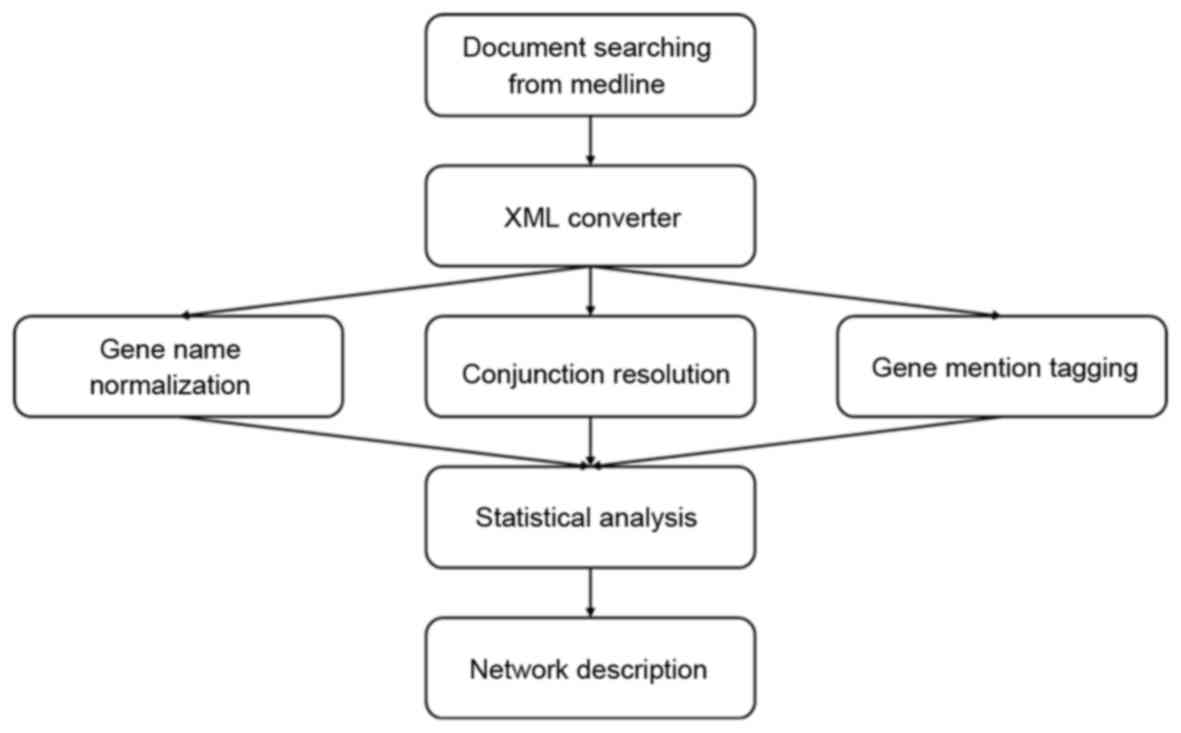
Introduction
Glucose-6-phosphate dehydrogenase (G6PD) is a
critical enzyme in the pentose phosphate pathway (PPP). G6PD serves
an important role in nucleic acid synthesis via generation of
ribose 5-phosphate, and in the cellular response to oxidative
stress through the generation of reduced nicotinamide
adenine-dinucleotide phosphate (NADPH) (1,2).
G6PD deficiency was the first prevalent enzyme deficiency
characterized by the World Health Organization in 1984 (3). It is an X-linked, hereditary genetic
defect involving mutations in the G6PD gene, which lead to specific
amino acid substitutions that alter enzymatic function (4). Globally, >400 million people
suffer from G6PD deficiency (5).
The most frequent clinical manifestations of G6PD deficiency are
neonatal jaundice and acute hemolytic anemia, which are initiated
by exogenous oxidative agents, including drugs, infection or
ingestion of fava beans (4,6).
As G6PD activity is essential for preventing
reactive oxygen species (ROS)/serum starvation-mediated cell death,
dysregulation of G6PD activity may lead to the development of
disease (7). G6PD deficiency
predisposes human foreskin fibroblasts to retarded growth and
accelerated cellular senescence (8). Knocking out G6PD is embryonically
lethal, and leads to cellular sensitivity to
H2O2 and additional oxidative agents
(9,10). By contrast, overexpression of G6PD
in NIH 3T3 cells leads to anchorage-independent growth (11). Decreased G6PD activity may
predispose humans to type 1 and type 2 diabetes (12,13).
In addition, G6PD has been associated with hypertension (14).
Increased G6PD activity has been reported in various
neoplasms, including hepatocellular cancer, leukemia, colorectal
cancer, breast tumors, renal cell carcinomas, and gastrointestinal
cancers (15–20). A previous study demonstrated that
silencing of G6PD in human A375 malignant melanoma cells
downregulated cyclin D1, cyclin E, p53, S100 calcium-binding
protein A4 and the apoptosis factor Fas, and upregulated the
apoptosis-inhibiting factors B-cell lymphoma (Bcl) 2 and Bcl-extra
large (21). G6PD regulates
apoptosis and the proliferation of A375 cells, potentially via the
signal transducer and activator of transcription (STAT) 3/5
signaling pathway (22).
MicroRNAs (miRNAs) are a class of single-stranded
molecules, 20 to 22 nucleotides in length, which constitute
approximately 0.01% of total RNA. By binding to specific sites
within the 3′-untranslated region, miRNAs inhibit the translation
of multiple mRNA targets (22).
miRNAs have been reported to serve critical roles in embryonic
development, cell proliferation, the cell cycle, apoptosis,
neovascularization, inflammation, tumorigenesis and various
additional diseases (23–33). Therefore, determining whether any
miRNAs regulate G6PD may provide an insight into G6PD-associated
biological processes and diseases.
miRNAs function to promote, as well as suppress
tumor development. For instance, overexpression of the oncogenic
miRNA-155 in transgenic mice leads to B cell proliferation, which
develops into B cell leukemia and aggressive lymphoma (23). By contrast, tumor suppressive
miRNAs prevent tumors from developing by negatively regulating
oncogenes and/or genes controlling proliferation or apoptosis. The
expression of let-7, which was one of the first miRNAs
discovered, is downregulated in lung cancer (34). Overexpression of let-7 is
capable of inhibiting cancer cell growth in vitro and in
vivo, while downregulation of let-7 impairs the survival
of various types of tumor (25,26,35).
In addition, the expression levels of several miRNAs have been
reported to correlate with tumor metastasis. For instance, the
expression level miRNA-422a is negatively correlated with
metastasis of hepatocellular carcinoma (HCC), and miRNA-422a is
significantly downregulated in HCC (33). Overexpression of miR-422a in HCC
tumor cells significantly inhibits tumor growth and liver
metastasis in xenograft tumor models (33).
In addition to cancer, miRNAs are involved in
various additional biological processes and diseases. An miRNA
cluster at chromosome 14q32 participates in neovascularization, and
inhibition of 14q32 miRNAs, including miR-329, miR-487b, miR-494
and miR-495 promotes neovascularization and recovery of blood flow
following ischemia (30,33). In addition, a number of miRNAs
influence energy metabolism. By repressing cardiac peroxisome
proliferator-activated receptor-Δ, miR-199a facilitates a metabolic
shift from a predominant reliance on fatty acid utilization toward
an increased reliance on glucose metabolism, which leads to lethal
heart failure (36). Inhibition of
miRNA binding improved cardiac function and restored mitochondrial
fatty acid oxidation in animal models, thus highlighting a novel
potential therapy for heart failure (36).
To the best of the author's knowledge no systematic
studies investigating the correlation between G6PD, G6PD-associated
genes, and miRNAs regulating G6PD have been conducted to date.
Therefore, 2,253 articles retrieved from the National Center for
Biotechnology Information (NCBI) PubMed library were used to
investigate the networks of G6PD-associated genes in the present
study. A total of 98 genes and 7 signaling pathways were
demonstrated to be significantly correlated with G6PD. In addition,
33 miRNAs that may modulate the expression or activity of G6PD were
identified.
Materials and methods
Data selection, extraction and
filtering for the natural language processing (NLP) analysis of
human G6PD
The PubMed library was first searched for articles
regarding G6PD deficiency that were published between January 1980
and June 2014, using the following keywords: ‘Glucosephosphate
Dehydrogenase Deficiency’[Mesh] AND ‘1980/01/01’[Date-Publication]:
‘2014/06/01’[Date-Publication]. All abstracts were downloaded as.
