Introduction
Liver fibrosis and its end-stage disease (cirrhosis)
are major health problems worldwide. Liver fibrosis results in
liver failure and portal hypertension, which frequently require
liver transplantation at later stages. The activation of hepatic
stellate cells (HSCs) serves an important role in the process of
liver fibrosis. HSCs are nonparenchymal, quiescent cells that store
vitamin A and maintain a normal basement membrane-type matrix in an
undamaged liver. However, liver injury induces an ‘activation’
process in HSCs that results in vitamin A loss, proliferation, and
the synthesis of a ‘fibrotic’ matrix that is rich in type I
collagen (1).
CCAAT enhancer binding protein-α (C/EBP-α) is a
transcription factor expressed only in certain tissues, including
the liver (2,3). It was previously demonstrated that
C/EBP-α may induce HSC apoptosis in vitro (4) and in vivo (5). In addition, C/EBP-α results in
differential effects on apoptosis in hepatocytes and HSCs in
vitro and in vivo, exerting little hepatocyte toxicity
(6). These previous results
suggested that C/EBP-α may be an effective anti-liver fibrosis
therapy that results in little hepatocyte damage.
Epigenetic mechanisms, including DNA methylation and
histone tail modifications, are primary contributors to the
function of C/EBP-α in numerous types of tumor, including lung
cancer (7), acute myeloid leukemia
(8), head and neck squamous cell
carcinoma (9) and pancreatic
cancer (10). However, there are a
limited number of studies on the epigenetic modifications of
C/EBP-α in fibrosis. Therefore, the present study investigated
whether the acetylation of C/EBP-α may induce HSC-T6 cellular
apoptosis, and the associated mechanism involved. The effects of
trichostatin A (TSA), suberoylanilide hydroxamic acid (SAHA) and
nicotinamide on the HSC line HSC-T6 and the hepatocyte line BRL-3A
were investigated, and it was observed that HSC-T6 cells were more
sensitive to TSA compared with BRL-3A cells. Therefore, TSA was
selected to further investigate its effect on apoptosis in HSC-T6
cells and the acetylation of C/EBP-α. The results of the present
study will improve understanding of the C/EBP-α mechanism in
hepatic fibrosis.
Materials and methods
Materials
Anti-C/EBP-α (cat. no. sc-61-G) was obtained from
Santa Cruz Biotechnology, Inc. (Dallas, Texas, USA), horseradish
peroxidase (HRP)-conjugated goat anti-mouse immunoglobulin (Ig)G
(cat. no. SA0001-1) and goat anti-rabbit IgG (cat. no. SA0001-1)
were obtained from ProteinTech Group, Inc. (Chicago, IL, USA).
Anti-β-actin (A5441) was purchased from Sigma-Aldrich (Merck KGaA,
Darmstadt, Germany). Antibodies against Caspase-3 (cat. no. #9662,
which recognizes the procaspase and cleaved Caspase forms), cleaved
Caspase-3 (cat. no. #9661, which detects only the cleaved form),
Caspase-8 (cat. no. #4790, which recognizes the procaspase and
cleaved Caspase forms), Caspase-9 (cat. no. #9508, which recognizes
the procaspase and cleaved Caspase forms), and acetylated lysine
(cat. no. #9441) were purchased from Cell Signaling Technology,
Inc. (Danvers, MA, USA). Anti-Caspase-12 (cat. no. 3282-100, which
recognizes the procaspase and cleaved Caspase forms) was purchased
from BioVision, Inc. (Milpitas, CA, USA). The Cell Counting Kit-8
(CCK-8) assay kit was purchased from Dojindo Molecular
Technologies, Inc. (Kumamoto, Japan). NE-PER nuclear and
cytoplasmic extraction reagents and the Pierce
Co-Immunoprecipitation (co-IP) kit were purchased from Thermo
Fisher Scientific, Inc. (Waltham, MA, USA). TRIzol reagent was
obtained from Sangon Biotech Co., Ltd. (Shanghai, China). The
PrimeScript RT Reagent kit (Perfect Real Time) and SYBR Premix Ex
Taq kit (Perfect Real Time) were obtained from Takara Biotechnology
Co., Ltd. (Dalian, China). TSA and nicotinamide were purchased from
Sigma-Aldrich (Merck KGaA). SAHA was obtained from Cayman Chemical
Company (Ann Arbor, MI, USA). Epigallocatechin gallate (EGCG) was
obtained from YuanYe Biotechnology Co., Ltd. (Shanghai, China).
