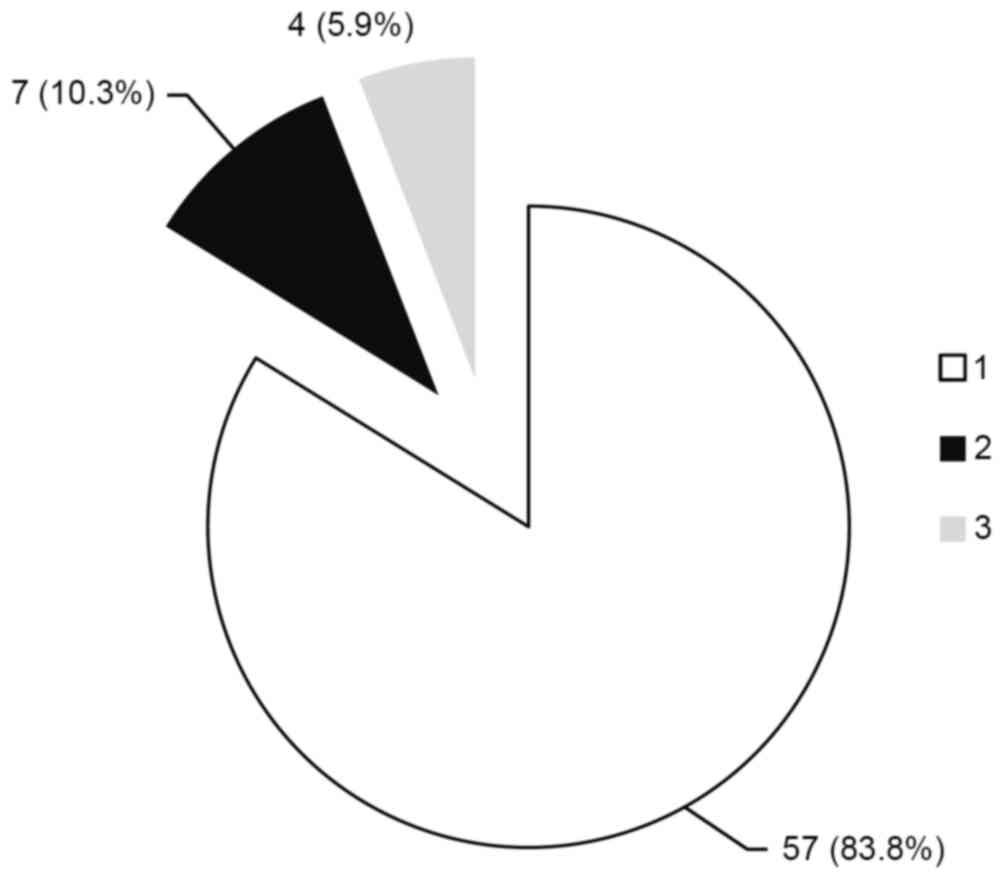
Phylogenetic analysis of the ORF1 region identified
that genotype 1 was dominant (57 patients; 83.8%; Fig. 6), followed by genotypes 2 (7
patients; 10.3%; Fig. 6) and 3 (4
patients; 5.9%; Fig. 6)
respectively. Alignment of representative ORF1 sequences revealed
marked variability in these sequences, and several conserved
regions belonging to TTV group 1 were identified among genotypes 1,
2, and 3 (Fig. 7). The overall
mean distance was 0.31. Certain unique nt substitutions and
deletions were detected in the Indonesian genotype 1 isolates,
which differentiated these isolates from Japanese genotype 1
isolates (TA278, AB017610) (Fig.
7). Strains with high similarity were clustered together with
significant bootstrap values. The phylogenetic tree demonstrated
clear differences among the genotypes, and the sequences YOGHttv1
and YOGHttv63 were identical (Fig.
8).
Discussion
GBV-C, previously known as hepatitis G virus, was
discovered in 1995 and is an enveloped single-stranded RNA
positive-sense virus (9.4 kb) belonging to the Flaviviridae
family. Following its discovery, multiple researchers have
attempted to determine the properties of this virus and its
association with diseases including hepatitis (24) and non-Hodgkin's lymphoma (25). However, no convincing evidence
supporting an association between GBV-C infection and any disease
exists. In fact, some previous studies have demonstrated beneficial
effects of GBV-C in patients infected with HIV or HCV. GBV-C
co-infection was associated with an improved prognosis and reduced
mortality among HIV-infected patients. GBV-C RNA positivity was
also associated with liver function improvement among HCV infected
patients (26–30).
