Introduction
Aortic dissection (AD) is symbolized by a tear in
the tunica intima of the aorta and consequently causing blood to
flow between layers of aortic wall and forcing the layers apart. It
remains a life-threatening disease with high mortality, despite of
significant improvements in the diagnosis and surgical repair
(1). Degenerative remodeling
within the medial layer (2) is
considered as the most important pathogenesis factor. It is
characterized by the loss of smooth muscle cells (SMCs) (3), destruction of the extracellular
matrix (ECM) and a combination of excessive destruction and
insufficient repair. Surgical repair or endovascular strategy
exists to treat AD at present. However, the result is not always
satisfactory. Although effective in preventing rupture, surgical
procedure is invasive and associated with a high mortality and
morbidity (4). Endovascular
aneurysm repair (EVAR) are minimally invasive interventions, yet
they have anatomical and clinical limitations and drawbacks such as
endoleaks and graft migrations (5). Therefore, an alternative that reduces
surgical invasion and the risk of rupture is needed.
It has been demonstrated that stem cell therapy
could not only enhance the stability of the aneurysm sac, but also
reduce the inflammatory process and reinforce arterial layers.
Mesenchymal stem cells (MSCs) are multipotent stem cells and can
home at sites of injury and contribute to tissue repair, and can be
easily harvested from bone marrow, adipose tissue. In vitro,
MSCs have been shown to differentiate into SMC-like cells upon
PDGF-BB stimulation (6), while
in vivo to contribute to healing of injured arteries
(7). Previous study showed that
MSCs could attenuate aortic aneurysm growth in model mice (8). It later had been demonstrated on
animals that MSCs stabilize already-formed aortic aneurysms and MSC
is a potential therapeutic intervention (9). MSCs attenuate aortic aneurysms on
mice and thus offers a promising insight into biologic therapies
for future medical treatment of aortic disease in human (10). Our previous animal study (11) demonstrated that bone marrow cells
are activated and recruited to diseased aortic wall when AD
occurred. Our findings highlight the protective role of bone marrow
cells in response to aortic stress and aortic inflammation. Another
study for aortic tissues (12)
confirmed that stem cells are more abundant in dissected aortic
tissue, and differentiation into SMCs within the diseased aortic
wall indicate stem cells a potential contributor to aortic
repair.
Given the current studies on MSCs' active role in
aortic aneurysm and dissection (13–15),
we hypothesis that MSCs in AD patient might have some deficits thus
patients consequently manifest with insufficient repair and thus
the repair-destruction balance is broken. As a matter of fact, some
researchers suggested that the dysregulation of MSCs' activity may
contribute to the disease (16).
Thus, we profiled MSCs' gene expression both from AD patients and
healthy donors (HD) by transcriptome sequencing/RNA sequencing
(RNA-seq) in this study, aiming to discover genes that may play a
crucial role in MSCs' possible protective effect on AD. As a
technology of next generation sequencing (NGS), RNA-seq is widely
used to detect differentially expressed genes (DEGs) between two
gene expression patterns. The DEGs from AD-MSCs and HD-MSCs were
then selected, validated, and subjected to bioinformatic analyses,
including gene ontology analysis, pathway analysis, and network
analysis. Analyzing the potential molecular markers and the
possible relationship among the DEGs in MSCs will help give further
insight into MSCs' role and mechanism in AD.
Materials and methods
Patient and donor samples
The MSCs of AD patient (AD group, n=9; mean age,
55.0±9.6 years) were collected from their sternum bone marrow
during surgery while the MSCs of HDs (HD group, n=6; mean age,
49.5±12.2 years) are harvested from their ilia. Three samples of
each group were used for RNA-seq and all the samples were used for
quantitative PCR (qRT-PCR) verification. No significant difference
in age was found between the AD and HD groups (t=0.9775, df=13,
P=0.3461). Neither the AD group nor normal control had any history
of Marfan syndrome, bicuspid aortic valve or any other aortic
pathology. All patients had acute dissections with onset no earlier
than 14 days before surgery. All of them were confirmed to have
Stanford type B AD by preoperative examination or surgery. All the
patients with hypertension (n=9) were taking antihypertensives for
at least 3 months before operation. Among them, 5 were taking
calcium channel blockers, 4 were taking diuretics, and 2 were
taking angiotensin-converting enzyme inhibitors. Neither statins
nor other relevant medications were taken. A detailed sample
description is recorded in Table
I.
 | Table I.Patient information. |
Table I.
Patient information.
| Laboratory
code | Sex | Age | Group | RNA-seq | qRT-PCR |
|---|
| 1 | M | 62 | HD | Yes | Yes |
| 2 | F | 46 | HD | Yes | Yes |
| 3 | F | 34 | HD | Yes | Yes |
| 4 | F | 55 | AD | Yes | Yes |
| 5 | F | 59 | AD | Yes | Yes |
| 6 | M | 71 | AD | Yes | Yes |
| 7 | F | 37 | HD | No | Yes |
| 8 | M | 60 | HD | No | Yes |
| 9 | F | 58 | HD | No | Yes |
| 10 | M | 48 | AD | No | Yes |
| 11 | F | 59 | AD | No | Yes |
| 12 | F | 59 | AD | No | Yes |
| 13 | M | 57 | AD | No | Yes |
| 14 | M | 36 | AD | No | Yes |
| 15 | F | 51 | AD | No | Yes |
The study was approved by the Ethics Committee of
Changzheng Hospital, and all patients gave informed consent. All
samples used in this study were prepared in parallel based on
published methods.
Isolation of MSCs
The general procedures are referred to published
literatures. Density gradient centrifugation was applied to bone
marrow from both the AD patients and HD by using Ficoll
(Ficoll-Paque Premium 1.073; GE Healthcare Bio-Sciences AB,
Piscataway, NJ, USA). Detailed procedures are followed by
manufacturer's protocol.
The mononuclear cells obtained after centrifugation
were plated in non-coated 10-cm culture dish in low-glucose
Dulbecco's modified Eagle's medium (DMEM-LG; Invitrogen, Carlsbad,
CA, USA) supplemented with 5% UltraGRO™−Advanced Cell
Culture Supplement (AventaCell BioMedical Co., Ltd., Atlanta, GA,
USA) with no penicillin or streptomycin. The cells were cultured at
37°C in a humidified atmosphere of 5% CO2. Non-adherent
cells were removed from the culture dish after 2 days, and the
medium was changed every other day until the cultured MSCs reached
90% confluence (passage 0). Then, MSCs were removed from the dish
by treatment with 0.05% trypsin (Invitrogen) for 30 sec at 37°C and
then replated in another culture dish at a density of 2,000
cells/cm2 (passage 1). When 90% confluence was obtained,
the cells were trypsinized and replated in another fresh culture
dish (passage 2). These processes were repeated up to passage 2,
when MSCs were used for all experiments.