Cell culture
The immortalized rat liver stellate cell line HSC-T6
was established by Dr. Scott L. Friedman and generously provided by
his student, Professor Lie-Ming Xu (Shanghai University of
Traditional Chinese Medicine, Shanghai, China). The rat hepatocyte
cell line BRL-3A was purchased from the American Type Culture
Collection (Manassas, VA, USA). The two cell lines were cultured in
Dulbecco's modified Eagle's medium supplemented with 10% fetal
bovine serum (both from Gibco; Thermo Fisher Scientific, Inc.) and
standard antibiotics in a 95% air and 5% CO2 humidified
atmosphere at 37°C.
Construction of the C/EBP-α
replication-defective recombinant adenovirus (Ad-C/EBP-α)
The plasmid PDC316 was constructed in the lab
previously (4). Adenovirus
construction, amplification, purification and titer detection were
performed as described previously (4).
CCK-8 assay
HSC-T6 and BRL-3A cells were seeded onto 96-well
plates at a density of 1×105 cells/well. Cells were
treated with varying concentrations of TSA, SAHA and nicotinamide
for 48 h at 37°C (Fig. 1). Cell
viability was measured using the CCK-8 kit. Control cells were
treated with 10% dimethyl sulfoxide for 48 h. Following the
addition of the test compounds, 10 µl CCK-8 was added to each well,
incubated for 1–3 h at 37°C, and optical density was read at a
wavelength of 450 nm in a VMax kinetic microplate reader (Molecular
Devices, LLC, Sunnyvale, CA, USA).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
Following treatment with TSA for the indicated times
(0, 1, 2, 4, 8, 12, 24, 36 and 48 h) at concentration of 0.1
µmol/l, total RNA from HSC-T6 cells was extracted using TRIzol
reagent, according to the manufacturer's protocol. Briefly, total
RNA (1 µg) was reverse-transcribed using a PrimeScript RT kit
(Takara Biotechnology Co., Ltd.) with oligodT primer and six random
primers, to obtain the cDNA. The cDNA was amplified by PCR. PCR was
performed in a Rotor Gene thermal cycler (RG-3000; Qiagen GmbH,
Hilden, Germany) under conditions optimized for efficient
amplification of the respective genes and the reference gene.
Amplification was performed as follows: Following the initial
activation step at 95°C for 5 min, 40 cycles of denaturation at
95°C for 5 sec and annealing at 60°C for 30 sec were performed.
qPCR (11) experiments were
conducted in three biological replicates. The following primer
sequences were used: Rat C/EBP-α, 5′CGG TGG ATA AGA ACA GCA ACGA3′
(sense) and 5′GCG GTC ATT GTC ACT GGT CAAC3′ (antisense); and rat
β-actin, 5′AGG ATG CAG AAG GAG ATT ACT GC3′ (sense) and 5′AAA ACG
CAG CTC AGT AAC AGT GC 3′ (antisense).
Western blot analysis
Following treatment at 37°C with 0.1 µmol/l TSA for
the indicated times (0, 1, 2, 4, 8, 12, 24, 36 and 48 h), or
treatment with TSA and/or Ad-C/EBP-α with a range of dosages,
HSC-T6 cells were collected, washed in cold PBS, and subsequently
lysed in ice-cold radioimmunoprecipitation assay buffer [10 µM
Tris-HCl (pH 8.0), 100 mM NaCl, 1 mM EDTA, 1% Nonidet P-40, 0.5%
sodium deoxycholate, 0.1% SDS, and 10 µl/ml protease inhibitor
cocktail] on ice for 45 min. Lysates were cleared by centrifugation
at 13,000 × g for 30 min at 4°C, and total protein concentration
was determined using a Bicinchoninic Acid Protein Assay kit.
Extracted cellular proteins (10 µg) from each sample were denatured
in SDS containing sample buffer, applied to a 10–12% SDS-PAGE gel
and transferred to nitrocellulose membranes. Following blocking in
5% non-fat milk for 1 h at room temperature, the membranes were
incubated overnight at 4°C with primary antibodies against
anti-C/EBP-α (1:200), anti-Caspase-3 (1:1,000), anti-Caspase-8
(1:1,000), anti-Caspase-9 (1:1,000) or anti-Caspase-12 (1:1,000),
respectively. β-actin (1:1,000) was used as a loading control.