GBV-C is predominantly transmitted via the
parenteral route. Thus, for epidemiological reasons, GBV-C is of
particular interest in patients undergoing hemodialysis who are at
risk of parenterally transmitted infection. Some previous studies
have used GBV-C as a tool to detect patient-to-patient transmission
in hemodialytic settings (31,32).
The prevalence of GBV-C infection is greater among
patients undergoing hemodialysis compared with blood donors or
healthy individuals in the same region. In several countries, the
reported prevalence of GBV-C markers ranged from 0.2–24.6% in blood
donors (33–35) and ranged from 3.9–26.5% in patients
undergoing hemodialysis (1–5,36).
In the present study, the overall prevalence of GBV-C infection was
57.1%, which is similar to the prevalence of 55% reported in
Yogyakarta in 1996 (7). This
suggests that the prevalence of GBV-C has not changed over the last
decade. However, the prevalence was greater than that observed in
Surabaya, Indonesia (6). The
prevalence of GBV-C infection was greater in Indonesian patients
undergoing hemodialysis compared with blood donors, possibly due to
the high prevalence in the general population, the lack of
screening of GBV-C in blood banks, or patient-to-patient
transmission.
Owing to its parenteral route of transmission, blood
transfusions are hypothesized to be the main risk factor for GBV-C
infection (37). Therefore,
patients undergoing hemodialysis who commonly require blood
transfusions are at increased risk of GBV-C infection. However, the
results of previous studies that investigated the association
between blood transfusion and GBV-C infection in patients
undergoing hemodialysis are inconsistent. Certain studies have
demonstrated that a history of blood transfusion or a history of
multiple blood transfusions are the main risk factors for GBV-C
infection (38), while other
studies reported negative associations (1,35,39,40).
The present study observed no association between the history of
blood transfusion and GBV-C infection, possibly as a consequence of
the limited sample size and the prevalence of other blood-borne
infections, including HBV and HCV. Substantial proportions of
GBV-C-negative patients were co-infected with HBV (17.4%) or HCV
(79.7%), and this high prevalence of co-infection might mask
clinically relevant associations. Further studies regarding
patients infected with GBV-C only are required to address this
issue.
Previous studies have demonstrated that a longer
duration of hemodialysis is a risk factor for GBV-C infection
(4), supporting the involvement of
patient-to-patient transmission in the high prevalence of GBV-C
infection within a hemodialysis unit. However, no association
between the duration of hemodialysis treatment and GBV-C infection
was observed in the present study, similar to earlier studies
(3,5,38).
This may be due to the high prevalence of co-infection with other
viruses, particularly HCV. HCV co-infection was common in
GBV-C-positive and GBV-C-negative patients. The history of GBV-C
infection was not assessed using E2 antibodies in the present
study; which only measured active GBV-C infection based on viral
RNA amplification. Accordingly, the present study potentially
underestimated the prevalence of GBV-C. The length of hemodialysis
treatment was associated with HCV infection in a previous study
regarding the same hemodialysis unit as the present study (17). The other risk factors analyzed in
the present study were not correlated with GBV-C infection.
GBV-C and HCV have similar genomic structures and
share the same mode of transmission. The prevalence of GBV-C is
high in HCV-infected patients (41,42).
This high prevalence of co-infection was demonstrated in the
present study and in several earlier studies of patients undergoing
hemodialysis (4,5) due to the shared mode of transmission.
However, it is unclear whether these viruses are transmitted
simultaneously or separately. By comparing the sequence alignment
and phylogenetic analysis with the HCV sequences from a previous
study (8), it was demonstrated
that GBV-C and HCV generally infect patients at different times.
Only one pair of strains isolated from two patients (no. 112 and
134) were identical in terms of the GBV-C 5′-UTR/E1 region and HCV
NS5B sequences, suggesting simultaneous transmission of GBV-C and
HCV from one patient to another patient (data not shown). These
results indicate that GBV-C and HCV are transmitted
independently.