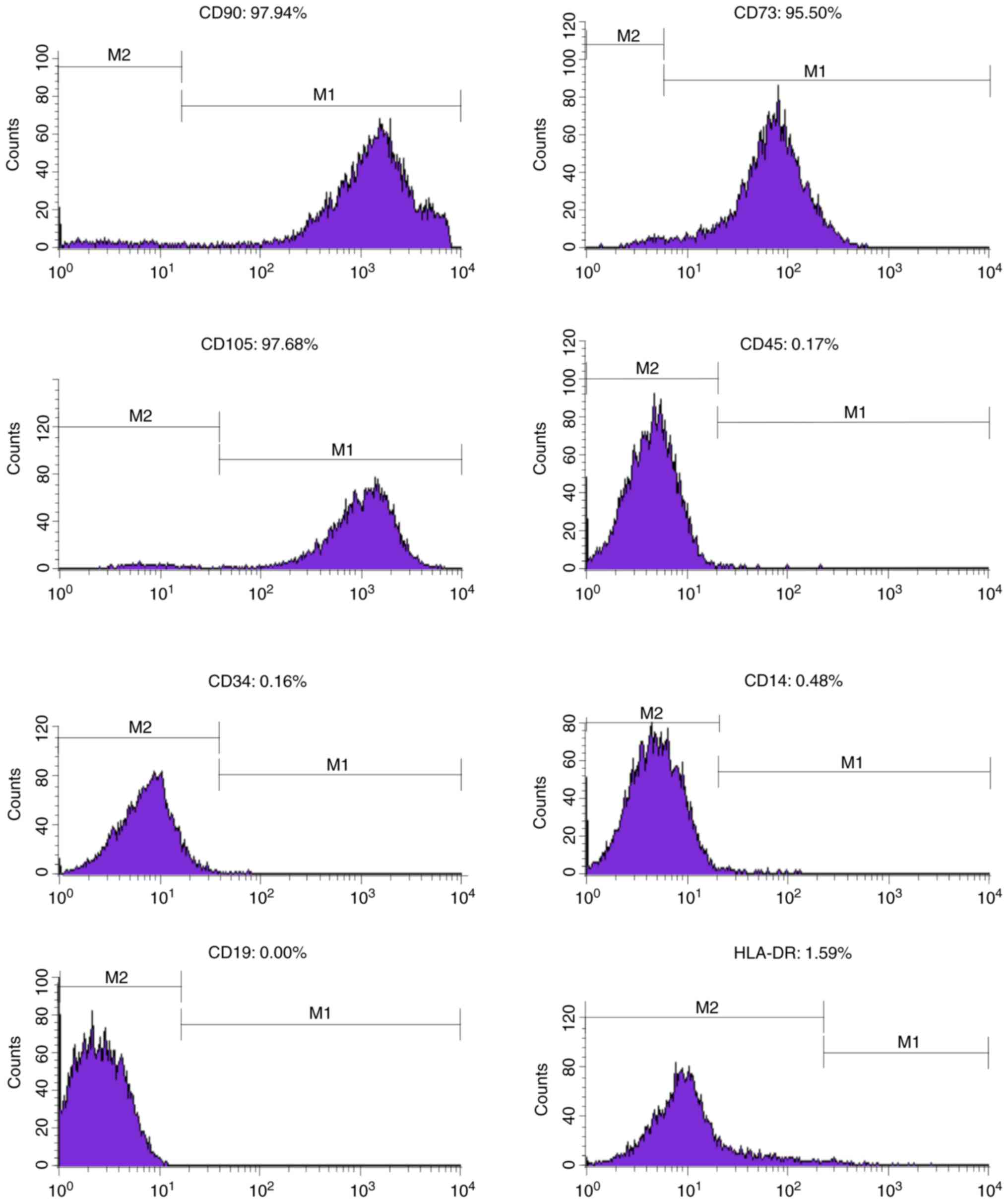
Identification of MSCs
To confirm the multipotentiality of MSCs used in our
research, experiments were performed in accordance with the minimal
criteria for defining multipotent MSCs proposed by the
International Society for Cellular Therapy (ISCT) (17).
The cultured plastic-adherent cells expressing the
markers CD73 (sic passim; eBioScience, San Diego, CA, USA), CD90
and CD105 but not expressing the markers CD14, CD19, CD34, CD45 and
HLA-DR were able to differentiate into adipocytes, osteoblasts and
chondrocyte induced by products of Stem Cell Technologies
(Vancouver, BC, Canada), which are respectively
MesenCult™ Adipogenic Differentiation Medium (human) and
MesenCult™ Osteogenic Stimulatory kit (human) and
MesenCult™−ACF Chondrogenic Differentiation Medium.
Manufacturer's manuals were referred.
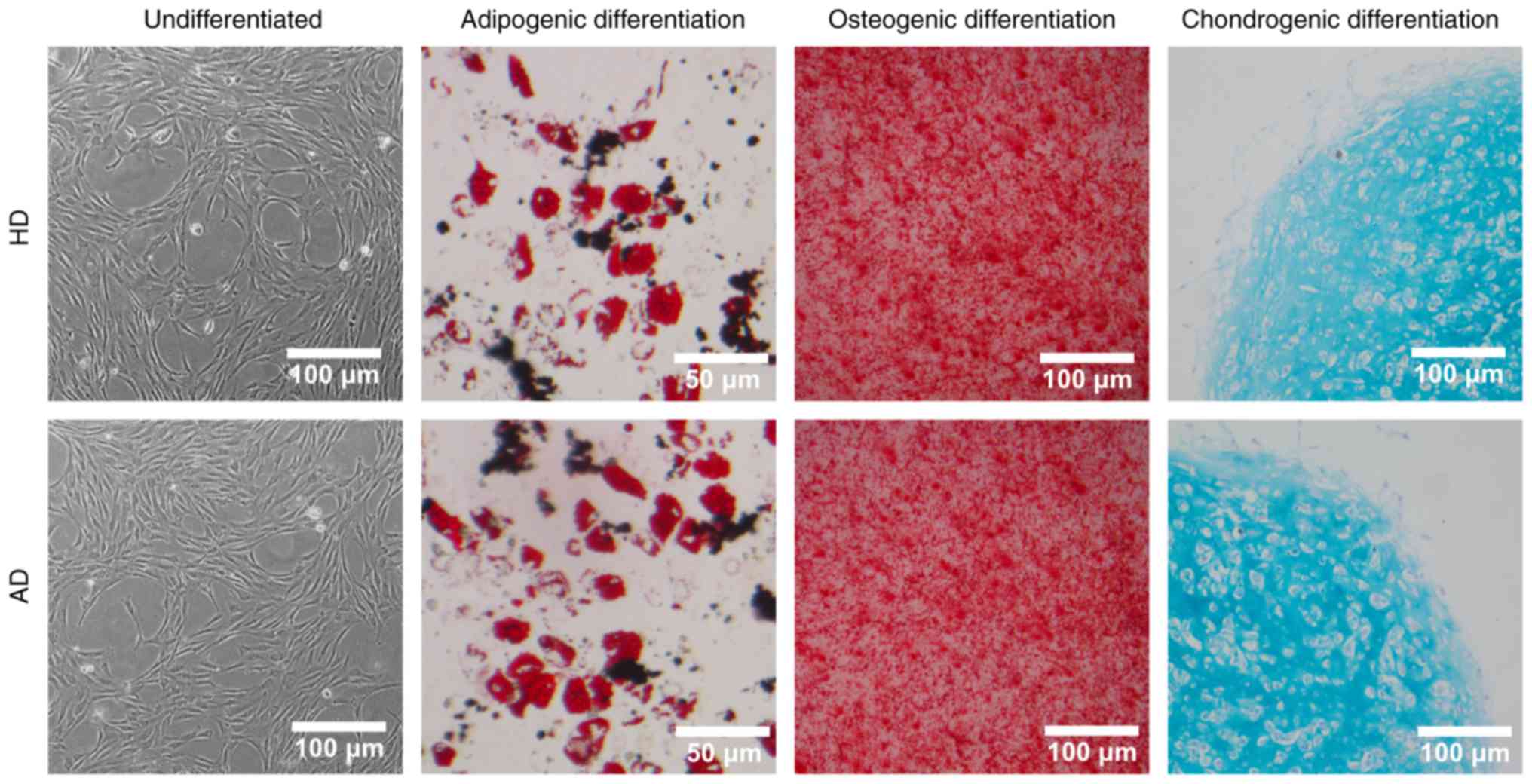
To determine whether the expanded MSC cultures
maintained multipotency differentiation characteristics, we tested
both HD-MSCs and AD-MSC for differentiation into adipogenic,
osteogenic and chondrogenic cell lines. MSCs cultured in adipogenic
differentiation medium showing lipid droplets were stained by Oil
Red O staining. Osteogenic differentiation was demonstrated by
calcium deposition, which was stained by Alizarin Red S.