Membranes were subsequently incubated for 1 h at 4°C with the
appropriate HRP-conjugated secondary antibody. Protein bands were
visualized using an Enhanced Chemiluminescence Assay kit (Thermo
Fisher Scientific, Inc.). ImageJ 1.50e (National Institutes of
Health, Bethesda, MD, USA) was used for densitometric analysis.
Nuclear and cytoplasmic protein
extraction
HSC-T6 cells (~1×106) were harvested
following treatment with 0.1 µmol/l TSA for 12 h at 37°C. Cells
were washed in cold PBS, and nuclear and cytoplasmic proteins were
extracted using NE-PER nuclear and cytoplasmic extraction reagents,
according to the manufacturer's protocol. Proteins were stored at
−80°C until ready for use in subsequent experiments.
Co-IP analysis of C/EBP-α and the
acetylated lysine antibody in HSC-T6 cells
Following treatment with TSA at a concentration of
0.1 µmol/l for 12 h at 37°C, co-IP analysis of C/EBP-α and the
acetylated lysine antibody was performed in HSC-T6 cells using a
co-IP kit, according to the manufacturer's protocol. C/EBP-α and
the acetylated lysine antibody were coupled to AminoLink Plus
Coupling Resin. HSC-T6 cell lysate was prepared with IP lysis/wash
buffer on ice for 5 min. Purified HSC-T6 cell lysate was incubated
with C/EBP-α, acetylated lysine antibody-specific MAb-conjugated
agarose resin overnight at 4°C. Following elution, proteins samples
were prepared for western blot analysis using antibodies against
C/EBP-α or acetylated lysine as aforementioned. A sample of the
whole input lysate served as a positive control, and normal mouse
IgG (Beyotime Institute of Biotechnology, Haimen, China) was used
as negative control.
Statistical analysis
All experiments were repeated three independent
times. Statistical analysis was performed using SPSS software,
version 11.0 (SPSS, Inc., Chicago, IL, USA). Data were expressed as
the mean ± standard deviation. Comparisons were performed using
Student's t-test. For multiple comparisons, repeated measures
analysis of variance and Dunnett's test were used. P<0.05 was
considered to indicate a statistically significant difference.
Results
TSA inhibits the proliferation of
HSC-T6 cells to a greater extent compared with the proliferation of
BRL-3A cells, following treatment with the three histone
deacetylase inhibitors (HDACIs)
HSC-T6 and BRL-3A cells exhibited differential
responses to the three HDACIs. HSC-T6 cells were more sensitive to
TSA compared with BRL-3A cells. Proliferation was inhibited by 0.05
µmol/l and 0.2 µmol/l TSA. The rate of proliferation inhibition was
67.54% in HSC-T6 cells and 22.21% in BRL-3A cells (P<0.01).
However, compared with treatment with TSA, BRL-3A cells were more
sensitive to SAHA and nicotinamide (Fig. 1). The ideal medicine to treat liver
fibrosis is one that is able to inhibit HSCs while exerting a small
effect on hepatocytes. Therefore, TSA was selected for the
subsequent experiments in HSC-T6 cells.
Treatment with TSA alone or in
combination with Ad-C/EBP-α induces the expression of apoptotic
markers in HSC-T6 cells
Varying doses of TSA alone or in combination with
100 multiplicity of infection (MOI) Ad-C/EBP-α were used to detect
the expression of apoptotic markers of Caspase-3, −8, −9 and −12
levels in HSC-T6 cells (Fig. 2).
It was observed that TSA alone and in combination with 100 MOI
Ad-C/EBP-induced apoptosis in HSC-T6 cells. Treatment with TSA
induced apoptosis through Caspase-9 and Caspase-12, whereas
Ad-C/EBP-α induced apoptosis primarily through the Caspase-9
pathway. However, activated Caspase-8 was not detected. These
results suggested that TSA may enhance C/EBP-α-induced apoptosis of
HSCs.
Inherent C/EBP-α protein expression in
HSC-T6 cells varies, although mRNA does not alter
In order to further investigate the effect of TSA on
C/EBP-α expression in HSC-T6 cells, they were first treated with
TSA at concentration of 0.1 µmol/l, and subsequently harvested for
RT-qPCR and western blot analyses at 0, 1, 2, 4, 8, 12, 24, 36 and
48 h. RT-qPCR analysis demonstrated that C/EBP-α mRNA expression
did not alter significantly, however it was observed that C/EBP-α
protein expression increased and reached a peak between 4 and 12 h,
prior to decreasing gradually, indicating that C/EBP-α may be
processed post-transcriptionally (Fig.