GBV-C and HCV co-infection is not correlated with
severity of hepatic disease progression (30,43),
as demonstrated by the low or normal liver enzyme concentrations.
The present study also demonstrated that GBV-C was not associated
with hepatic pathogenic effects, because GBV-C viremia was not
associated with ALT or GGT. In fact, previous studies have
demonstrated a beneficial effect of GBV-C infection in HCV-infected
patients (30,41).
GBV-C is distributed globally, and 7 genotypes have
been identified to date (8). The
genotypes are widespread, with distinct geographical distributions.
Genotype 1 was first described in Africa, and the other genotypes
were discovered in Europe (genotype 2), Japan (genotype 3),
Southeast Asia (genotype 4), South Africa (genotype 5), Indonesia
(genotype 6), and China (genotype 7) (8,44–49).
Genotype 4 was reported to be dominant among Indonesian blood
donors (55.5%), but genotype 6 was dominant in patients with
chronic liver disease (60%) and patients undergoing hemodialysis
(83.3%) (6). Thus, genotype 6
appears to be more common among hemodialysis patients compared with
other populations. The present study provided data demonstrating
that genotype 6 is the most common genotype among patients
undergoing hemodialysis. The predominance of genotype 6 may reflect
an outbreak of GBV-C infection from a common source. The present
study also suggests the involvement of patient-to-patient
transmission because several isolates were identical, including
some displaying identical 5′-UTR and E1 region sequences. It is
feasible that genotype 6 has adapted to be easily transmitted among
patients with impaired immunity, including patients undergoing
hemodialysis.
Genotype 6 was the predominant genotype observed,
based on phylogenetic analyses of the 5′-UTR and E1 region.
However, the results were inconsistent, owing to the different
proportions of each genotype classified by the 5′UTR or E1 region
sequences (Fig. 1). A total of 20
isolates (23.3%) classified as genotype 6 based on phylogenetic
analysis of the 5′-UTR were reclassified as genotype 4 based on
phylogenetic analysis of the E1 region. In addition, 1 isolate
classified into genotype 6 based on the 5′-UTR analysis was
reclassified into genotype 3 based on the E1 region. However,
analyses of the 5′-UTR and E1 region were consistent for genotype
2. There are some possible explanations for these results. For
example, the 5′-UTR is more conserved than the E1 region, so
analyses based on the E1 region show lower discrimination than
analyses based on the 5′-UTR. The possibility of co-infection with
≥2 GBV-C genotypes might also be increased in patients undergoing
hemodialysis due to frequent contact with contaminated blood or
blood products. In addition, based on full genome analysis, Feng
et al (8) proposed that
GBV-C genotypes could be classified into 5 groups by combining
genotypes 4, 6, and 7 into one group, due to their genetic
similarities. Thus, genotypes 4 and 6 might represent a single
genotype. Genotype 2 was previously revealed to be the most common
GBV-C genotype in HIV-infected patients in Yogyakarta, Indonesia
(9). This suggests that the
difference in genotype distribution in this population is
predominantly associated with transmission via drug injection.
TTV is a human non-enveloped single-stranded
circular DNA virus, first described in 1997 by Nishizawa et
al (50), and is a member of
the Anelloviridae family. TTV infection was previously
reported to be associated with a number of diseases, including
hepatitis, based on epidemiological data (24,25,50,51).
However, there is no strong evidence linking TTV infection to any
specific disease. This virus is globally distributed and the
prevalence of TTV infection is high in various populations,
including patients with liver diseases, patients with HIV, drug
users and healthy individuals (51–54).
In the present study, the prevalence of TTV infection among
patients undergoing hemodialysis, a high-risk population, was
determined.
The overall prevalence of TTV infection, based on
the 5′-UTR and ORF1 sequences, was 100%. A previous nationwide
study of TTV infection in Indonesian healthy individuals revealed a
prevalence of 95% (15). Thus, the
prevalence of TTV infection among patients undergoing hemodialysis
patients in the present study was marginally greater than that
observed in the general population. However, on Java Island, where
Yogyakarta is located, TTV was detected based on the 5′-UTR in 100%
of healthy individuals (15). TTV
was detected by amplification of the ORF1 in 68 (42.2%) patients,
which was similar to a previous study where the prevalence was
reported to be 42% in healthy individuals (15). This suggests that the prevalence of
TTV is similarly high among hemodialysis patients and healthy
individuals. As with GBV-C, the high prevalence of TTV infection
was not associated with any of the risk factors analyzed in the
present study. Furthermore, TTV was not associated with hepatic
injury.