Histological sections of chondrogenic pellet were stained with
Alcian Blue and Nuclear Fast Red. Undifferentiated AD-MSCs and
HD-MSCs were used as controls.
Proliferation assay
Cell proliferation was assessed with the Cell
Counting kit-8 (CCK-8) assay (Beyotime Institute of Biotechnology,
Haimen, China). Cells were seeded onto 96-well plates
(1×103 cells/well) and then OD at 450 nm was measured
from the 1st day to the 7th day after addition of 10 µl of CCK-8
solution to each well and a sequential incubation for 1 h at 37°C.
The assay was performed in triplicate.
Cell cycle assay
The cell cycle was analyzed using propidium iodide
(PI; Sigma-Aldrich, St. Louis, MO, USA) as described by Nicoletti
et al (18). The cell cycle
was blocked by reducing FBS to 0.1% for 24 h, and then the
concentration of FBS was returned to 10%. Three days later, the
cells were harvested for cell cycle analysis. First, the cells were
washed and fixed overnight in cold ethanol (70%). Then, the fixed
cells were washed and reconstituted in 250 µl of buffer (0.1% NP40,
0.2 mg/ml RNase, and 0.2 mg/ml PI) and incubated for 30 min at 4°C.
Ten thousand events were collected from each sample using a
FACSCalibur flow cytometer (BD Biosciences, San Jose, CA, USA). All
experiments were performed in triplicate. The data were analyzed
using CellQuest software (BD Biosciences).
RNA-seq
Total RNA from MSC cultures in passage 2 was
obtained using EZNA® Total RNA kit I (Omega Bio-Tek,
Inc., Norcross, GA, USA) according to the manufacturer's
instructions.
The RNA-seq was commercially commissioned to Jia
Laboratory (Life Science Institute, Zhejiang University, Hangzhou,
China). The data were generated by Hiseq2500 through NGS in fast
mode as single end. After sequencing completing,
configureBclToFastq.pl, a perl script from illumina®,
was run to get reads data in fastq format. Then we used Tophat to
map reads against hg19 reference transcript and genome. Through our
laboratory pipeline we counted each sample's reads mapped to each
gene of hg19, and got the result of difference expression of genes
through edgeR package (dispersion=0.04, other parameters used as
default).
Quantitative PCR (qRT-PCR)
qRT-PCR analyses were performed using 500 ng of mRNA
treated with EZNA® Total RNA kit I and reverse
transcribed with ReverTra Ace® qPCR RT Master mix with
gDNA Remover (Toyobo Co., Ltd., Osaka, Japan). Each reaction was
performed with 10 µl of EvaGreen qPCR Mastermix (Applied Biological
Materials Inc., Richmond, BC, Canada), 5 µl of cDNA (100 ng of
cDNA), 0.5 µl each primer (10 µM) and 4 µl of ddH2O. The
quantitative determination of mRNA levels was performed using The
Infinite® 200 PRO NanoQuant (Tecan Trading AG,
Männedorf, Switzerland). The reactions were performed in CFX
Connect™ Real-Time PCR Detection system (Bio-Rad
Laboratories, Inc., Philadelphia, PA, USA) using the following
program: 95°C for 10 min, followed by 40 cycles at 95°C for 10 sec,
60°C for 20 sec and 72°C for 15 sec, and then a final extension at
65°C to 95°C with increment of 0.5°C for 5 sec. Dissociation curve
analysis was used to demonstrate equal amplification efficiency of
a specific PCR product for all primers used in this study; all
primers demonstrated equal amplification efficiency and specific
PCR products through dissociation curve analysis. The determination
of fold expression change was calculated using Livak's
ΔΔCT method. Expression levels were estimated in
triplicate, and GAPDH were used as normalization genes. The primers
of tested genes are listed in Table
II.
 | Table II.qRT-PCR primers for the 9 selected
DEGs. |
Table II.
qRT-PCR primers for the 9 selected
DEGs.
| Gene | Forward primer
(5′–3′) | Reverse primer
(5′–3′) |
|---|
| GAPDH |
GTCAACGGATTTGGTCGTATTG |
TGGAAGATGGTGATGGGATTT |
| ABCA4 |
GGTTCCTGGACAGCTTCTCC |
CCAGACTGGCCTTGGAGAAG |
| CXCL1 |
TCCTGCTCCTGGTAGCCG |
TCCGCCCATTCTTGAGTGTG |
| CXCL5 |
GTCCTTCGAGCTCCTTGTGC |
CGTTCTTCAGGGAGGCTACC |
| EMX2 |
ACCTTCTACCCCTGGCTCAT |
GGCGTGTTCCAGCCTTAGAA |
| HTR7 |
TGGTGATCTCCGTGTGCTTC |
CTGATCACGCACAGGGTCAT |
| IGFBP2 |
TTCCGGGAGAAGGTCACTGA |
GAGGTTGTACAGGCCATGCT |
| NCAM1 |
CTGGAGGACTTCTACCCGGA |
TGGTTCCCCTCCCAAGTGTA |
| SERPINB7 |
GCCTTCACCAAGAGCGAAAC |
CTCAGGCAGCAGAACGTACA |
| SNAP25 |
GGGGCAATAATCAGGACGGA |
CCCATATCCAGGGCCATGTG |
Statistical analysis
Parametric data were expressed as mean ± standard
deviation (normally distributed) or median with inter-quartile
range (not normally distributed) and evaluated by Student's t-test
or ANOVA with Tukey's honest significant difference method for
comparisons between groups if possible. Wilcoxon test (also known
as Mann-Whitney test) or Kruskal-Wallis Rank Sum test was likewise
performed for non-parametric data. Shapiro-Wilk test and Bartlett
test are performed for normality and homogeneity of variance
respectively. All statistical analyses are conducted using R [R
Core Team (2017). R: A language and environment for statistical
computing. R Foundation for Statistical Computing, Vienna, Austria;
https://www.R-project.org/] or GraphPad
Prism™ software (GraphPad Software, Inc., La Jolla, CA,
USA). Probability values of <5% were considered significant. All
experiments were conducted in triplicate. Basic analysis of RNA-seq
data is done by Jia Laboratory and advanced analysis (including
KEGG/GO enrichment and PPI analysis) is done by Guangzhou RiboBio
Co., Ltd. (Guangzhou, China).