3).
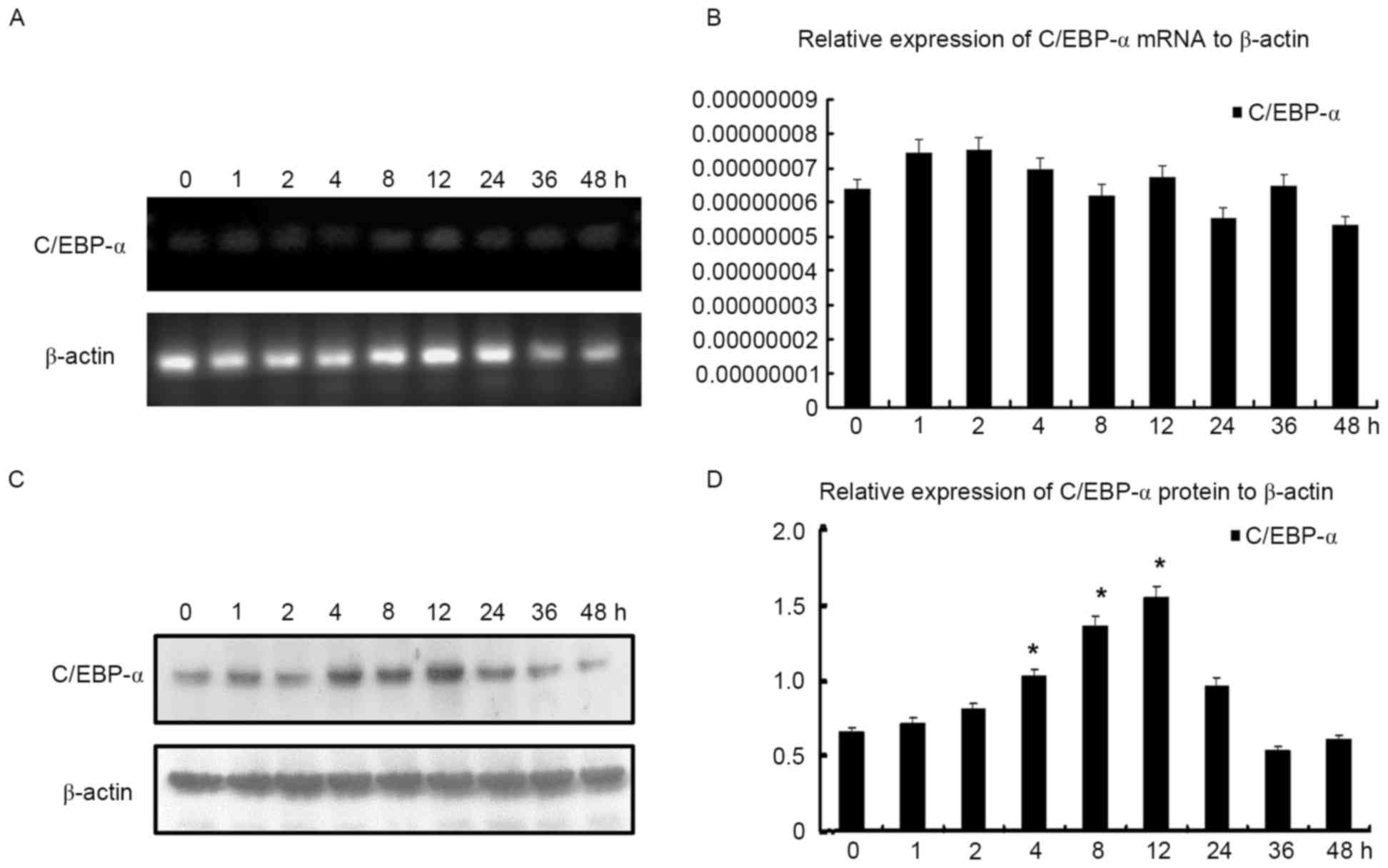 | Figure 3.C/EBP-α mRNA and protein levels
following treatment with TSA at the indicated times. (A)
Representative image of C/EBP-α mRNA level following treatment with
TSA for 0, 1, 2, 4, 8, 12, 24, 36 and 48 h, demonstrated by agarose
gel electrophoresis. (B) C/EBP-α mRNA level following treatment
with TSA at 0, 1, 2, 4, 8, 12, 24, 36 and 48 h as determined by
quantitative polymerase chain reaction analysis. (C) C/EBP-α
protein level following treatment with TSA at 0, 1, 2, 4, 8, 12,
24, 36 and 48 h, analyzed by western blotting. (D) Densitometric
analysis of the protein expression of C/EBP-α. *P<0.05 vs.