The genotypic distribution of TTV in the present
study was similar to that in healthy individuals. In healthy
individuals, genotype 1 was predominant (53%), followed by
genotypes 3 (28%) and 2 (18%). These genotypes belong to TTV group
1. TTV group 2 was detected in <2% of subjects (15). In the present study, genotype 1 was
predominant (83.8%), followed by genotypes 2 (10.3%) and 3 (5.9%).
These data suggest that the increase in the prevalence of genotype
1 may be due to patient-to-patient transmission. However, it is
also possible that the patients were infected with TTV prior to
starting hemodialysis, owing to the very high prevalence of TTV in
the general population. This hypothesis is supported by a study of
sex workers in Papua, Indonesia, which identified that the
genotypic distribution of TTV reflected the birth place of the
subjects rather than their work environment, and that the infection
was more likely to occur during the early period of life rather
than via sexual transmission (16). There was substantial genetic
diversity of TTV in the present study and only one pair of
sequences was identical, suggesting nosocomial infection was
unlikely to be responsible for the high prevalence of TTV among
patients undergoing hemodialysis.
Although the prevalence rates of GBV-C and TTV
infection were high in the present study, screening for GBV-C and
TTV infection is not mandatory. GBV-C and TTV do not appear to have
any pathogenic properties and do not appear to cause liver disease
or other clinical disorders. Furthermore, GBV-C may have beneficial
effects in patients co-infected with HCV or HIV. Screening programs
are mandatory for other blood-borne viruses, particularly HBV, HCV
and HIV. It has previously been reported that the high prevalence
of HBV and HCV infection in patients undergoing hemodialysis were
associated with nosocomial infection, owing to the failure of
hemodialysis units to adhere to strict infection-control procedures
(17). Strict adherence to
infection-control procedures prevents cross-infection of
blood-borne viruses between patients (32).
In conclusion, prevalence rates of GBV-C and TTV
infection are high among hemodialysis patients in Yogyakarta,
Indonesia. Nosocomial transmission may be involved in infection due
to inconsistent implementation of infection-control procedures
within hemodialysis units. Hemodialysis units in Indonesia should
implement strict infection-control procedures designed to prevent
the transmission of blood-borne viruses.
Acknowledgements
The authors would like to thank Dr Widya
Wasityastuti and Dr Laura Navika Yamani for their valuable
assistance with the laboratory work. This work was partly supported
by Grant-in-Aid for Scientific Research (B) (grant no.
16H05826).
References
|
1
|
Eslamifar A, Hamkar R, Ramezani A, Ahmadi
F, Gachkar L, Jalilvand S, Adibi L, Atabak S, Khameneh A, Ghadimi R
and Aghakhani A: Hepatitis G virus exposure in dialysis patients.
Int Urol Nephrol. 39:1257–1263. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hammad AM and Zaghloul MH: Hepatitis G
virus infection in Egyptian children with chronic renal failure
(single centre study). Ann Clin Microbiol Antimicrob. 8:362009.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hosseini-Moghaddam SM, Keyvani H, Samadi
M, Alavian SM, Mahdavimazdeh M, Daneshvar S and Razzaghi Z: GB
virus type C infection in hemodialysis patients considering
co-infection with hepatitis C virus. J Med Virol. 80:1260–1263.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ozdarendeli A, Toroman ZA, Kalkan A, Kilic
SS, Ozden M and Doymaz MZ: Prevalence and genotypes of hepatitis G
virus among hemodialysis patients in Eastern Anatolia, Turkey. Med
Princ Pract. 14:102–106. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Filho Ramos R, Carneiro MA, Teles SA, Dias
MA, Cardoso DD, Lampe E, Yoshida CF and Martins RM: GB virus
C/hepatitis G virus infection in dialysis patients and kidney
transplant recipients in Central Brazil. Mem Inst Oswaldo Cruz.