HTML text without images and converted into. XML documents. The
genes and proteins reported were compiled into a data pool, and
gene mention recognition was performed using A Biomedical Named
Entity Recognizer version 1.5 tool (http://pages.cs.wisc.edu/~bsettles/abner/) (37), and conjunction resolution was
conducted to obtain specialized descriptions of the extracted
genes. For instance, ‘CASP 3/7 gene’ was divided into the CASP 3
genes and CASP 7 genes. In order to normalize the variety of gene
aliases identified, all genes were unified according to the Entrez
gene official symbols (http://www.ncbi.nlm.nih.gov/gene) (38–42).
The process of NLP is illustrated in Fig. 1.
Statistical analysis of selected
data
For all publications retrieved from the PubMed
database (N) the number of genes mentioned in the PubMed database
for G6PD (m) and associated genes (n) were recorded. The frequency
at which each gene appeared was calculated. The rate at which each
gene was mentioned was assumed to be associated with the likelihood
that it was correlated with G6PD, and the number of articles in
which a given gene was associated with G6PD (k) was recorded. By
employing the hypergeometric distribution, the probability of a
frequency greater than k under completely random conditions was
calculated using the following formula:
p=1–∑i=0k–1p(i|n,m,N)
where,
p(i|n,m,N)=n!(N–n)!m!(N–m)!(n–i)!i!(n–m)!(N–n–m+i)!N!
The G6PD-gene associations where the p-value was
equal to <0.05, were subsequently summarized and subjected to a
linked database (http://www.r-project.org/; http://www.bioconductor.org/packages/2.4/bioc/html/KEGGSOAP.html;
http://www.genmapp.org/; http://mips.helmholtz-muenchen.de/proj/ppi) for
further analysis.
Gene ontology (GO) analysis
Enrichment of gene sets was analyzed using the
GOstats software package (http://www.bioconductor.org/packages/release/bioc/html/GO
stats.html) (39). GO analysis
was implemented through the GSEABase package version 1.4.0 from The
R Project for Statistical Computing (http://www.r-project.org/) (43). Genes were classified according to
biological process, cellular component and molecular function.
Pathway and gene network analysis
The G6PD-associated genes were mapped to the Kyoto
Encyclopedia of Genes and Genomes pathway database (KEGG;
http://www.kegg.jp/) using the GenMAPP version C3
(http://www.genmapp.org/), and the enrichment
p-value was calculated for each pathway (44,45).
Based on the protein interactions, regulated genes and protein
modifications listed in the KEGG database, the G6PD-associated
genes were integrated into the following three different
interaction associations: Enzyme-enzyme interactions, representing
two enzymes catalyzing successive reaction steps; protein-protein
interactions, representing binding and modification; transcription
factor-target gene product interactions, representing gene
expression interactions. In addition, the KEGG database was used to
investigate the differentially expressed genes associated with
metabolism and disease signaling pathways. Briefly, the results of
the signaling pathway analysis was downloaded from the KEGG
database, and the interactions between genes were analyzed using
the KEGGSOAP package version 2.4 (http://www.bioconductor.org/packages/2.4/bioc/html/KEGGSOAP.html)
derived from The R Project for Statistical Computing (http://www.r-project.org/) (46,47).
Protein-protein interaction data was downloaded from the MIPS
Mammalian Protein-Protein Interaction Database (http://mips.helmholtz-muenchen.de/proj/ppi/) (48). The frequency of the co-citation of
each gene was calculated employing co-citation matrices to identify
a specific gene term and its variants included within an abstract.
Statistical analysis was performed using the same methods described
for the NLP analysis of G6PD, and the subsequent network was
visualized using the open-source Medusa software program version
3.0 (https://sites.google.com/site/medusa3visualization)
(49). Genes were classified
according to associated signaling pathways. Genes involved in
various signaling pathways were assigned to the single pathway with
the smallest enrichment P-value. Following integration of the
PubMed articles, the collection of protein-protein interaction and
conjunction resolution information, and additional results from the
Search Tool for the Retrieval of Interacting Genes/Proteins tool
version 2.0 (http://string-db.org/) (50), a network of G6PD-associated genes
was established.
Prediction of miRNAs targeting
G6PD
The miRNAs targeting G6PD were identified using four
independent software programs: TargetScan 6.2 (http://www.targetscan.org/) (51); miRanda (version 5.0; http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/);
starBase (version, 2.0; http://starbase.sysu.edu.cn/browseIntersectTargetSite.php)
(52) and PicTar (http://pictar.mdc-berlin.de/cgi-bin/PicTar_vertebrate.cgi?species=vertebrate).
Results
Natural language processing analysis
of G6PD
A search of the PubMed library for articles
discussing G6PD deficiency that were published between January 1980
and June 2014, identified 2,253 primary G6PD-associated studies
(Fig. 1). Each gene mentioned in
these articles was recorded, and the top five most cited genes were
glutathione-disulfide reductase (GSR), insulin (INS), catalase
(CAT), superoxide dismutase (SOD1) and adenosine deaminase (ADA;
Table I). Each gene mentioned in
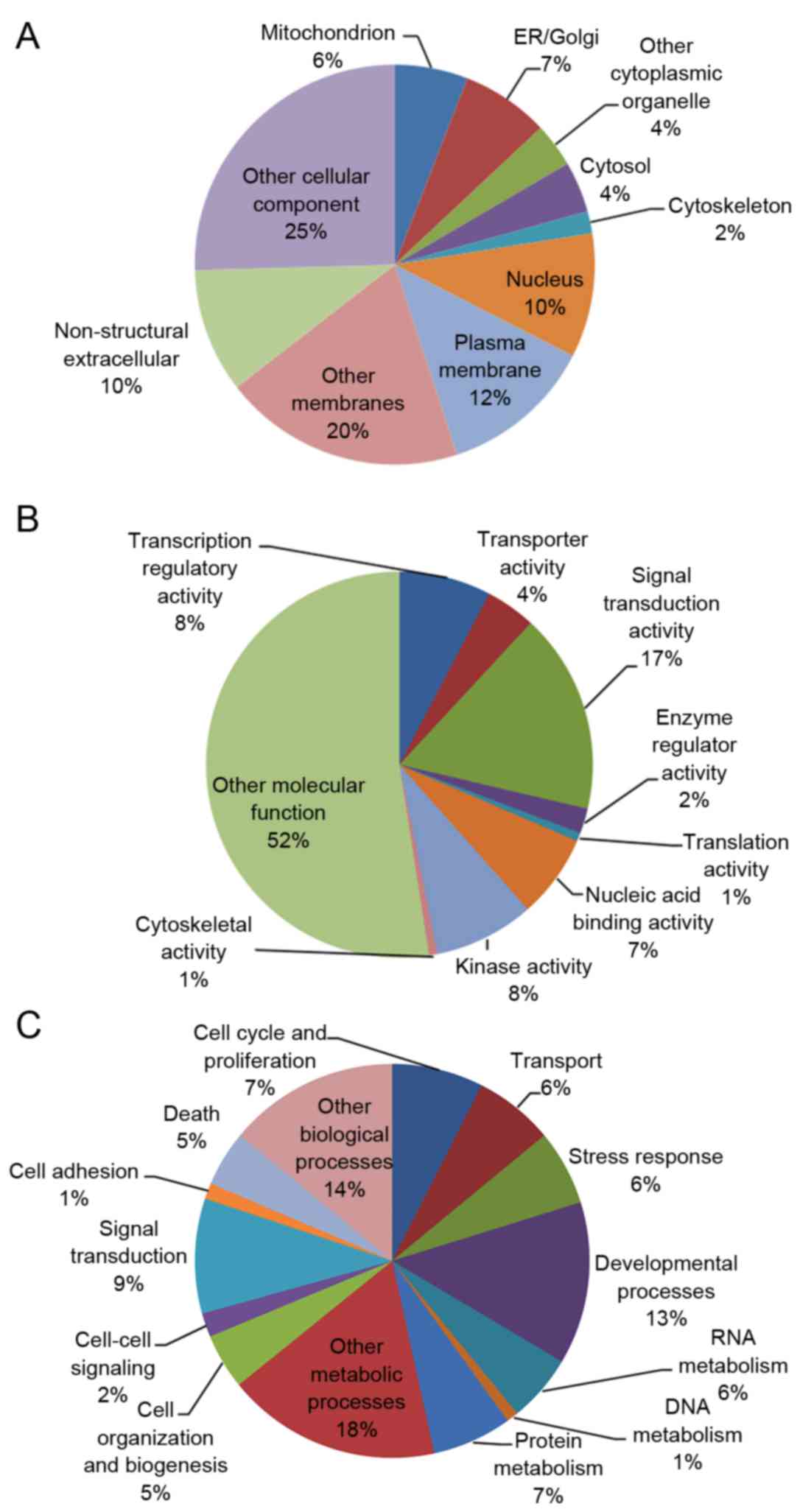
these articles was categorized using GO analysis (Table II), and 98 G6PD-associated genes
were identified and categorized according to cellular component,
molecular function and biological process. The most highly
represented categories of cellular components were ‘other cellular
component’ (25%), ‘other membranes’ (20%) and the plasma membrane
(12%; Fig. 2A). The most highly
represented categories of molecular function were ‘other molecular
function’ (52%), ‘signal transduction’ (17%) and ‘kinase activity
(8%; Fig. 2B). The most highly
represented categories of biological processes were ‘other
metabolic processes’ (18%), ‘other biological processes’ (14%) and
developmental processes (13%; Fig.