Results
In vitro characterization of MSC
cultures
After isolation and adherence to cell culture
dishes, MSCs were morphologically spindle-shaped, as is shown in
Fig. 1. The identity of the MSCs
was determined by confirming the panel of surface markers (positive
for CD90, CD73 and 105; negative for CD45, CD34, CD14, CD19 and
HLA-DR in Fig. 2) and multipotency
capacity of differentiation into adipogenic, osteogenic and
chondrogenic cells (Fig. 1).
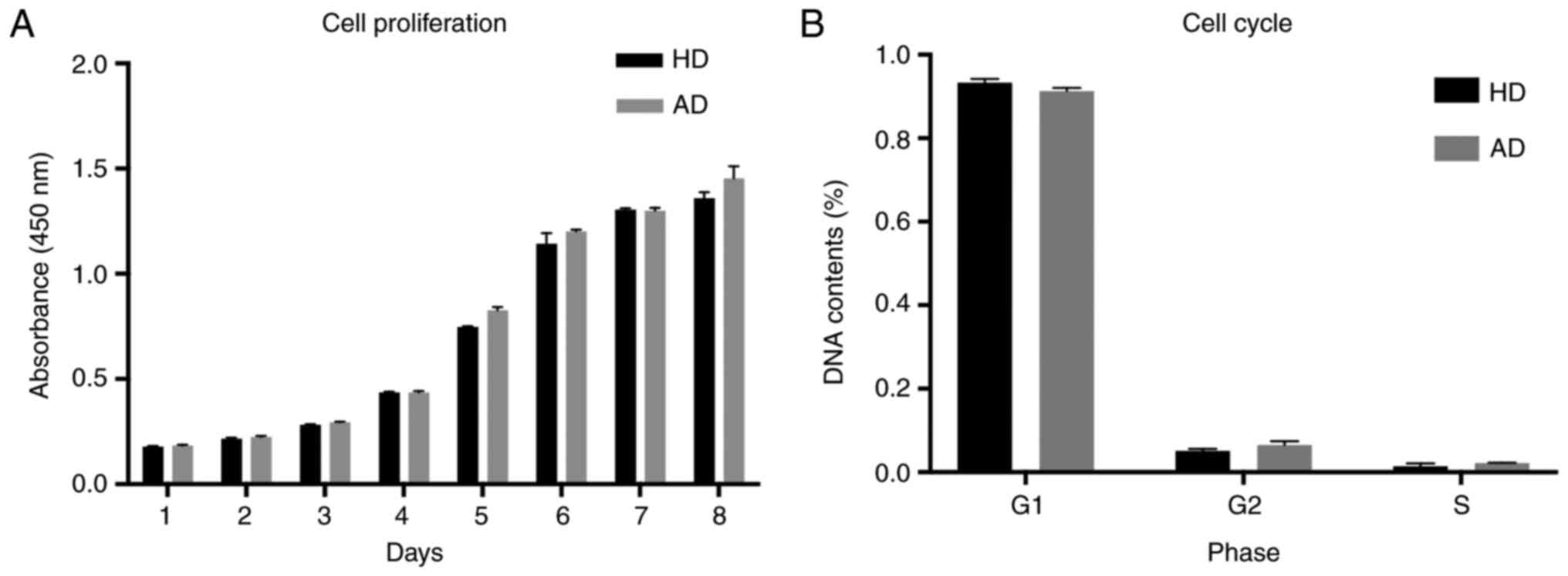
AD-MSCs displayed conserved
proliferation and cell cycle profiles compared with HD-MSCs
To verify whether AD-MSCs presented changes in their
proliferation potential and had cell cycle arrest, we performed
cell proliferation and flow cytometry assays and compared the
results with those of HD-MSCs. As observed in Fig. 3, no significant alterations were
observed for proliferation and cell cycle profiles of all MSC
cultures. These results indicate that MSC cultures conserved their
proliferation capacity despite the disease condition.
AD-MSCs molecular profile
Although AD-MSCs conserved their proliferation
profile, the molecular pattern of these cells could differ from
that of HD-MSCs. Thus, we determined the global gene expression
pattern for AD-MSCs and compared it with that for HD-MSCs. We
performed a comparative transcriptome analysis using an expression
profiling sequencing.
In this assay, 3 samples from different AD-MSCs were
compared with 3 from HD-MSCs. After the second passage, total RNA
was obtained from each MSC culture, and then RNA-seq was conducted.
Using a ≥2-fold change (FC) and <0.05 P-value as a cut-off to
define overexpression or downregulation, 201 genes were found to be
differentially expressed in all RNA-seq. Notably, 93 of these 201
genes were overexpressed in AD-MSCs, while 108 of these genes were
downregulated. The number of DEGs varies along with the alteration
of filters: a stricter filter results in a smaller DEGs profile.
Table III shows some common
filters and corresponding DEGs size. As is shown in Fig. 4, the volcano plot (Fig. 4A) intuitively exhibited the
distribution of DEGs and the hierarchical clustering in the heatmap
(Fig. 4B) of these DEGs suggests
that a common molecular signature exists for all AD-MSCs compared
to HD-MSCs. The filter for volcano plot and heatmap in Fig. 4 was P<0.01 and FC>4 on
account of the readability. A stricter filter results in a small
number of DEGs, which makes the plot easier to recognize.
 | Table III.The number of DEGs varies in
correspondence with the alteration of filters. |
Table III.