99:639–643. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Handajani R Soetjipto, Lusida MI,
Suryohudoyo P, Adi P, Setiawan PB, Nidom CA, Soemarto R, Katayama
Y, Fujii M and Hotta H: Prevalence of GB virus C/Hepatitis G virus
infection among various populations in Surabaya, Indonesia, and
identification of novel groups of sequence variants. J Clin
Microbiol. 38:662–668. 2000.PubMed/NCBI
|
|
7
|
Tsuda F, Hadiwandowo S, Sawada N, Fukuda
M, Tanaka T, Okamoto H, Miyakawa Y and Mayumi M: Infection with GB
virus C (GBV-C) in patients with chronic liver disease or on
maintenance hemodialysis in Indonesia. J Med Virol. 49:248–252.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Feng Y, Zhao W, Feng Y, Dai J, Li Z, Zhang
X, Liu L, Bai J, Zhang H, Lu L and Xia X: A novel genotype of GB
virus C: Its identification and predominance among injecting drug
users in Yunnan, China. PLoS One. 6:e211512011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Anggorowati N, Yano Y, Subronto YW, Utsumi
T, Heriyanto DS, Mulya DP, Rinonce HT, Widasari DI, Lusida MI
Soetjipto and Hayashi Y: GB virus C infection in Indonesian
HIV-positive patients. Microbiol Immunol. 57:298–308. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Afkari R, Pirouzi A, Mohsenzadeh M, Azadi
M and Jafari M: Molecular detection of TT virus and SEN virus
infections in hemodialysed patients and blood donors in south of
Iran. Indian J Pathol Microbiol. 55:478–480. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Irshad M, Mandal K, Singh S and Agarwal
SK: Torque teno virus infection in hemodialysis patients in North
India. Int Urol Nephrol. 42:1077–1083. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Massau A, Martins C, Nachtigal GC, Araújo
AB, Rossetti ML, Niel C and Da Silva CM: The high prevalence of
Torque teno virus DNA in blood donors and haemodialysis patients in
southern Brazil. Mem Inst Oswaldo Cruz. 107:684–686. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rivanera D, Lozzi MA, Idili C and Lilli D:
Prevalence of TT virus infection in Italian dialysis patients.
Pathol Biol (Paris). 57:97–100. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jahromi AS, Erfanian S, Farjam MR,
Moghaddam M and Madani A: Molecular epidemiology and clinical
importance of TT virus infection in haemodialysis patients, South
of Iran. Life Sci J. 11:182–185. 2014.
|
|
15
|
Muljono DH, Nishizawa T, Tsuda F,
Takahashi M and Okamoto H: Molecular epidemiology of TT virus (TTV)
and characterization of two novel TTV genotypes in Indonesia. Arch
Virol. 146:1249–1266. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mulyanto Hijikata M, Matsushita M,
Ingkokusmo G, Widjaya A, Sumarsidi D, Kanai K, Ohta Y and Mishiro
S: TT virus (TTV) genotypes in native and non-native prostitutes of
Irian Jaya, Indonesia: Implication for non-occupational
transmission. Arch Virol. 145:63–72. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rinonce HT, Yano Y, Utsumi T, Heriyanto
DS, Anggorowati N, Widasari DI, Lusida MI, Soetjipto Prasanto H,
Hotta H and Hayashi Y: Hepatitis B and C virus infection among
hemodialysis patients in Yogyakarta, Indonesia: Prevalence and
molecular evidence for nosocomial transmission. J Med Virol.
85:1348–1361. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Muerhoff AS, Simons JN, Erker JC, Desai SM
and Mushahwar IK: Identification of conserved nucleotide sequences
within the GB virus C 5′-untranslated region: Design of PCR primers
for detection of viral RNA. J Virol Methods. 62:55–62. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Okamoto H, Takahashi M, Kato N, Fukuda M,
Tawara A, Fukuda S, Tanaka T, Miyakawa Y and Mayumi M:
Sequestration of TT virus of restricted genotypes in peripheral
blood mononuclear cells. J Virol. 74:10236–10239. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Okamoto H, Takahashi M, Nishizawa T, Ukita
M, Fukuda M, Tsuda F, Miyakawa Y and Mayumi M: Marked genomic
heterogeneity and frequent mixed infection of TT virus demonstrated
by PCR with primers from coding and noncoding regions. Virology.