control. C/EBP-α, CCAAT enhancer binding protein-α; TSA,
trichostatin A. |
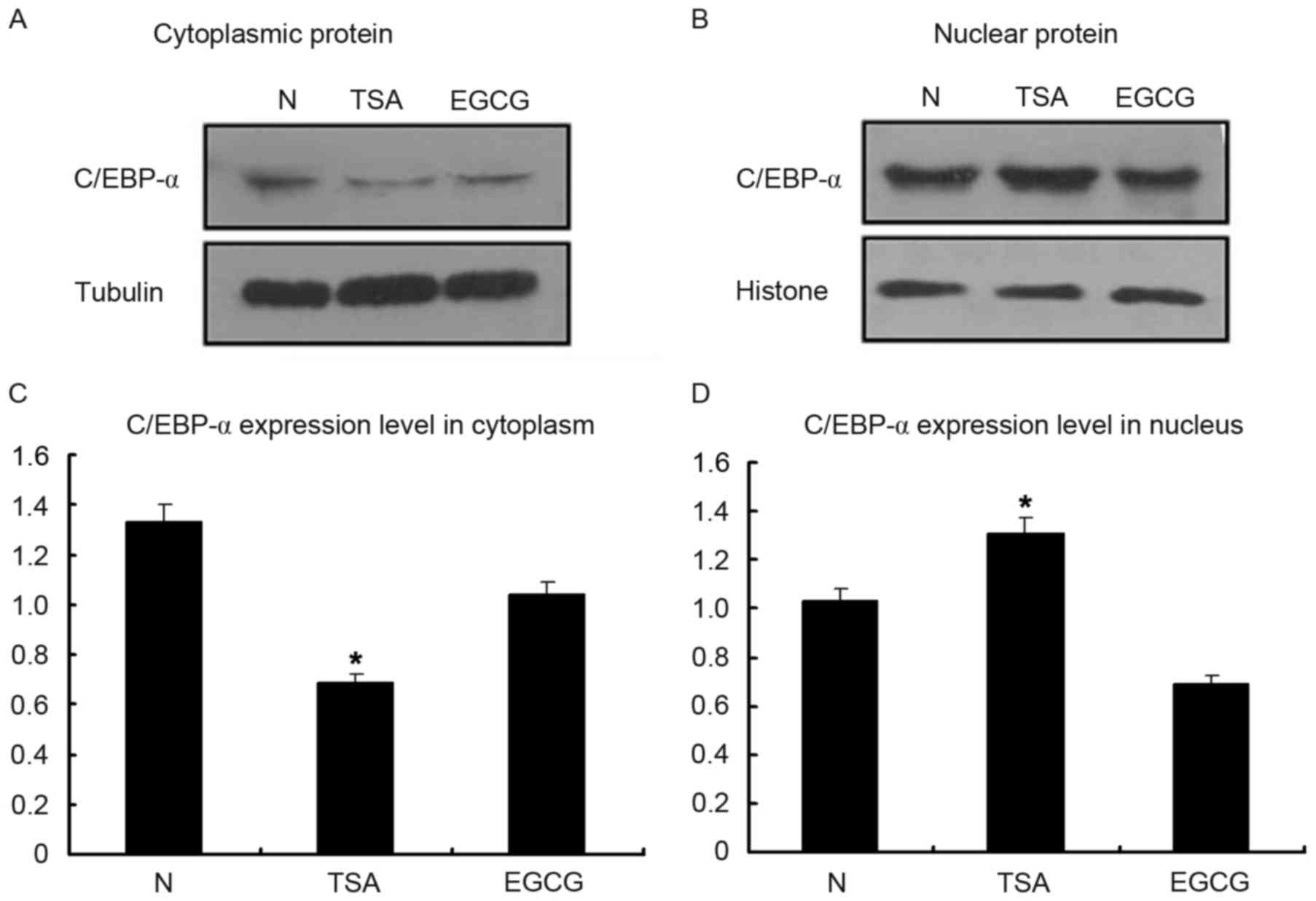
Inherent C/EBP-α protein expression
increases primarily in the nuclei of HSC-T6 cells
Following treatment with TSA for 12 h at a
concentration of 0.1 µmol/l, cells were harvested to examine the
nuclear and cytoplasmic proteins. C/EBP-α protein expression was
decreased in the cytoplasm following treatment with EGCG, which
served as the negative control (Fig.