The number of DEGs varies in
correspondence with the alteration of filters.
| Filters | No. of DEGs | Up-regulated | Down-regulated |
|---|
| P<0.05,
FC>2 | 201 | 93 | 108 |
| P<0.05,
FC>4 | 56 | 30 | 26 |
| P<0.01,
FC>2 | 60 | 28 | 32 |
| P<0.01,
FC>4 | 30 | 12 | 18 |
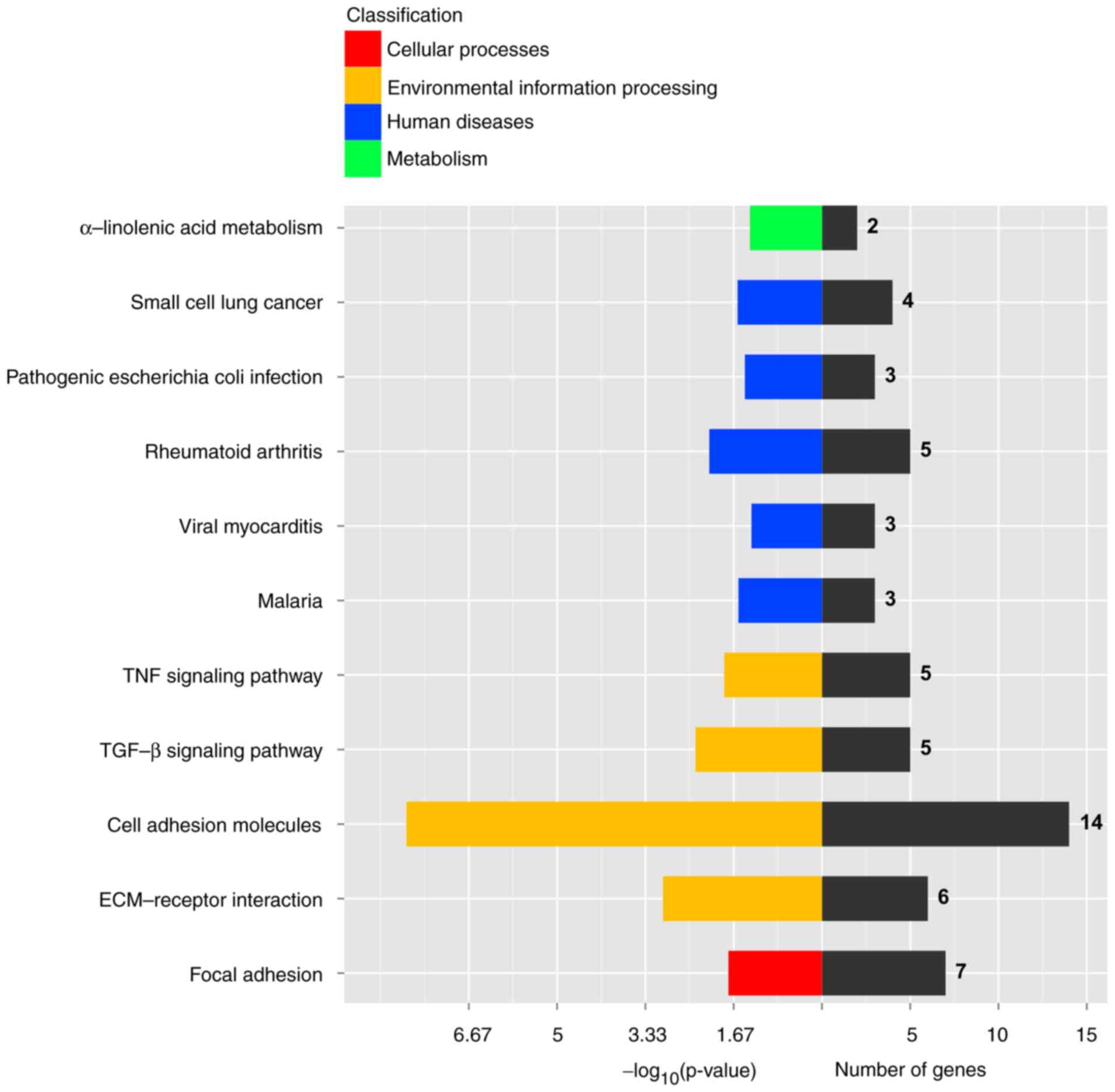
Further analysis of these results showed that the
genes found altered in AD-MSCs function in important signaling
pathways involved in MSCs' function. As is shown in KEGG analysis
(Fig. 5), DEGs are involved
primarily in cell adhesion molecules (CAMs), ECM-receptor
interaction, focal adhesion and TGF-β signaling etc. GO
analysis (Fig. 6) revealed DEGs
are enriched mainly in multicellular organismal development, system
development, organ development, anatomical structure morphogenesis
etc. Both KEGG and GO analysis illustrated AD-MSCs may have some
deficits in development or adhesion function.
qRT-PCR confirmed the molecular
signature of AD-MSCs
To confirm the obtained RNA-seq results, qRT-PCR of
some overexpressed and downregulated genes was performed. In this
case, the analysis was performed with a larger sample size: 6
AD-MSCs samples and 9 HD-MSCs samples. The age is normally
distributed (HD-MSCs: W=0.87117, P=0.2309; AD-MSCs: W=0.93065,
P=0.4875) and balanced between two groups (HD-MSCs: n=6, mean age,
49.5±12.3 years; AD-MSCs: n=9, 55.0±9.6 years; t=0.9282, df=8.988,
P=0.3776). The overexpressed genes, i.e., CXCL1, CXCL5, HTR7 and
SNAP25, and the downregulated genes, i.e., EMX2, NCAM1 and
IGFBP2 were used in this analysis. Detailed information about
the 9 genes are listed in Table
IV. These genes were chosen because all of them had been
previously described as related to MSCs functions or AD
pathogenesis. For example, both CXCL/CXCR signal axis and
IGFBP/IGF-signaling pathway are highly related to MSCs' function,
which will be discussed later. As is illustrated in Fig. 7, qRT-PCR validation for 9 selected
DEGs were consistent with the RNA-seq results (Fig. 7A), so was western blotting
validation for 2 selected DEGs (Fig.
7C). The hierarchical clustering analysis using the 9-gene
panel, acting as unsupervised learning, could even spontaneously
divided MSC samples into HD and AD groups correctly (Fig. 7B).
 | Table IV.Detailed information of the 9
selected DEGs, including expression level (reads), FC and P-values
acquired via RNA sequencing assay. |
Table IV.
Detailed information of the 9
selected DEGs, including expression level (reads), FC and P-values
acquired via RNA sequencing assay.
| Gene ID | HD1 | HD2 | HD3 | AD1 | AD2 | AD3 | logFC | P-value | FC |
|---|
| SNAP25 |
8 |
9 |
8 | 58 | 518 | 117 | 4.41 | 0.00 | 21.19 |
| CXCL5 |
8 |
5 |
6 | 176 | 45 | 20 | 3.32 | 0.00 | 10.00 |
| CXCL1 |
2 | 57 | 39 | 792 | 151 | 54 | 3.30 | 0.01 | 9.87 |
| HTR7 | 49 | 148 | 27 | 440 | 414 | 539 | 2.37 | 0.00 | 5.17 |
| IGFBP2 | 1,489 | 888 | 2,118 | 331 | 490 | 214 | −2.56 | 0.00 | 0.17 |
| NCAM1 | 189 | 575 | 160 | 43 | 54 | 97 | −2.58 | 0.00 | 0.17 |
|
SERPINB7 | 812 | 316 | 495 | 54 | 88 | 114 | −3.26 | 0.00 | 0.10 |
| SCN9A |
7 | 638 | 68 | 10 |
1 | 33 | −4.20 | 0.00 | 0.05 |
| EMX2 | 544 | 14 | 509 | 11 | 36 | 11 | −4.77 | 0.00 | 0.04 |
Analysis of PPI network and functional
annotation of genetic biomarkers for AD-MSC
We queried STRING database of 30 most statistically
significantly DEGs of AD by applying a stricter filter (P<0.01
and FC>4). As a result, functional modules were shown in
Fig. 8. Some genes are not shown
because no interactions were observed in the database. Our data
implied that CAMs (e.g., NCAM1) that were interacted with
chemokines (CXCL1), might be account for the MSCs'
functional changes. Another interaction network is collagen-related
molecules. Collagen, as well as elastin, is the main component of
aorta. This network implies MSCs may have some effect on collagen
in AD pathogenesis.