2C).
 | Table I.Top ten significant
glucose-6-phosphate dehydrogenase-associated genes. |
Table I.
Top ten significant
glucose-6-phosphate dehydrogenase-associated genes.
| Entrez gene ID | Official gene
symbol | Gene
definition | PubMed count | P-value |
|---|
| 2936 | GSR | Glutathione
reductase | 44 |
<1.00×10−3 |
| 3630 | INS | Insulin | 40 |
<1.00×10−3 |
| 847 | CAT | Catalase | 38 |
4.80×10−9 |
| 2908 | NR3C1 | Nuclear receptor
subfamily 3, group C, member 1 (glucocorticoid receptor) | 23 |
<1.00×10−3 |
| 6647 | SOD1 | Superoxide
dismutase 1, soluble | 17 |
<1.00×10−3 |
| 3251 | HPRT1 | Hypoxanthine
phosphoribosyltransferase 1 | 7 |
<1.00×10−3 |
| 2157 | F8 | Coagulation factor
VIII, procoagulant component | 6 |
3.96×10−5 |
| 100 | ADA | Adenosine
deaminase | 3 |
1.21×10−3 |
| 10327 | AKR1A1 | Aldo-keto reductase
family 1, member A1 (aldehyde reductase) | 3 |
1.75×10−5 |
| 3439 | IFNA1 | Interferon, α
1 | 3 |
2.15×10−3 |
 | Table II.Gene ontology analysis of significant
genes obtained from the natural language processing analysis. |
Table II.
Gene ontology analysis of significant
genes obtained from the natural language processing analysis.
| A, Cellular
component |
|---|
|
|---|
| Term | Genes | Count | P-value |
|---|
| Mitochondrion | CAT, NOS1, HK1,
MPO, BCL2, SOD1, GCK, CYB5R3, ATP5A1, GSR | 10 |
1.50×10−2 |
| Endoplasmic
reticulum/Golgi | NOX4, UGT1A1, SGK1,
BCL2, PRKG1, PTGS2, HSPA5, EPHX1, ATP7B, POR, CYB5R3, NOS3 | 12 |
1.42×10−2 |
| Additional
cytoplasmic organelles | CAT, TNF, MPO, GLA,
ATP6AP1, XDH | 6 |
2.13×10−2 |
| Cytosol | CCND1, NR3C1, HK1,
BCL2, HSP90AA1, NADK, MDH1 | 7 |
1.32×10−3a |
| Plasma
membrane | EGF, L1CAM, SLC6A8,
NOX4, VASP, NOS1, ATF4, TNF, UGT1A1, INSR, FPR1, BCL2, EGFR | 21 |
1.00×10−2 |
| Non-structural
extracellular | FST, EGF, IL1B,
CSF3, F8, TNF, PTH, IFNA1, LPL, LTF, IL6, SOD1 | 17 |
8.03×10−4a |
| Additional cellular
components | CCND1, L1CAM, IL1B,
AR, NR3C1, SORD, VASP, NOS1, ATF4, TNF, NASP, ESR1 | 43 |
7.67×10−3a |
|
| B, Molecular
function |
|
| Term | Genes | Count | P-value |
|
| Transcription
regulatory activity | AR, NR3C1, ATF4,
ESR1, CREM, BCL2, STAT3, PGR, CREB1, SMARCA5, CDKN2A | 11 |
2.15×10−2 |
| Kinase
activity | CCND1, INSR, HK1,
PGK1, SGK1, PDGFRB, EGFR, PRKG1, GCK, NADK, CDKN2A, IKBKG | 12 |
9.20×10−4a |
| Additional
molecular functions | TKT, CCND1, FST,
EGF, L1CAM, SLC6A8, AR, CAT, GSS, NR3C1, SORD, NOX4 | 75 |
2.08×10−3a |
|
| C, Biological
process |
|
| Term | Genes | Count | P-value |
|
| Cell cycle and
proliferation | CCND1, FST, EGF,
IL1B, AR, CAT, NOX4, TNF, NASP, ESR1, PTEN, ADA | 23 |
4.13×10−8a |
| Transport | IL1B, SLC6A8, NOS1,
TNF, NASP, SGK1, ADA, BCL2, SLC2A1, CREB1, NOX1, LTF | 22 |
1.75×10−2 |
| Stress
response | CCND1, IL1B, CAT,
F8, TNF, MPO, IFNA1, SGK1, ADA, BCL2, STAT3, HSPB1 | 19 |
3.75×10−6 |
| Developmental
processes | CCND1, FST, EGF,
L1CAM, IL1B, AR, NR3C1, NOX4, CSF3, VASP, TNF, INSR | 41 |
1.21×10−8a |
| Additional
metabolic processes | TKT, IL1B, AR, CAT,
GSS, NR3C1, SORD, NOX4, NOS1, ATF4, TNF, UGT1A1 | 54 |
3.37×10−12a |
| Cell-to-cell
signaling | IL1B, TF, AR,
CREB1, IL6, GCK | 6 |
7.59×10−3a |
| Signal
transduction | CCND1, FST, EGF,
L1CAM, IL1B, AR, CAT, NR3C1, TNF, INSR, PTEN, PLCG1 | 29 |
8.84×10−3a |
| Death | NR3C1, TNF, PTEN,
SGK1, ADA, BCL2, HPRT1, IL6, SOD1, IL2, IL10, ALDH1A1 | 14 |
7.66×10−5a |
| Additional
biological processes | TKT, CCND1, FST,
L1CAM, IL1B, TF, AR, GSS, NR3C1, NOX4, CSF3, NOS1 | 43 |
3.94×10−2a |
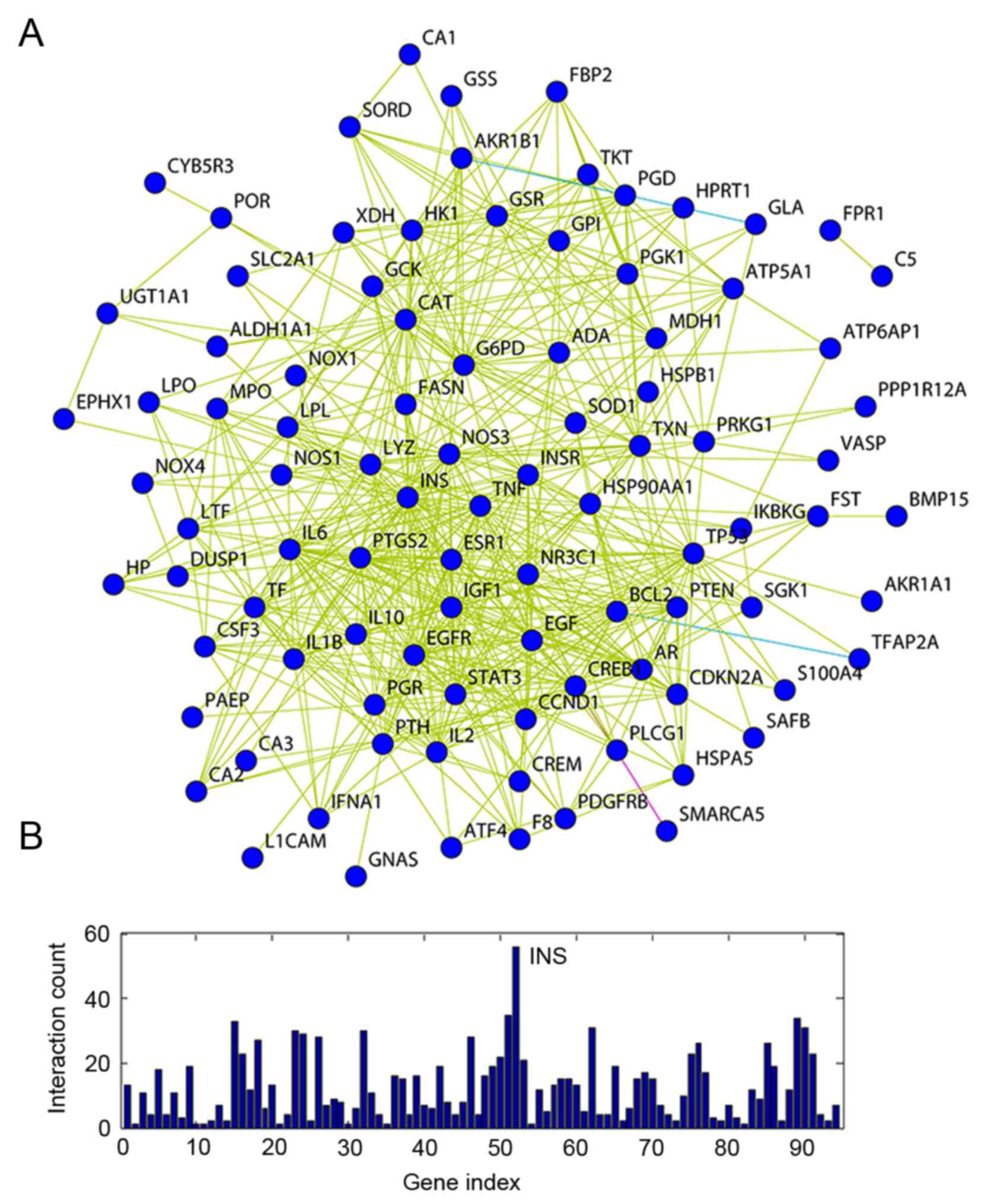
A gene network was constructed to identify a
correlation between these 98 G6PD-associated genes, and the
stability of gene regulatory networks (Fig. 3A). The most highly connected hub
genes critically contribute to the stability of the network. The
most highly connected gene identified in the present study was INS,
which encodes insulin that regulates glucose absorption (Fig. 3B).