259:428–436. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Okamoto H, Akahane Y, Ukita M, Fukuda M,
Tsuda F, Miyakawa Y and Mayumi M: Fecal excretion of a nonenveloped
DNA virus (TTV) associated with posttransfusion non-A-G hepatitis.
J Med Virol. 56:128–132. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Okamoto H, Nishizawa T, Kato N, Ukita M,
Ikeda H, Iizuka H, Miyakawa Y and Mayumi M: Molecular cloning and
characterization of a novel DNA virus (TTV) associated with
posttransfusion hepatitis of unknown etiology. Hepatol Res.
10:1–16. 1998. View Article : Google Scholar
|
|
23
|
Kimura M: A simple method for estimating
evolutionary rate of base substitutions through comparative studies
of nucleotide sequences. J Mol Evol. 16:111–120. 1980. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Reshetnyak VI, Karlovich TI and Ilchenko
LU: Hepatitis G virus. World J Gastroenterol. 14:4725–4734. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chang CM, Stapleton JT, Klinzman D,
Mclinden JH, Purdue MP, Katki HA and Engels EA: GBV-C infection and
risk of NHL among U.S. adults. Cancer Res. 74:5553–5560. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bhattarai N and Stapleton JT: GB virus C:
The good boy virus? Trends Microbiol. 20:124–130. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Giret MT and Kallas EG: GBV-C: State of
the art and future prospects. Curr HIV/AIDS Rep. 9:26–33. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sahni H, Kirkwood K, Kyriakides TC,
Stapleton J, Brown ST and Holodniy M: OPTIMA Study Team: GBV-C
viremia and clinical events in advanced HIV infection. J Med Virol.
86:426–432. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ernst D, Greer M, Akmatova R, Pischke S,
Wedemeyer H, Heiken H, Tillmann HL, Schmidt RE and Stoll M: Impact
of GB virus C viraemia on clinical outcome in HIV-1-infected
patients: A 20-year follow-up study. HIV Med. 15:245–250. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Feng Y, Liu L, Feng YM, Zhao W, Li Z,
Zhang AM, Song Y and Xia X: GB virus C infection in patients with
HIV/hepatitis C virus coinfection: Improvement of the liver
function in chronic hepatitis C. Hepat Mon. 14:e141692014.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kao JH, Huang CH, Chen W, Tsai TJ, Lee SH,
Hung KY and Chen DS: GB virus C infection in hemodialysis patients:
Molecular evidence for nosocomial transmission. J Infect Dis.
180:191–194. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ross RS, Viazov S, Clauberg R, Wolters B,
Fengler I, Eveld K, Scheidhauer R, Husing J, Philipp T, Kribben A
and Roggendorf M: Lack of de novo hepatitis C virus infections and
absence of nosocomial transmissions of GB virus C in a large cohort
of German haemodialysis patients. J Viral Hepat. 16:230–238. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Xiao W, Lin F, Sun P, Ma L and Li C:
Detection of GB virus C/hepatitis G markers in Chinese voluntary
blood donors. Braz J Infect Dis. 18:352–353. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Alhetheel A and El-Hazmi MM: Hepatitis G
virus in Saudi blood donors and chronic hepatitis B and C patients.