4). However, it was observed that C/EBP-α protein expression
was increased in the nuclear fraction following treatment with TSA,
verifying that C/EBP-α predominantly functions in the nucleus.
C/EBP-α lysine acetylation increases
following treatment with TSA, as determined by co-IP analysis
Following treatment with TSA for 12 h at
concentration of 0.1 µmol/l, cells were harvested to examine the
lysine acetylation of C/EBP-α by co-IP analysis. As presented in
Fig. 5, immunoprecipitation of
cell lysates with the anti-acetylated lysine antibody
coprecipitated with the C/EBP-α antibody, and the C/EBP-α antibody
additionally coprecipitated with the anti-acetylated lysine
antibody. Western blot analysis was performed to detect the
expected bands; compared with untreated cells, lysine acetylation
increased following treatment with TSA.
 | Figure 5.C/EBP-α lysine acetylation, as
determined by co-immunoprecipitation analysis in HSC-T6 cells. (A)
Cell lysates were subjected to immunoprecipitation with the C/EBP-α
antibody, followed by immunoblotting with anti-acetylated lysine,
following treatment with TSA. (B) Cell lysates were subjected to
immunoprecipitation with the acetylated lysine antibody, followed
by immunoblotting with C/EBP-α, following treatment with TSA.
C/EBP-α, CCAAT enhancer binding protein-α; TSA, trichostatin A; WB,
western blotting; NC, negative control; PC, positive control; IP,
immunoprecipitation. |
Discussion
The exponential growth in the amount of research
into HDACs has been driven by the ability of HDACIs to modulate
transcriptional activity. HDACIs are able to block angiogenesis and
the cell cycle, in addition to promoting apoptosis and
differentiation (12. They belong to a heterogeneous class of
compounds that includes derivatives of short chain fatty acids,
hydroxamic acids, cyclic tetrapeptides, and benzamides. Of the
hydroxamic acids, TSA and SAHA are commonly used HDACIs. It has
been reported that TSA exerts a potent antifibrogenic effect on
HSCs (13). It is possible that
TSA induces or maintains the expression of genes essential for the
quiescent phenotype of stellate cells. Alternatively, gene products
induced by TSA may interfere with the signaling pathways involved
in stellate cell activation. Thirdly, TSA may directly repress the
transcription of genes that are induced during the activation
process (14). In the present
study, three HDACIs were applied to determine their inhibitory
effects on the growth of HSC-T6 and BRL-3A cells. The results
demonstrated that HSC-T6 cells were more sensitive to TSA compared
with BRL-3A cells; therefore, TSA was selected for use in the
subsequent experiments.
The mechanisms underlying apoptosis are complex and
involve an energy-dependent cascade of molecular events. Protein
expression of Caspase-3, −8, −9, and −12 was evaluated in the
present study, in order to investigate the apoptotic pathways in
HSC-T6 and BRL-3A cells. A previous study demonstrated that
Caspase-9 was activated in Ad-C/EBP-α-induced apoptosis (6). In the present study, it was observed
that TSA induced HSC-T6 cellular apoptosis through the Caspase-9
and −12 pathways. However, the Caspase-12 pathway may be inhibited
by co-action with Ad-C/EBP-α.
Post-translational modifications (PTMs) are
important for regulating the functions of a number of eukaryotic
proteins. The lysine side chain is thus a target of numerous PTMs.
Lysine acetylation is now known to occur in >80 transcription
factors, multiple nuclear regulators and various cytoplasmic
proteins (14). Lysine residues
are targets for acetylation and ubiquitination, and one of these
modifications appears to prevent the other. Deacetylation by HDACs,
in numerous cases, is a prerequisite for subsequent ubiquitination.
Conversely, acetylation may protect a protein from ubiquitination
and degradation (15). Whether
C/EBP-α ubiquitination is involved requires further
investigation.