The signaling pathways that correlated with each
G6PD-associated gene were retrieved from the KEGG database, which
implicated 47 pathways involved G6PD expression and activity. Out
of these 7 were significantly associated with G6PD (P<0.01;
Table III). G6PD-associated
genes were most significantly correlated with the cytokine-cytokine
receptor interaction pathway. In addition, G6PD-associated genes
were involved in regulating the mitogen-activated protein kinase
(MAPK) signaling pathway, as well as the focal adhesion, Janus
kinase (Jak)-signal transducer and activator of transcription
(STAT), p53, long-term depression and apoptosis signaling pathways
(Table III).
 | Table III.Signaling pathways significantly
represented by glucose-6-phosphate dehydrogenase-associated
genes. |
Table III.
Signaling pathways significantly
represented by glucose-6-phosphate dehydrogenase-associated
genes.
| Pathway ID | Pathway name | Genes | Count | Enrichment
P-value |
|---|
| 4060 | Cytokine-cytokine
receptor interaction | EGF, IL1B, CSF3,
TNF, IFNA1, PDGFRB, EGFR, IL6, IL2, IL10 | 10 |
5.28×10−3a |
| 4010 | MAPK signaling
pathway | EGF, IL1B, ATF4,
TP53, TNF, PDGFRB, EGFR, HSPB1, DUSP1, IKBKG | 10 |
6.51×10−3a |
| 4510 | Focal adhesion | CCND1, EGF, VASP,
PTEN, PDGFRB, BCL2, EGFR, PPP1R12A, IGF1 | 9 |
2.75×10−3a |
| 4630 | Jak-STAT signaling
pathway | CCND1, CSF3, IFNA1,
STAT3, IL6, IL2, IL10 | 7 |
8.11×10−3a |
| 4910 | Insulin signaling
pathway | INSR, HK1, INS,
FBP2, GCK, FASN | 6 |
1.67×10−2 |
| 4020 | Calcium signaling
pathway | NOS1, PLCG1,
PDGFRB, EGFR, NOS3, GNAS | 6 |
4.95×10−2 |
| 4115 | p53 signaling
pathway | CCND1, TP53, PTEN,
IGF1, CDKN2A | 5 |
3.30×10−3a |
| 4730 | Long-term
depression | NOS1, PRKG1, NOS3,
IGF1, GNAS | 5 |
3.98×10−3a |
| 4210 | Apoptosis | IL1B, TP53, TNF,
BCL2, IKBKG | 5 |
9.85×10−3a |
| 4540 | Gap junction | EGF, PDGFRB, EGFR,
PRKG1, GNAS | 5 |
1.13×10−3 |
| 4620 | Toll-like receptor
signaling pathway | IL1B, TNF, IFNA1,
IL6, IKBKG | 5 |
1.72×10−2 |
| 4660 | T cell receptor
signaling pathway | TNF, PLCG1, IL2,
IL10, IKBKG | 5 |
2.23×10−2 |
| 4920 | Adipocytokine
signaling pathway | TNF, STAT3, SLC2A1,
IKBKG | 4 |
1.75×10−2 |
| 4370 | VEGF signaling
pathway | PLCG1, HSPB1,
PTGS2, NOS3 | 4 |
2.54×10−2 |
| 4640 | Hematopoietic cell
lineage | IL1B, CSF3, TNF,
IL6 | 4 |
4.07×10−2 |
| 4612 | Antigen processing
and presentation | IFNA1, CREB1,
HSPA5, HSP90AA1 | 4 |
4.22×10−2 |
In addition to the notable PPP, G6PD-associated
genes were observed to be involved in the regulation of fructose
and mannose, glycerolipid, nitrogen, galactose, starch, sucrose,
amino and nucleotide sugars, glutathione and drug metabolic
pathways (3 or 4 counts for each metabolic pathway; P<0.05;
Table IV). Furthermore,
G6PD-associated genes were significantly correlated with a number
of cancers, autoimmune diseases and oxidative stress-associated
disorders (>3 counts for each disease; P<0.05; Table V). These results suggest that the
aberrant regulation of G6PD signaling and/or metabolic pathways may
be associated with the pathogenesis of these diseases.
 | Table IV.Metabolic pathways significantly
represented by glucose-6-phosphate dehydrogenase-associated
genes. |
Table IV.
Metabolic pathways significantly
represented by glucose-6-phosphate dehydrogenase-associated
genes.
| Metabolic
pathway | Genes | Count | Enrichment
P-value |
|---|
| Pentose phosphate
pathway | TKT, FBP2, GPI,
PGD | 4 |
5.42×10−4a |
| Fructose and
mannose metabolism | AKR1B1, SORD, HK1,
FBP2 | 4 |
1.37×10−3a |
| Glycerolipid
metabolism | AKR1B1, GLA, LPL,
AKR1A1 | 4 |
4.72×10−3a |
| Starch and sucrose
metabolism | UGT1A1, HK1, GCK,
GPI | 4 |
7.32×10−3a |
| Nitrogen
metabolism | CA1, CA2, CA3 | 3 |
5.22×10−3a |
| Galactose
metabolism | AKR1B1, HK1,
GLA | 3 |
5.87×10−3a |
| Glutathione
metabolism | GSS, GSR, PGD | 3 |
3.83×10−2 |
| Drug
metabolism-other enzymes | UGT1A1, HPRT1,
XDH | 3 |
4.03×10−2 |
| Amino sugar and
nucleotide sugar metabolism | HK1, GCK,
CYB5R3 | 3 |
2.60×10−2 |
 | Table V.Diseases significantly represented by
glucose-6-phosphate dehydrogenase-associated genes. |
Table V.
Diseases significantly represented by
glucose-6-phosphate dehydrogenase-associated genes.