Discussion
The exact etiological mechanisms are not fully
established. As far as we know, AD is characterized by chronic
inflammation especially atherosclerotic changes and develops as a
result of ECM destruction within the aortic wall, where the
infiltrating macrophage release matrix metalloproteinases (MMPs),
inflammatory cytokines and chemokines, leading to a loss of elastin
in the aortic wall (19). In human
histological studies, increasing aneurysm diameter and rupture was
associated with a higher density of inflammatory cells in the
adventitia. Various stimuli have been linked to chronic
inflammation observed in AD. Therefore, the control of inflammation
may be an alternative strategy for treatment of AD. A number of
experimental investigations and clinical studies have attempted to
treat aortic disease using various drugs and factors to control the
inflammation, for example, angiotensin converting enzyme inhibitor
and statin (20,21), doxycycline (22,23),
nonsteroidal anti-inflammatory drugs (24) and c-jun N-terminal kinase inhibitor
(25). However, these
pharmacotherapies have still not been established for clinical
application because of their side effects caused by systemic
administration of these agents. Another disadvantage of using these
agents is that special equipment might be required to deliver them
locally for the treatment. Cell therapy with MSCs seem to be free
of severe systemic adverse reactions and the homing capacity makes
MSCs a ‘targeted drug’. As is mentioned above, cell therapy could
not only enhance the stability of the aneurysm sac, but also reduce
the inflammatory process and developing new reinforced arterial
layers in animals. Many researches have been focusing on cellular
therapies to attempt stabilizing AD and repairing the lesion, and
MSCs are considered as an attractive cell source in cell therapy
and tissue engineering (26). As
for cell therapy for aortic disease, MSCs were originally thought
to differentiate into endothelial and SMCs (27,28)
and involved in vascular repair processes, neoangiogenesis and
stabilization of injuries (29,30).
But MSCs' anti-inflammatory effect seems to account for more.
Inflammatory reaction within the aortic wall leads to weakness and
degeneration of the vessel. The anti-inflammatory and angiogenic
capacity, the ability to release a range of growth factors
(especially vascular and fibroblast related), and the regulatory
role in MMPs secretion makes MSCs ideal candidates for the
treatment of AD. Recent studies have highlighted the critical
importance of MSCs as essential constituents for the aortic
aneurysm or dissection niche through inducing chemokine and
cytokine secretion (31) and wound
healing (32,33). More and more studies are providing
insights into aberrant microenvironment of AD pathology, and MSCs
have been recognized as a crucial element for AD microenvironment
(14). It has been demonstrated
(34) that the capacity of MSCs to
protect against aneurysm formation by immunomodulation on
CD4+ T cell and IL-17.
Since most studies have demonstrated MSCs could
protect or repair AD, we could reasonably hypothesize that there
might be some deficits with AD-MSCs compared with HD-MSCs.
Therefore, we compared bone marrow derived AD-MSCs with HD-MSCs in
this study, to further understand MSCs' role in AD and to provide
possible treatment targets. The reason why MSC samples in our study
were harvested from bone marrow instead of aortic tissue was mostly
technical restriction. As is known, MSCs can be easily harvested
from bone marrow, adipose tissue and umbilical cord blood but the
derivation from aorta is not reported, probably for its
insufficiency in aortic wall. As a matter of fact, our previous
study on animal has demonstrated that bone marrow-derived MSCs is
recruited to diseased aortic tissue. From our perspective, it is
reasonable to test bone marrow-derived MSCs at the second passage
as an alternative. The reason why we didn't deploy the protocol of
FACS cell sorting direct from bone marrow was mainly for technical
barriers, too. As for FACS cell sorting, either positive selection
or negative selection is faced with unsolvable problems. Due to the
lack of cell specific surface marker of MSCs, we currently have to
use a panel of markers to identify MSCs. Negative selection can
only eliminate unwanted cells while positive selection would
activate the downstream pathway of the surface maker, altering the
expression profile. We finally adapted the protocol of primary
culturing cells to passage 2 instead of, when all criteria by ISCT
(morphology, surface antigen and differentiation capacity) were
met. MSCs' basic features including morphology and proliferation
showed no significant difference between AD and HD group. But then
RNA-seq revealed a molecular profile consisted of 201 DEGs between
AD-MSCs and HD-MSCs, suggesting a specific AD-MSCs molecular
signature. Based on published literature, we finally chose 9 most
significant DEGs, naming ABCA4, CXCL1, CXCL5, EMX2, HTR7,
IGFBP2, NCAM1, SERPINB7 and SNAP25, for qRT-PCR
verification with an enlarged sample size and then made an
unsupervised hierarchical clustering analysis to see if this 9-gene
molecular profile is also capable of distinguishing AD-MSCs from
HD-MSCs. As is shown in Fig. 7B,
the 9-gene molecular profile we selected can separate AD-MSCs from
HD-MSCs. Either the 209-DEGs or 9-DEGs profile suggests a molecular
signature that could even be a possible molecular panel of markers
to identify AD-MSCs.
Among the DEGs, a significant target gene revealed
by DEGs is CXCL1/5. CXCL/CXCR signal axis is considered as
responsible for important functions of MSCs' like migration and
immunomodulatory (35,36). It has been reported that
CXCR4 has been reported to enhance the migration of bone
marrow MSCs in vitro in a rat abdominal aortic aneurysms
model (37,38), and our findings suggested other
genes belonging to CXCL/CXCR family contribute to MSCs' function in
AD pathogenesis. IGFBP2 was another differently expressed
gene. Previous researchers (39,40)
have found that IGFBP/IGF-signaling pathway are responsible for
MSCs' migration immunomodulatory and differentiation. Besides,
IGFBP2 together with CXCL1/5 is responsible for
cellular communication related to MSCs' repairing function. What's
more, the DEGs may even form complicated interaction networks and
conjointly contribute to the MSCs' role in AD pathogenesis.
KEGG and GO analysis visualization (Figs. 5 and 6) suggested alteration in pathway and
function of AD-MSCs compared with HD-MSC. KEGG prompted adhesion
while GO indicated development. In that view, we believe MSCs in AD
patients have some deficits in adhesion dysfunction, no matter at
intercellular level or cell-matrix level. The deficit might
contribute to AD pathogenesis but needs further investigation in
vitro and in vivo. Cell adhesion, especially the
adherence to ECM, is critical for determining cellular fates, such
as proliferation, migration and differentiation both in
vitro and in vivo (41). MSCs can produce various ECM
components and thus promote the reconstruction of the aorta.
Finally, we must put out limitations of our study.