J Infect Dev Ctries. 8:110–115. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Odeh RA, Yasin S, Nasrallah G and Babi Y:
Rates of infection and phylogenetic analysis of GB virus-C among
Kuwaiti and Jordanian blood donors. Intervirology. 53:402–407.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kelishadi M, Mojerloo M, Moradi A, Bazouri
M, Hashemi P, Samadi S, Saeedi A and Tabarraei A: GB virus C
viremia and anti-E2 antibody response among hemodialysis patients
in Gorgan, Iran. Jundishapur J Microbiol. 7:e131222014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Alter HJ, Nakatsuji Y, Melpolder J, Wages
J, Wesley R, Shih JW and Kim JP: The incidence of
transfusion-associated hepatitis G virus infection and its relation
to liver disease. N Engl J Med. 336:747–754. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hinrichsen H, Leimenstoll G, Stegen G,
Schrader H, Fölsch UR and Schmidt WE: Prevalence of and risk
factors for hepatitis G (HGV) infection in haemodialysis patients:
A multicentre study. Nephrol Dial Transplant. 17:271–275. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Fabrizi F, De Vecchi AF, Lunghi G, Finazzi
S, Bisegna S and Ponticelli C: Epidemiology of GB virus c/hepatitis
g virus infection in patients on peritoneal dialysis. Perit Dial
Int. 22:405–410. 2002.PubMed/NCBI
|
|
40
|
Huang JJ, Lee WC, Ruaan MK, Wang MC, Chang
TT and Young KC: Incidence, transmission, and clinical significance
of hepatitis G virus infection in hemodialysis patients. Eur J Clin
Microbiol Infect Dis. 20:374–379. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Berzsenyi MD, Bowden DS and Roberts SK: GB
virus C: Insights into co-infection. J Clin Virol. 33:257–266.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ghanbari R, Ravanshad M, Hosseini SY,
Yaghobi R and Shahzamani K: Genotyping and infection rate of GBV-C
among iranian HCV-infected patients. Hepat Mon. 10:80–87.
2010.PubMed/NCBI
|
|
43
|
Januszkiewicz-Lewandowska D, Wysocki J,
Rembowska J, Lewandowski K, Nowak T, Pernak M and Nowak J:
Hepatitis G virus co-infection may affect the elimination of
hepatitis C virus RNA from the peripheral blood of hemodialysis
patients. Acta Virol. 45:261–263. 2001.PubMed/NCBI
|
|
44
|
Muerhoff AS, Dawson GJ and Desai SM: A
previously unrecognized sixth genotype of GB virus C revealed by
analysis of 5′-untranslated region sequences. J Med Virol.
78:105–111. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Muerhoff AS, Leary TP, Sathar MA, Dawson
GJ and Desai SM: African origin of GB virus C determined by
phylogenetic analysis of a complete genotype 5 genome from South
Africa. J Gen Virol. 86:1729–1735. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Naito H, Win KM and Abe K: Identification
of a novel genotype of hepatitis G virus in Southeast Asia. J Clin
Microbiol. 37:1217–1220. 1999.PubMed/NCBI
|
|
47
|
Sathar MA, Soni PN, Pegoraro R, Simmonds
P, Smith DB, Dhillon AP and Dusheiko GM: A new variant of GB virus
C/hepatitis G virus (GBV-C/HGV) from South Africa. Virus Res.
64:151–160. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Smith DB, Basaras M, Frost S, Haydon D,
Cuceanu N, Prescott L, Kamenka C, Millband D, Sathar MA and
Simmonds P: Phylogenetic analysis of GBV-C/hepatitis G virus. J Gen
Virol. 81:769–780. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Tucker TJ, Smuts H, Eickhaus P, Robson SC
and Kirsch RE: Molecular characterization of the 5′ non-coding
region of South African GBV-C/HGV isolates: Major deletion and
evidence for a fourth genotype. J Med Virol. 59:52–59. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Nishizawa T, Okamoto H, Konishi K,
Yoshizawa H, Miyakawa Y and Mayumi M: A novel DNA virus (TTV)
associated with elevated transaminase levels in posttransfusion
hepatitis of unknown etiology. Biochem Biophys Res Commun.
241:92–97. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Asim M, Singla R, Gupta RK and Kar P:
Clinical & molecular characterization of human TT virus in
different liver diseases. Indian J Med Res. 131:545–554.
2010.PubMed/NCBI
|
|
52
|
Alzahrani AJ, Dela Cruz DM, Obeid OE,
Bukhari HA, Al-Qahtani AA and Al-Ahdal MN: Molecular detection of
hepatitis B, hepatitis C, and torque teno viruses in drug users in
Saudi Arabia. J Med Virol. 81:1343–1347. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Devalle S and Niel C: Distribution of TT
virus genomic groups 1–5 in Brazilian blood donors, HBV carriers,
and HIV-1-infected patients. J Med Virol. 72:166–173. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Vasilyev EV, Trofimov DY, Tonevitsky AG,
Ilinsky VV, Korostin DO and Rebrikov DV: Torque teno virus (TTV)
distribution in healthy Russian population. Virol J. 6:1342009.
View Article : Google Scholar : PubMed/NCBI
|