| Disease | Genes | Count | Enrichment
P-value |
|---|
| Prostate
cancer | CCND1, EGF, AR,
ATF4, TP53, PTEN, PDGFRB, BCL2, EGFR, INS, CREB1, HSP90AA1, IGF1,
IKBKG | 14 |
2.81×10−11a |
| Glioma | CCND1, EGF, TP53,
PTEN, PLCG1, PDGFRB, EGFR, IGF1, CDKN2A | 9 |
3.79×10−7a |
| Melanoma | CCND1, EGF, TP53,
PTEN, PDGFRB, EGFR, IGF1, CDKN2A | 8 |
8.66×10−6a |
| Small cell lung
cancer | CCND1, NOS1, TP53,
PTEN, BCL2, PTGS2, NOS3, IKBKG | 8 |
3.61×10−5a |
| Amyotrophic lateral
sclerosis | CAT, NOS1, TP53,
TNF, BCL2, SOD1, NOS3 | 7 |
1.46×10−5a |
| Pancreatic
cancer | CCND1, EGF, TP53,
EGFR, STAT3, CDKN2A, IKBKG | 7 |
8.63×10−5a |
| Non-small cell lung
cancer | CCND1, EGF, TP53,
PLCG1, EGFR, CDKN2A | 6 |
1.35×10−4a |
| Alzheimer's
disease | IL1B, NOS1, TNF,
LPL, NOS3, ATP5A1 | 6 |
3.72×10−2 |
| Bladder cancer | CCND1, EGF, TP53,
EGFR, CDKN2A | 5 |
3.65×10−4a |
| Type II diabetes
mellitus | TNF, INSR, HK1,
INS, GCK | 5 |
6.20×10−4a |
| Endometrial
cancer | CCND1, EGF, TP53,
PTEN, EGFR | 5 |
9.91×10−4a |
| Colorectal
cancer | CCND1, TP53,
PDGFRB, BCL2, EGFR | 5 |
8.13×10−3a |
| Type I diabetes
mellitus | IL1B, TNF, INS,
IL2 | 4 |
3.69×10−3a |
| Chronic myeloid
leukemia | CCND1, TP53,
CDKN2A, IKBKG | 4 |
2.54×10−2 |
| Graft-vs.-host
disease | IL1B, TNF, IL6,
IL2 | 4 |
3.10×10−3a |
| Autoimmune thyroid
disease | IFNA1, IL2,
IL10 | 3 |
4.23×10−2 |
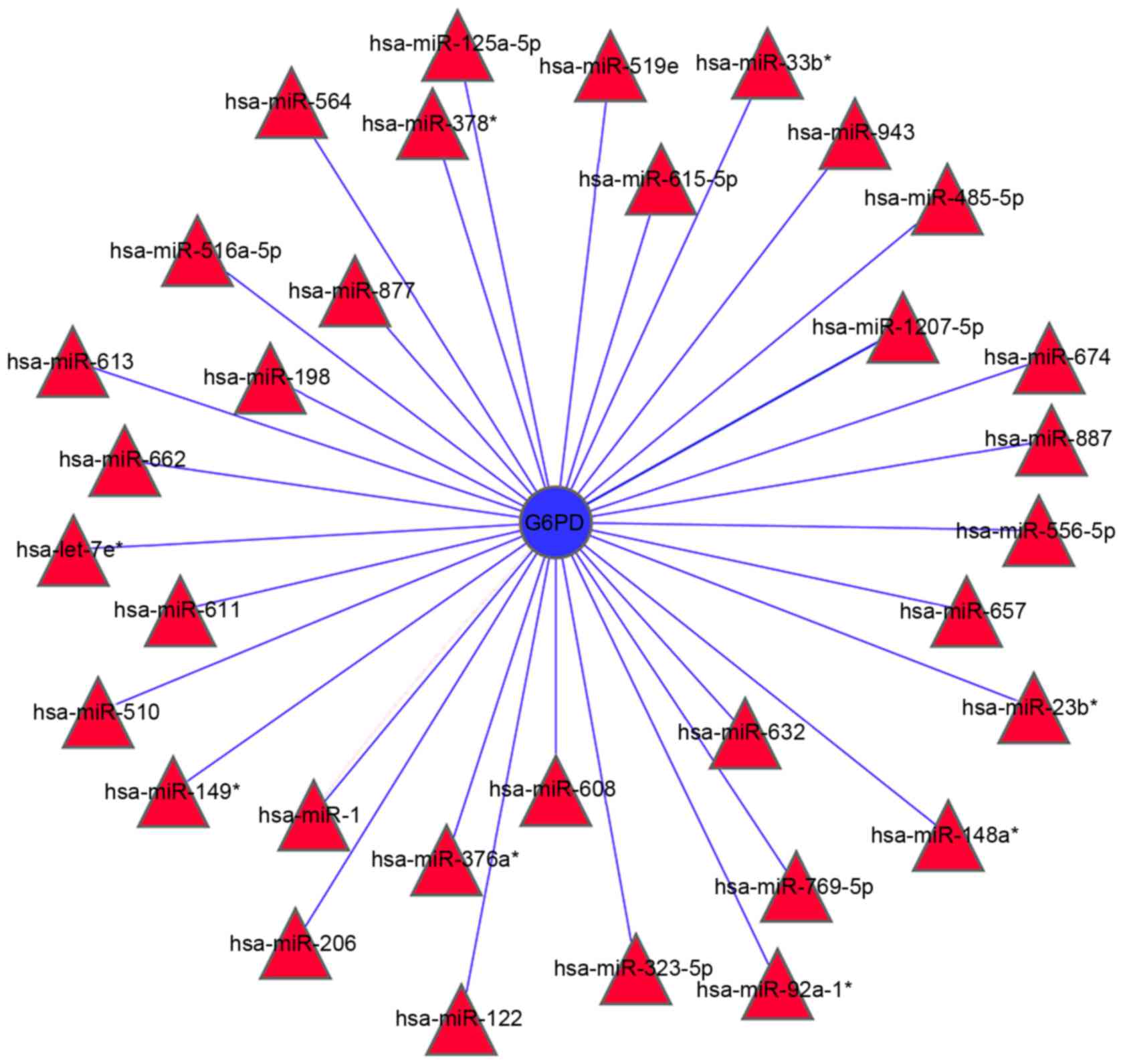
Analysis of miRNAs targeting G6PD. Four independent
software programs were used to predict the miRNA-targets of G6PD.
The searches yielded 33 miRNAs that were predicted by at least two
software programs to target G6PD (Fig.
4). Out of these, three miRNAs, including miR-1207-5P, miR-1
and miR-125a-5p, were predicted to target G6PD by all four software
programs, thus indicating that these miRNAs are most likely to be
involved in the regulation of G6PD expression.
Discussion
A systematic analysis of G6PD-associated genes may
provide useful information regarding the role of G6PD in healthy
and pathological processes. In the present study, all genes
reportedly associated with G6PD in published articles retrieved
from the PubMed library were systematically analyzed. NLP and
database searching strategies were employed to analyze the
association between the identified genes and G6PD, as well as the
signaling pathways that they may be involved in, and their
potential connection to a number of clinical diseases.
A total of 98 G6PD-associated genes were identified,
and out of the 47 implicated signaling pathways, seven were highly
likely to be regulated by G6PD-associated genes. These results
highlight the mechanisms by which G6PD dysregulation may contribute
to cancer or autoimmune diseases. In addition, four separate
databases were used to identify 33 miRNAs that were predicted to
regulate G6PD. To the best of the author's knowledge, this is the
first study to perform systematic analysis of miRNAs that may
target G6PD.
Out of the 98 G6PD-associated genes that were
identified, the five most frequently cited genes were GSR, INS,
CAT, SOD1 and ADA. The products encoded by these genes are involved
in maintaining homeostasis of NADPH. GSR utilizes NADPH, derived
from the PPP (of which G6PD catalyzes the first committed step),
for recycling oxidized glutathione disulfide to reduced glutathione
(Fig. 5) (53,54).
In addition, NADPH is required and consumed during fatty acid
synthesis. Reduced consumption of NADPH has been observed when
fatty acid synthesis is inhibited by AMP-activated protein kinase
(AMPK) via inhibition of acetyl-coenzyme A carboxylase (55). The INS gene encodes insulin, which
increases cell permeability to fatty acids and accelerates the
pentose phosphate cycle for NADPH production in the liver (56,57).
The CAT gene encodes catalase, a crucial scavenger of hydrogen
peroxide in human erythrocytes, which is dependent on the
availability of PPP-derived NADPH (58). NADPH is known to be tightly bound
to mammalian catalase, and prevents inactivation of catalase by
H2O2 (Fig.
5) (59). Significantly lower
SOD and G6PD activities have been reported in cystic echinococcosis
of the liver and Parkinson's disease (57,60).
Reduced SOD activity in erythrocytes, potentially as a result of
inactivation of SOD by diminished reductive equivalents, has been
detected in 26 carriers of G6PD defects (Fig. 5) (61).