Firstly, the sample size should have been larger. Given the
constraints of funding and time, we finally adapted the sample size
of 3+3 for RNA-seq and 6+9 for qRT-PCR. We believe the sample size
is acceptable, for it at least ensures a repeat in triplicates.
Besides, the between-group Pearson correlation coefficient (r) is
significantly larger than within-group r (t=4.0153, df=10.895,
P=0.002071). Quality control has also confirmed the validity of our
RNA-seq data. Secondly, the uncertainty of the causality. We
currently cannot indisputably attribute these changes to either
etiologic factors or disease results. We could only say these
changes are associated with/related to AD in a certain way. Further
validations of selected DEGs are needed, especially in vivo
experiments. If alterations like mortality/morbidity or
pathological changes on aortic walls are observed on KO mice, we
then could say this gene indeed plays a part in AD. This is mainly
due to constraints of time, funding and patient source. Thirdly, we
did not strictly distinguish AD and aneurysm in our research. On
the one hand, they are quite similar in pathogenesis and AD can
result directly from aneurysm progression; and on the other hand,
limitations come from current researches, that is, no very specific
animal models for each disease. Additionally, no changes in MMP-2/9
are observed in our RNA-seq result, unfavourably. Actually, MMP-2/9
is one of our expected targets according to existing publications.
A possible explanation, from our perspectives, might be that
MMP-2/9 take effect as secretory factors in cell or tissue matrix,
mainly at intercellular level. That's probably why didn't see a
significant change on MMP-2/9.
In conclusion, the current study suggested there is
a common molecular signature for AD-MSCs, and this signature is
capable of distinguishing AD-MSCs from HD-MSCs. Moreover, changes
in the expressed proteins suggested that AD-MSCs signaling and
function alterations, especially adhesion and development related,
could be an important factor in MSCs' effect on AD process. Our
study creates a detailed transcriptome picture of the MSCs
alteration in AD. And the molecular profile provides candidate
genes for further study to definitively confirm their functions in
MSCs for AD.
Acknowledgements
This study was funded by the National Natural
Science Foundation of China (81570440 by Qu Lefeng and 81470576 by
Liao Mingfang) and the Shanghai Science and Technology Talent
Project (15YF1400500 by Zou Sili). We thank Professor Luyang Yu and
all the staffs of Yu Laboratory, College of Life Sciences, Zhejiang
University, for providing necessary technical support.
References
|
1
|
Hagan PG, Nienaber CA, Isselbacher EM,
Bruckman D, Karavite DJ, Russman PL, Evangelista A, Fattori R,
Suzuki T, Oh JK, et al: The International Registry of Acute Aortic
Dissection (IRAD): New insights into an old disease. JAMA.
283:897–903. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Schlatmann TJ and Becker AE: Pathogenesis
of dissecting aneurysm of aorta. Comparative histopathologic study
of significance of medial changes. Am J Cardiol. 39:21–26. 1977.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
López-Candales A, Holmes DR, Liao S, Scott
MJ, Wickline SA and Thompson RW: Decreased vascular smooth muscle
cell density in medial degeneration of human abdominal aortic
aneurysms. Am J Pathol. 150:993–1007. 1997.PubMed/NCBI
|
|
4
|
De Bruin JL, Baas AF, Buth J, Prinssen M,
Verhoeven EL, Cuypers PW, van Sambeek MR, Balm R, Grobbee DE and
Blankensteijn JD: DREAM Study Group: Long-term outcome of open or
endovascular repair of abdominal aortic aneurysm. N Engl J Med.
362:1881–1889. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
SVN Task Force for Clinical Practice
Guideline: 2009 Clinical practice guideline for patients undergoing
endovascular repair of abdominal aortic aneurysms (AAA). J Vasc
Nurs. 27:48–63. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nabel EG, Yang Z, Liptay S, San H, Gordon
D, Haudenschild CC and Nabel GJ: Recombinant platelet-derived
growth factor B gene expression in porcine arteries induce intimal
hyperplasia in vivo. J Clin Invest. 91:1822–1829. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Phinney DG and Prockop DJ: Concise review:
Mesenchymal stem/multipotent stromal cells: The state of
transdifferentiation and modes of tissue repair-current views. Stem
Cells. 25:2896–2902. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hashizume R, Yamawaki-Ogata A, Ueda Y,
Wagner WR and Narita Y: Mesenchymal stem cells attenuate
angiotensin II-induced aortic aneurysm growth in apolipoprotein
E-deficient mice. J Vasc Surg. 54:1743–1752. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Schneider F, Saucy F, de Blic R, Dai J,
Mohand F, Rouard H, Ricco JB, Becquemin JP, Gervais M and Allaire
E: Bone marrow mesenchymal stem cells stabilize already-formed
aortic aneurysms more efficiently than vascular smooth muscle cells
in a rat model. Eur J Vasc Endovasc Surg. 45:666–672. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Davis JP, Salmon M, Pope NH, Lu G, Su G,
Sharma AK, Ailawadi G and Upchurch GR Jr: Attenuation of aortic
aneurysms with stem cells from different genders. J Surg Res.
199:249–258. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zou S, Ren P, Zhang L, Azares AR, Zhang S,
Coselli JS, Shen YH and LeMaire SA: AKT2 promotes bone marrow
cell-mediated aortic protection in mice. Ann Thorac Surg.
101:2085–2096. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shen YH, Hu X, Zou S, Wu D, Coselli JS and
LeMaire SA: Stem cells in thoracic aortic aneurysms and
dissections: Potential contributors to aortic repair. Ann Thorac
Surg. 93:1524–1533. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xie J, Jones TJ, Feng D, Cook TG, Jester
AA, Yi R, Jawed YT, Babbey C, March KL and Murphy MP: Human
adipose-derived stem cells suppress elastase-induced murine
abdominal aortic inflammation and aneurysm expansion through
paracrine factors. Cell Transplant. 26:173–189. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sharma AK, Salmon MD, Lu G, Su G, Pope NH,
Smith JR, Weiss ML and Upchurch GR Jr: Mesenchymal stem cells
attenuate NADPH oxidase-dependent high mobility group box 1
production and inhibit abdominal aortic aneurysms. Arterioscler
Thromb Vasc Biol. 36:908–918. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
del Moral Riera L, Largo C, Ramirez JR,
Vega Clemente L, Heredero Fernández A, de Cubas Riera L,
Garcia-Olmo D and Garcia-Arranz M: Potential of mesenchymal stem
cell in stabilization of abdominal aortic aneurysm sac. J Surg Res.