In conclusion, the results of the present study
suggested that TSA may increase C/EBP-α expression by increasing
its lysine acetylation in HSCs. However, the probable mechanism
requires further investigation. There are few reports on C/EBP-α
and its acetylation in hepatic fibrosis. However, it is essential
to understand this mechanism in order to treat hepatic
fibrosis.
Acknowledgements
The present study was supported by the PhD Start-up
Fund of the Natural Science Foundation of Guangdong Province (grant
no. 2015A030310031), the Health and Family Planning System of
Scientific Research Project of Shen Zhen (grant no. 201401048),
Perking University, Shenzhen Hospital Science and Research
Foundation (grant no. 201401), the National Natural Science
Foundation of China (grant no. 81470857, to X.P.L.), and the
Shanghai Natural Science Foundation (grant no. 134119b1100).
References
|
1
|
Reeves HL and Friedman SL: Activation of
hepatic stellate cells-A key issue in liver fibrosis. Front Biosci.
7:d808–d826. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mueller BU and Pabst T: C/EBP alpha and
the pathophysiology of acute myeloid leukemia. Curr Opin Hematol.
13:7–14. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lane MD, Tang QQ and Jiang MS: Role of the
CCAAT enhancer binding proteins (C/EBPs) in adipocyte
differentiation. Biochem Biophys Res Commun. 266:677–683. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang X, Huang G, Mei S, Qian J, Ji J and
Zhang J: Over-expression of C/EBP-alpha induces apoptosis in
cultured rat hepatic stellate cells depending on p53 and peroxisome
proliferator-activated receptor-gamma. Biochem Biophys Res Commun.
380:286–291. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mei S, Wang X, Zhang J, Qian J and Ji JL:
In vivo transfection of C/EBP-alpha gene could ameliorate
CCL(4)-induced hepatic fibrosis in mice. Hepatol Res. 37:531–539.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tao LL, Cheng YY, Ding D, Mei S, Xu JW, Yu
J, Ou-Yang Q, Deng L, Chen Q, Li QQ, et al: C/EBP-α ameliorates
CCl(4)-induced liver fibrosis in mice through promoting apoptosis
of hepatic stellate cells with little apoptotic effect on
hepatocytes in vitro and in vivo. Apoptosis. 17:492–502. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tada Y, Brena RM, Hackanson B, Morrison C,
Otterson GA and Plass C.: Epigenetic modulation of tumor suppressor
CCAAT/Enhancer binding protein a activity in lung cancer. J Natl
Cancer Inst. 98:396–406. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hackanson B, Bennett KL, Brena RM, Jiang
J, Claus R, Chen SS, Blagitko-Dorfs N, Maharry K, Whitman SP,
Schmittgen TD, et al: Epigenetic modification of CCAAT/enhancer
binding protein alpha expression in acute myeloid leukemia. Cancer
Res. 68:3142–3151. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bennett KL, Hackanson B, Smith LT,
Morrison CD, Lang JC, Schuller DE, Weber F, Eng C and Plass C:
Tumor suppressor activity of CCAAT/enhancer binding protein alpha
is epigenetically down-regulated in head and neck squamous cell
carcinoma. Cancer Res. 67:4657–4664. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kumagai T, Akagi T, Desmond JC, Kawamata
N, Gery S, Imai Y, Song JH, Gui D, Said J and Koeffler HP:
Epigenetic regulation and molecular characterization of C/EBP alpha
in pancreatic cancer cells. Int J Cancer. 124:827–833. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gridelli C, Rossi A and Maione P: The
potential role of histone deacetylase inhibitors in the treatment
of non-small-cell lung cancer. Crit Rev Oncol Hematol. 68:29–36.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Niki T, Rombouts K, De Bleser P, De Smet
K, Rogiers V, Schuppan D, Yoshida M, Gabbiani G and Geerts A: A
histone deacetylase inhibitor, trichostatin A, suppresses
myofibroblastic differentiation of rat hepatic stellate cells in
primary culture. Hepatology. 29:858–867. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang XJ and Seto E: Lysine acetylation:
Codified crosstalk with other posttranslational modifications. Mol
Cell. 31:449–461. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Glozak MA, Sengupta N, Zhang X and Seto E:
Acetylation and deacetylation of non-histone proteins. Gene.
363:15–23. 2005. View Article : Google Scholar : PubMed/NCBI
|