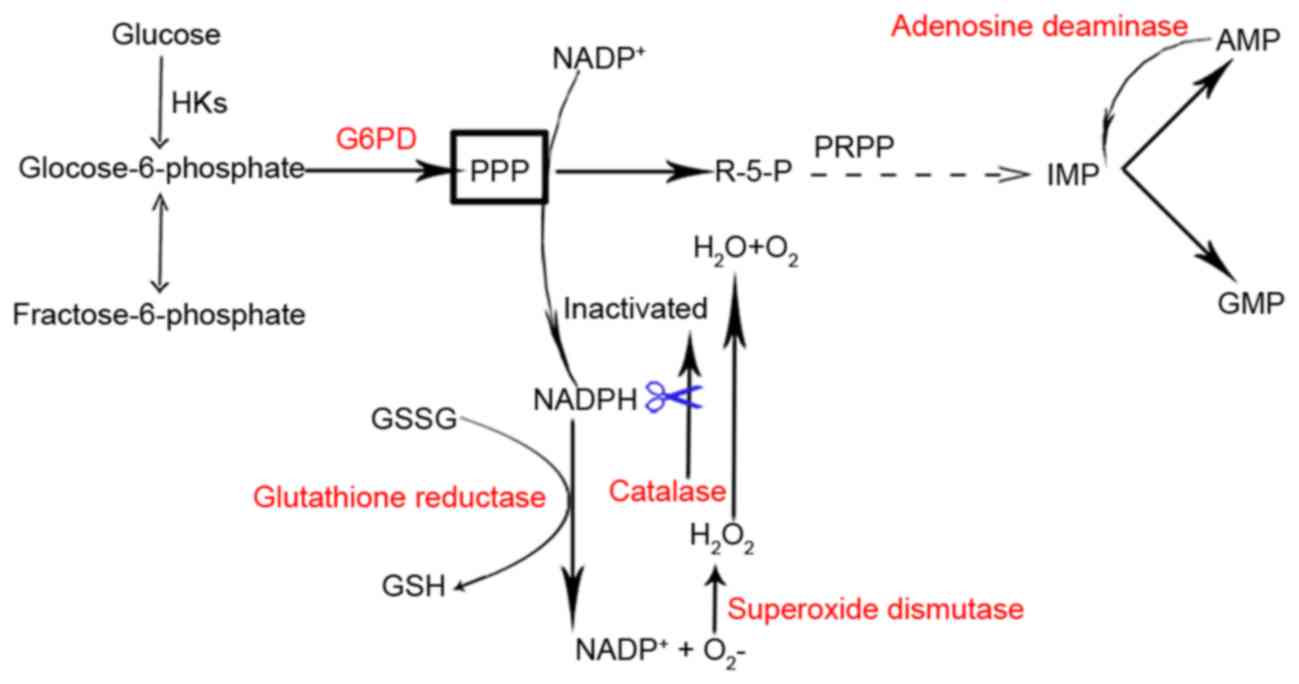 | Figure 5.A schematic indicating the enzymatic
reactions that involve the most cited G6PD-associated genes. As the
rate-limiting enzyme in the pentose phosphate pathway, G6PD
catalyzes the production of R-5-P and NADPH. R-5-Pis converted to
IMP, which leads to the production of AMP. This product is
converted to IMP when adenosine deaminase is present. NADPH serves
crucial roles in the reduction of GSSG to GSH by glutathione
reductase, and protects catalase from inactivation by
H2O2. HKs, hexokinases; G6PD,
guocose-6-phosphate dehydrogenase; R-5-P, ribose-5-phosphate; PPP,
pentose phosphate pathway; NADP+, oxidized nicotinamide
adenine-dinucleotide phosphate; NADPH, reduced nicotinamide
adenine-dinucleotide phosphate; PRPP, phosphoribosyl pyrophosphate;
IMP, inosine monophosphate; AMP, adenosine monophosphate; GMP,
guanosine monophosphate; GSSG, glutathione disulfide; GSH, reduced
glutathione. |
Signaling pathway analysis identified 47 pathways,
of which the cytokine-cytokine receptor interaction was most
significantly associated with G6PD-related genes. In addition,
G6PD-associated genes were demonstrated to be potentially involved
in regulating focal adhesion, the MAPK, Jak-STAT, p53, long-term
depression and apoptosis signaling pathways. Notably, G6PD itself
may be involved in regulating the MAPK and p53 signaling pathways.
By increasing the availability of glucose for glycolysis, AMPK
functions as a master sensor of cellular energy balance. AMPK is
activated when cells undergo energy-depleting stresses (62–65).
Due to insufficient NADPH, G6PD-deficiency leads to a switch of the
biosynthetic pathway for GSH, thus limiting AMPK activation
(62). p38 MAPK has been
implicated in a number of cellular processes, including the cell
cycle, apoptosis, differentiation, neoplasm metastasis,
angiogenesis and vasculogenesis (63). Transforming growth
factor-β-activated protein kinase 1-binding protein 1 (TAB1) is a
scaffold protein responsible for the autophosphorylation of MAPK
(64). Activation of AMPK has been
demonstrated to recruit p38 MAPK to the TAB1/AMPK-containing
macromolecular complex, thus facilitating p38 MAPK activation in
the ischemic heart (64).
It was previously reported that p53 might prevent
the inactive G6PD monomer from forming active dimers (65). In addition, p53 may be regulated by
G6PD. Following phosphorylation by ataxia telangiectasia mutated
(ATM), p53 regulates lipid metabolism through promoting
transcription of Lpin1, which encodes a protein that regulates
lipid metabolism (66,67). Notably, Lpin1 induction is
dependent on elevated ROS and ATM levels, and this process is
inhibited by the N-acetyl cysteine antioxidant (66). ATM is activated directly by
oxidative stress (67).
Considering that the ROS-ATM-p53 signaling pathway contributes to
the induction of Lpin1 and G6PD greatly influences ROS levels, it
is reasonable to hypothesize that G6PD may regulate p53 function.
Whether G6PD regulates MAPK and p53 signaling pathways remains to
be addressed in future studies.
Alteration of G6PD-regulated pathways may lead to
certain diseases. Dysregulation of G6PD has been reported to be
associated with neoplasms (68–70).
For example, increased G6PD expression levels correlated with
grading, metastasis and poor prognosis in human hepatocellular
carcinoma patients (68).
Alterations in G6PD levels may promote tumor cell proliferation or
apoptosis via the STAT3/5 pathway in a human melanoma xenograft
model, and G6PD regulated STAT3 activity via the NADPH oxidase
4-NADPH-ROS-proto-oncogene tyrosine protein kinase/protein-tyrosine
phosphatase 1D signaling pathway in A375 melanoma cells (69,70).
In the present study that G6PD-associated genes were highly
correlated with various cancers, including prostate cancer, glioma,
small cell lung cancer, non-small cell lung cancer, chromic myeloid
leukemia, pancreatic cancer and bladder cancer. In addition,
G6PD-associated genes were highly correlated with autoimmune
diseases, including amyotrophic lateral sclerosis,
graft-versus-host diseases, and autoimmune thyroid disease. One of
the G6PD-associated genes, interleukin 6 (IL6), has been reported
to be involved in autoimmune diseases (71). Activation of the IL6 inflammatory
loop may mediate trastuzumab resistance in human epidermal growth
factor receptor 2-positive breast cancer by promoting growth of the
cancer stem cell population (72).
Accumulating evidence suggests that miRNAs are
involved in various biological processes and human diseases, and
they are versatile regulators of gene expression in higher
eukaryotes and additional organisms. Aberrant expression of G6PD
leads to the development of a number of different diseases.
Therefore, miRNAs capable of targeting G6PD may be of therapeutic
interest. Previous studies have reported the potential for
particular miRNAs to influence G6PD expression (73,74).
Therefore, the aim of the present study was to identify all miRNAs
capable of binding G6PD. A total of 33 miRNAs were predicted to
target G6PD in at least two of four independent programs employed
in the current study. miR-1207-5P, miR-1 and miR-125a-5p were the
only three miRNAs predicted by all four software programs to
potentially target G6PD, indicating that these three miRNAs may be
potentially important regulators of G6PD. Notably, Alvarez
(75) validated G6PD as a direct
target of miR-1207-5p using a dual Firefly-Renilla
luciferase reporter assay.
In conclusion, the present study systematically
evaluated G6PD-associated genes and miRNAs that potentially target
G6PD. The results of the analysis identified signaling and
metabolic pathways regulated by G6PD-associated genes, as well as
miRNAs that are predicted to target G6PD. This may provide a
greater understanding of the role of G6PD in different pathological
processes, and contribute to the development of rational
therapeutic approaches for G6PD-associated diseases. However,
further experiments will be required to confirm the results. By
integrating genomics, transcriptomics, and proteomics analyses of
G6PD-associated disorders, a novel area of G6PD research may be
explored.
Acknowledgements
The present study was supported by grants from the
Natural Science Foundation of China (grant nos. 81160246, 81460421
and 31660246) and the Science and Technology Fund of Yunnan
Province (grant nos. 2013FB102 and 2016FB003).
References
|
1
|
Carson PE and Frischer H:
Glucose-6-phosphate dehydrogenase deficiency and related disorders
of the pentose phosphate pathway. Am J Med. 41:744–761. 1966.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lewis CA, Parker SJ, Fiske BP, McCloskey
D, Gui DY, Green CR, Vokes NI, Feist AM, Heiden MG Vander and
Metallo CM: Tracing compartmentalized NADPH metabolism in the
cytosol and mitochondria of mammalian cells. Mol Cell. 55:253–263.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Glucose-6-phosphate dehydrogenase
deficiency. WHO Working Group. Bull World Health Organ. 67:601–611.