195:325–333. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ciavarella C, Alviano F, Gallitto E, Ricci
F, Buzzi M, Velati C, Stella A, Freyrie A and Pasquinelli G: Human
vascular wall mesenchymal stromal cells contribute to abdominal
aortic aneurysm pathogenesis through an impaired immunomodulatory
activity and increased levels of matrix metalloproteinase-9. Circ
J. 79:1460–1469. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dominici M, Le Blanc K, Mueller I,
Slaper-Cortenbach I, Marini F, Krause D, Deans R, Keating A,
Prockop Dj and Horwitz E: Minimal criteria for defining multipotent
mesenchymal stromal cells. The international society for cellular
therapy position statement. Cytotherapy. 8:315–317. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nicoletti I, Migliorati G, Pagliacci MC,
Grignani F and Riccardi C: A rapid and simple method for measuring
thymocyte apoptosis by propidium iodide staining and flow
cytometry. J Immunol Methods. 139:271–279. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Longo GM, Xiong W, Greiner TC, Zhao Y,
Fiotti N and Baxter BT: Matrix metalloproteinases 2 and 9 work in
concert to produce aortic aneurysms. J Clin Invest. 110:625–632.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hackam DG, Thiruchelvam D and Redelmeier
DA: Angiotensin-converting enzyme inhibitors and aortic rupture: A
population-based case-control study. Lancet. 368:659–665. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Schouten O, van Laanen JH, Boersma E,
Vidakovic R, Feringa HH, Dunkelgrün M, Bax JJ, Koning J, van Urk H
and Poldermans D: Statins are associated with a reduced infrarenal
abdominal aortic aneurysm growth. Eur J Vasc Endovasc Surg.
32:21–26. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yamawaki-Ogata A, Hashizume R, Satake M,
Kaneko H, Mizutani S, Moritan T, Ueda Y and Narita Y: A doxycycline
loaded, controlled-release, biodegradable fiber for the treatment
of aortic aneurysms. Biomaterials. 31:9554–9564. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Baxter BT, Pearce WH, Waltke EA, Littooy
FN, Hallett JW Jr, Kent KC, Upchurch GR Jr, Chaikof EL, Mills JL,
Fleckten B, et al: Prolonged administration of doxycycline in
patients with small asymptomatic abdominal aortic aneurysms: Report
of a prospective (Phase II) multicenter study. J Vasc Surg.
36:1–12. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Walton LJ, Franklin IJ, Bayston T, Brown
LC, Greenhalgh RM, Taylor GW and Powell JT: Inhibition of
prostaglandin E2 synthesis in abdominal aortic aneurysms:
Implications for smooth muscle cell viability, inflammatory
processes, and the expansion of abdominal aortic aneurysms.
Circulation. 100:48–54. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yoshimura K, Aoki H, Ikeda Y, Fujii K,
Akiyama N, Furutani A, Hoshii Y, Tanaka N, Ricci R, Ishihara T, et
al: Regression of abdominal aortic aneurysm by inhibition of c-Jun
N-terminal kinase. Nat Med. 11:1330–1338. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bianco P, Riminucci M, Gronthos S and
Robey PG: Bone marrow stromal stem cells: Nature, biology, and
potential applications. Stem Cells. 19:180–192. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Matsumura G, Miyagawa-Tomita S, Shin'oka
T, Ikada Y and Kurosawa H: First evidence that bone marrow cells
contribute to the construction of tissue-engineered vascular
autografts in vivo. Circulation. 108:1729–1734. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Allaire E, Muscatelli-Groux B, Guinault
AM, Pages C, Goussard A, Mandet C, Bruneval P, Méllière D and
Becquemin JP: Vascular smooth muscle cell endovascular therapy
stabilizes already developed aneurysms in a model of aortic injury
elicited by inflammation and proteolysis. Ann Surg. 239:417–427.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sata M, Saiura A, Kunisato A, Tojo A,
Okada S, Tokuhisa T, Hirai H, Makuuchi M, Hirata Y and Nagai R:
Hematopoietic stem cells differentiate into vascular cells that
participate in the pathogenesis of atherosclerosis. Nat Med.
8:403–409. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Saiura A, Sata M, Hirata Y, Nagai R and
Makuuchi M: Circulating smooth muscle progenitor cells contribute
to atherosclerosis. Nat Med. 7:382–383. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ryu JH, Park M, Kim BK, Ryu KH and Woo SY:
Tonsil-derived mesenchymal stromal cells produce CXCR2-binding
chemokines and acquire follicular dendritic cell-like phenotypes
under TLR3 stimulation. Cytokine. 73:225–235. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Veréb Z, Póliska S, Albert R, Olstad OK,
Boratkó A, Csortos C, Moe MC, Facskó A and Petrovski G: Role of
human corneal stroma-derived mesenchymal-like stem cells in corneal
immunity and wound healing. Sci Rep. 6:262272016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li Q, Guo Y, Chen F, Liu J and Jin P:
Stromal cell-derived factor-1 promotes human adipose tissue-derived
stem cell survival and chronic wound healing. Exp Ther Med.
12:45–50. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sharma AK, Lu G, Jester A, Johnston WF,
Zhao Y, Hajzus VA, Saadatzadeh MR, Su G, Bhamidipati CM, Mehta GS,
et al: Experimental abdominal aortic aneurysm formation is mediated
by IL-17 and attenuated by mesenchymal stem cell treatment.
Circulation. 126 11 Suppl 1:S38–S45. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Iida Y, Xu B, Xuan H, Glover KJ, Tanaka H,
Hu X, Fujimura N, Wang W, Schultz JR, Turner CR and Dalman RL:
Peptide inhibitor of CXCL4-CCL5 heterodimer formation, MKEY,
inhibits experimental aortic aneurysm initiation and progression.
Arterioscler Thromb Vasc Biol. 33:718–726. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Anzai A, Shimoda M, Endo J, Kohno T,
Katsumata Y, Matsuhashi T, Yamamoto T, Ito K, Yan X, Shirakawa K,
et al: Adventitial CXCL1/G-CSF expression in response to acute
aortic dissection triggers local neutrophil recruitment and
activation leading to aortic rupture. Circ Res. 116:612–623. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tanios F, Pelisek J, Lutz B, Reutersberg
B, Matevossian E, Schwamborn K, Hösel V, Eckstein HH and Reeps C:
CXCR4: A potential marker for inflammatory activity in abdominal
aortic aneurysm wall. Eur J Vasc Endovasc Surg. 50:745–753. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Michineau S, Franck G, Wagner-Ballon O,
Dai J, Allaire E and Gervais M: Chemokine (C-X-C motif) receptor 4
blockade by AMD3100 inhibits experimental abdominal aortic aneurysm
expansion through anti-inflammatory effects. Arterioscler Thromb
Vasc Biol. 34:1747–1755. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ramos-Mozo P, Rodriguez C, Pastor-Vargas
C, Blanco-Colio LM, Martinez-Gonzalez J, Meilhac O, Michel JB, de
Ceniga Vega M, Egido J and Martin-Ventura JL: Plasma profiling by a
protein array approach identifies IGFBP-1 as a novel biomarker of
abdominal aortic aneurysm. Atherosclerosis. 221:544–550. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang J, Razuvaev A, Folkersen L, Hedin E,
Roy J, Brismar K and Hedin U: The expression of IGFs and IGF
binding proteins in human carotid atherosclerosis, and the possible
role of IGF binding protein-1 in the regulation of smooth muscle
cell proliferation. Atherosclerosis. 220:102–109. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Giancotti FG and Ruoslahti E: Integrin
signaling. Science. 285:1028–1032. 1999. View Article : Google Scholar : PubMed/NCBI
|