1989.PubMed/NCBI
|
|
4
|
Cappellini MD and Fiorelli G:
Glucose-6-phosphate dehydrogenase deficiency. Lancet. 371:64–74.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang YP, Zhou LS, Zhao YZ, Wang SW, Chen
LL, Liu LX, Ling ZQ, Hu FJ, Sun YP, Zhang JY, et al: Regulation of
G6PD acetylation by SIRT2 and KAT9 modulates NADPH homeostasis and
cell survival during oxidative stress. EMBO J. 33:1304–1320.
2014.PubMed/NCBI
|
|
6
|
WHO Working Group, . Glucose-6-phosphate
dehydrogenase deficiency. Bull World Health Organ. 67:601–611.
1989.PubMed/NCBI
|
|
7
|
Tian WN, Braunstein LD, Apse K, Pang J,
Rose M, Tian X and Stanton RC: Importance of glucose-6-phosphate
dehydrogenase activity in cell death. Am J Physiol.
276:C1121–C1131. 1999.PubMed/NCBI
|
|
8
|
Ho HY, Cheng ML, Lu FJ, Chou YH, Stern A,
Liang CM and Chiu DT: Enhanced oxidative stress and accelerated
cellular senescence in glucose-6-phosphate dehydrogenase
(G6PD)-deficient human fibroblasts. Free Radic Biol Med.
29:156–169. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pandolfi PP, Sonati F, Rivi R, Mason P,
Grosveld F and Luzzatto L: Targeted disruption of the housekeeping
gene encoding glucose 6-phosphate dehydrogenase (G6PD): G6PD is
dispensable for pentose synthesis but essential for defense against
oxidative stress. EMBO J. 14:5209–5215. 1995.PubMed/NCBI
|
|
10
|
Longo L, Vanegas OC, Patel M, Rosti V, Li
H, Waka J, Merghoub T, Pandolfi PP, Notaro R, Manova K and Luzzatto
L: Maternally transmitted severe glucose 6-phosphate dehydrogenase
deficiency is an embryonic lethal. EMBO J. 21:4229–4239. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kuo W, Lin J and Tang TK: Human
glucose-6-phosphate dehydrogenase (G6PD) gene transforms NIH 3T3
cells and induces tumors in nude mice. Int J Cancer. 85:857–864.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cappai G, Songini M, Doria A, Cavallerano
JD and Lorenzi M: Increased prevalence of proliferative retinopathy
in patients with type 1 diabetes who are deficient in
glucose-6-phosphate dehydrogenase. Diabetologia. 54:1539–1542.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Heymann AD, Cohen Y and Chodick G:
Glucose-6-phosphate dehydrogenase deficiency and type 2 diabetes.
Diabetes Care. 35:e582012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gaskin RS, Estwick D and Peddi R: G6PD
deficiency: Its role in the high prevalence of hypertension and
diabetes mellitus. Ethn Dis. 11:749–754. 2001.PubMed/NCBI
|
|
15
|
Rao KN, Elm MS, Kelly RH, Chandar N, Brady
EP, Rao B, Shinozuka H and Eagon PK: Hepatic hyperplasia and cancer
in rats: Metabolic alterations associated with cell growth.
Gastroenterology. 113:238–248. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Batetta B, Pulisci D, Bonatesta RR, Sanna
F, Piras S, Mulas MF, Spano O, Putzolu M, Broccia G and Dessì S:
G6PD activity and gene expression in leukemic cells from
G6PD-deficient subjects. Cancer Lett. 140:53–58. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Van Driel BE, Valet GK, Lyon H, Hansen U,
Song JY and Van Noorden CJ: Prognostic estimation of survival of
colorectal cancer patients with the quantitative histochemical
assay of G6PDH activity and the multiparameter classification
program CLASSIF1. Cytometry. 38:176–183. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Polat MF, Taysi S, Gul M, Cikman O, Yilmaz
I, Bakan E and Erdogan F: Oxidant/antioxidant status in blood of
patients with malignant breast tumour and benign breast disease.
Cell Biochem Funct. 20:327–331. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Langbein S, Frederiks WM, zur Hausen A,
Popa J, Lehmann J, Weiss C, Alken P and Coy JF: Metastasis is
promoted by a bioenergetic switch: New targets for progressive
renal cell cancer. Int J Cancer. 122:2422–2428. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang J, Yuan W and Chen Z, Wu S, Chen J,
Ge J, Hou F and Chen Z: Overexpression of G6PD is associated with
poor clinical outcome in gastric cancer. Tumour Biol. 33:95–101.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hu T, Zhang C, Tang Q, Su Y, Li B, Chen L,
Zhang Z, Cai T and Zhu Y: Variant G6PD levels promote tumor cell
proliferation or apoptosis via the STAT3/5 pathway in the human
melanoma xenograft mouse model. BMC Cancer. 13:2512013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Dong H, Lei J, Ding L, Wen Y, Ju H and
Zhang X: MicroRNA: Function, detection, and bioanalysis. Chem Rev.
113:6207–6233. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Costinean S, Zanesi N, Pekarsky Y, Tili E,
Volinia S, Heerema N and Croce CM: Pre-B cell proliferation and
lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155
transgenic mice. Proc Natl Acad Sci USA. 103:7024–7029. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang B, Pan X, Cobb GP and Anderson TA:
microRNAs as oncogenes and tumor suppressors. Dev Biol. 302:1–12.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li C, Feng Y, Coukos G and Zhang L:
Therapeutic microRNA strategies in human cancer. AAPS J.
11:747–757. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Burchard J, Zhang C, Liu AM, Poon RT, Lee
NP, Wong KF, Sham PC, Lam BY, Ferguson MD, Tokiwa G, et al:
microRNA-122 as a regulator of mitochondrial metabolic gene network
in hepatocellular carcinoma. Mol Syst Biol. 6:4022010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Brameier M, Herwig A, Reinhardt R, Walter
L and Gruber J: Human box C/D snoRNAs with miRNA like functions:
Expanding the range of regulatory RNAs. Nucleic Acids Res.
39:675–686. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Dumortier O, Hinault C and Van Obberghen
E: MicroRNAs and metabolism crosstalk in energy homeostasis. Cell
Metab. 18:312–324. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yates LA, Norbury CJ and Gilbert RJ: The
long and short of microRNA. Cell. 153:516–519. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Dimmeler S and Ylä-Herttuala S: 14q32
miRNA cluster takes center stage in neovascularization. Circ Res.
115:680–682. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sachdeva M, Mito JK, Lee CL, Zhang M, Li
Z, Dodd RD, Cason D, Luo L, Ma Y, van Mater D, et al: MicroRNA-182
drives metastasis of primary sarcomas by targeting multiple genes.
J Clin Invest. 124:4305–4319. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xie C, Han Y, Liu Y, Han L and Liu J:
miRNA-124 down-regulates SOX8 expression and suppresses cell
proliferation in non-small cell lung cancer. Int J Clin Exp Pathol.
7:6534–6542. 2014.PubMed/NCBI
|
|
33
|
Zhang J, Yang Y, Yang T, Yuan S, Wang R,
Pan Z, Yang Y, Huang G, Gu F, Jiang B, et al: Double-negative
feedback loop between microRNA-422a and forkhead box (FOX)G1/Q1/E1
regulates hepatocellular carcinoma tumor growth and metastasis.
Hepatology. 61:561–573. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Johnson CD, Esquela-Kerscher A, Stefani G,
Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J,
Shingara J, et al: The let-7 microRNA represses cell proliferation
pathways in human cells. Cancer Res. 67:7713–7722. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Welten SM, Bastiaansen AJ, de Jong RC, de
Vries MR, Peters EA, Boonstra MC, Sheikh SP, La Monica N,
Kandimalla ER, Quax PH and Nossent AY: Inhibition of 14q32
microRNAs miR-329, miR-487b, miR-494, and miR-495 increases
neovascularization and blood flow recovery after ischemia. Circ
Res. 115:696–708. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
El Azzouzi H, Leptidis S, Dirkx E, Hoeks
J, van Bree B, Brand K, McClellan EA, Poels E, Sluimer JC, van den
Hoogenhof MM, et al: The hypoxia-inducible microRNA cluster
miR-199a-214 targets myocardial PPARδ and impairs mitochondrial
fatty acid oxidation. Cell Metab. 18:341–354. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Settles B: ABNER: An open source tool for
automatically tagging genes, proteins and other entity names in
text. Bioinformatics. 21:3191–3192. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Falcon S and Gentleman R: Using GOstats to
test gene lists for GO term association. Bioinformatics.
23:257–258. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Morgan AA, Lu Z, Wang X, Cohen AM, Fluck
J, Ruch P, Divoli A, Fundel K, Leaman R, Hakenberg J, et al:
Overview of BioCreative II gene normalization. Genome Biol.
9:(Suppl 2): S32008. View Article : Google Scholar
|
|
40
|
Maglott D, Ostell J, Pruitt KD and
Tatusova T: Entrez Gene: Gene-centered information at NCBI. Nucleic
Acids Res. 39:D52–D57. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Arighi CN, Carterette B, Cohen KB,
Krallinger M, Wilbur WJ, Fey P, Dodson R, Cooper L, van Slyke CE,
Dahdul W, et al: An overview of the BioCreative 2012 Workshop Track
III: Interactive text mining task. Database (Oxford).
2013:bas0562013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Arighi CN, Lu Z, Krallinger M, Cohen KB,
Wilbur WJ, Valencia A, Hirschman L and Wu CH: Overview of the
BioCreative III workshop. BMC Bioinformatics. 12 Suppl 8:S12011.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Morgan MFS and Gentleman R: GSEABase: Gene
set enrichment data structures and methods. R package version
1.4.0. available at conductor.org. 2–June. 20112007.
|
|
44
|
Dahlquist KD, Salomonis N, Vranizan K,
Lawlor SC and Conklin BR: GenMAPP, a new tool for viewing and
analyzing microarray data on biological pathways. Nat Genet.
31:19–20. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kanehisa M, Goto S, Sato Y, Furumichi M
and Tanabe M: KEGG for integration and interpretation of
large-scale molecular data sets. Nucleic Acids Res. 40:D109–D114.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Sultana KZ, Bhattacharjee A and Jamil H:
Querying KEGG pathways in logic. Int J Data Min Bioinform. 9:1–21.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Pagel P, Kovac S, Oesterheld M, Brauner B,
Dunger-Kaltenbach I, Frishman G, Montrone C, Mark P, Stümpflen V,
Mewes HW, et al: The MIPS mammalian protein-protein interaction
database. Bioinformatics. 21:832–834. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Pavlopoulos GA, Hooper SD, Sifrim A,
Schneider R and Aerts J: Medusa: A tool for exploring and
clustering biological networks. BMC Res Notes. 4:3842011.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Franceschini A, Szklarczyk D, Frankild S,
Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C
and Jensen LJ: STRING v9.1: Protein-protein interaction networks,
with increased coverage and integration. Nucleic Acids Res.
41:D808–D815. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Agarwal V, Bell GW2, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. Aug 12–2015.(Epub ahead of print). doi: 10.7554/eLife.05005.
View Article : Google Scholar :
|
|
52
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42:D92–D97. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Préville X, Salvemini F, Giraud S,
Chaufour S, Paul C, Stepien G, Ursini MV and Arrigo AP: Mammalian
small stress proteins protect against oxidative stress through
their ability to increase glucose-6-phosphate dehydrogenase
activity and by maintaining optimal cellular detoxifying machinery.
Exp Cell Res. 247:61–78. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Rajasekaran NS, Connell P, Christians ES,
Yan LJ, Taylor RP, Orosz A, Zhang XQ, Stevenson TJ, Peshock RM,
Leopold JA, et al: Human alpha B-crystallin mutation causes
oxido-reductive stress and protein aggregation cardiomyopathy in
mice. Cell. 130:427–439. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Patra KC and Hay N: The pentose phosphate
pathway and cancer. Trends Biochem Sci. 39:347–354. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Ceddia RB, Bikopoulos GJ, Hilliker AJ and
Sweeney G: Insulin stimulates glucose metabolism via the pentose
phosphate pathway in Drosophila Kc cells. FEBS Lett. 555:307–310.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Heidarpour M, Mohri M, Borji H and
Moghdass E: Oxidant/antioxidant status in cattle with liver cystic
echinococcosis. Vet Parasitol. 195:131–135. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Gaetani GF, Rolfo M, Arena S, Mangerini R,
Meloni GF and Ferraris AM: Active involvement of catalase during
hemolytic crises of favism. Blood. 88:1084–1088. 1996.PubMed/NCBI
|
|
59
|
Kirkman HN, Rolfo M, Ferraris AM and
Gaetani GF: Mechanisms of protection of catalase by NADPH. Kinetics
and stoichiometry. J Biol Chem. 274:13908–13914. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Abraham S, Soundararajan CC, Vivekanandhan
S and Behari M: Erythrocyte antioxidant enzymes in Parkinson's
disease. Indian J Med Res. 121:111–115. 2005.PubMed/NCBI
|
|
61
|
Grieger M and Schulz S: Determination of
superoxide dismutase (sod) in erythrocytes in glucose-6-phosphate
dehydrogenase deficiency (G6PD-deficiency). Folia Haematol Int Mag
Klin Morphol Blutforsch. 110:427–436. 1983.(In German). PubMed/NCBI
|
|
62
|
Tang HY, Ho HY, Wu PR, Chen SH, Kuypers
FA, Cheng ML and Chiu DT: Inability to maintain GSH pool in
G6PD-deficient red cells causes futile AMPK activation and
irreversible metabolic disturbance. Antioxid Redox Signal.
22:744–759. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Boutros T, Chevet E and Metrakos P:
Mitogen-activated protein (MAP) kinase/MAP kinase phosphatase
regulation: Roles in cell growth, death, and cancer. Pharmacol Rev.
60:261–310. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Li J, Miller EJ, Ninomiya-Tsuji J, Russell
RR III and Young LH: AMP-activated protein kinase activates p38
mitogen-activated protein kinase by increasing recruitment of p38
MAPK to TAB1 in the ischemic heart. Circ Res. 97:872–879. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Jiang P, Du W, Wang X, Mancuso A, Gao X,
Wu M and Yang X: p53 regulates biosynthesis through direct
inactivation of glucose-6-phosphate dehydrogenase. Nat Cell Biol.
13:310–316. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Assaily W, Rubinger DA, Wheaton K, Lin Y,
Ma W, Xuan W, Brown-Endres L, Tsuchihara K, Mak TW and Benchimol S:
ROS-mediated p53 induction of Lpin1 regulates fatty acid oxidation
in response to nutritional stress. Mol Cell. 44:491–501. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Guo Z, Kozlov S, Lavin MF, Person MD and
Paull TT: ATM activation by oxidative stress. Science. 330:517–521.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Kowalik MA, Guzzo G, Morandi A, Perra A,
Menegon S, Masgras I, Trevisan E, Angioni MM, Fornari F, Quagliata
L, et al: Metabolic reprogramming identifies the most aggressive
lesions at early phases of hepatic carcinogenesis. Oncotarget.
7:32375–32393. 2016.PubMed/NCBI
|
|
69
|
Hu T, Zhang C, Tang Q, Su Y, Li B, Chen L,
Zhang Z, Cai T and Zhu Y: Variant G6PD levels promote tumor cell
proliferation or apoptosis via the STAT3/5 pathway in the human
melanoma xenograft mouse model. BMC Cancer. 13:2512013. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Cai T, Kuang Y, Zhang C, Zhang Z, Chen L,
Li B, Li Y, Wang Y, Yang H, Han Q and Zhu Y: Glucose-6-phosphate
dehydrogenase and NADPH oxidase 4 control STAT3 activity in
melanoma cells through a pathway involving reactive oxygen species,
c-SRC and SHP2. Am J Cancer Res. 5:1610–1620. 2015.PubMed/NCBI
|
|
71
|
Liao SL, Lai SH, Tsai MH and Weng YH:
Cytokine responses of TNF-α, IL-6, and IL-10 in G6PD-deficient
infants. Pediatr Hematol Oncol. 31:87–94. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Korkaya H, Kim GI, Davis A, Malik F, Henry
NL, Ithimakin S, Quraishi AA, Tawakkol N, D'Angelo R, Paulson AK,
et al: Activation of an IL6 inflammatory loop mediates trastuzumab
resistance in HER2+ breast cancer by expanding the
cancer stem cell population. Mol Cell. 47:570–584. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Hu T, Chang YF, Xiao Z, Mao R, Tong J,
Chen B, Liu GC, Hong Y, Chen HL, Kong SY, et al: miR-1 inhibits
progression of high-risk papillomavirus-associated human cervical
cancer by targeting G6PD. Oncotarget. 7:86103–86116.
2016.PubMed/NCBI
|
|
74
|
Coda DM, Lingua MF, Morena D, Foglizzo V,
Bersani F, Ala U, Ponzetto C and Taulli R: SMYD1 and G6PD
modulation are critical events for miR-206-mediated differentiation
of rhabdomyosarcoma. Cell Cycle. 14:1389–1402. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Alvarez ML: Faster experimental validation
of microRNA targets using cold fusion cloning and a dual
firefly-Renilla luciferase reporter assay. Methods Mol Biol.
1182:227–243. 2014. View Article : Google Scholar : PubMed/NCBI
|