Introduction
In 1985, George Smith was the first to employ a
phage display system (1). He
expressed, by inserting exogenous DNA into phage gene III, various
foreign peptides on the capsid of the filamentous bacteriophage f1.
Peptides and proteins of interest were obtained from a library of
random insert genes in a phage display vector by ≥1 rounds of
selection, thus facilitating the analysis of associations between
genotype and phenotype (1). The
common principle of all phage display systems is that exogenous
peptide-coding sequences are inserted into a phage capsid protein
gene, which allows the expression of foreign proteins or peptides
fused to the capsid protein of the phage particle (2–4).
Exogenous peptides or proteins of interest are subsequently
selected from a large library of phage particles by techniques such
as affinity elutriation (5,6).
Phage display systems are an efficient and rapid tool in the
investigation of protein sequences and have also been particularly
important in proteomics.
Numerous bacteriophage species have been employed in
phage display systems, including f1, fd, T4, M13 and T7, of which
the latter two examples are considered to be efficient display
vectors. The most commonly employed phage display system is M13 as
it contains nonessential regions that allow exogenous gene
insertions. Exogenous peptides, retaining their normal function,
are expressed in the M13 coat proteins (7,8) and
the phage retains the ability to accumulate at high concentrations
in hosts. Although a few of peptide repertoires successfully
expressed in a cDNA library of filamentous phage (9,10),
M13 phage display is associated with limitations in the
construction of the cDNA library. The formation of fusion proteins
consisting of the coat proteins and the expressed peptides, and the
secretion of phage into the periplasm, are also potential problems
in M13 phage display. However, these problems associated with M13
phage display may be avoided by using the T7 phage display
system.
Rapid development in the research of pathogenic
microorganisms and proteomics has been achieved through use of the
T7 phage display system. Various virus vaccines, such as those
against influenza virus and hepatitis B virus, require frequent
development, and the methods of diagnosis and treatment of
infectious diseases also require continuous improvement. By using a
T7 phage display library, one study identified the mechanism
underlying the anti-inflammatory effect of hydrostatin-SN1, a
peptide present in the venom gland of Hydrophis cyanocinctus
(11). In addition, a
reconstructive T7 phage combined with a magnetic separation
technique was applied for the detection of viable bacterial cells
(12). The favorable
characteristics of the T7 phage result in a wide range of potential
applications. The present study primarily describes the structure
and biological properties of the T7 phage and its application
through phage display system to various research areas, including
the mechanism of protein interactions in biology, the discovery of
novel pathogenic antigens, vaccine development, and cancer
diagnosis and therapy.
T7 bacteriophage: Structure and biological
properties
Bacteriophage T7 was defined in 1945 as one of the
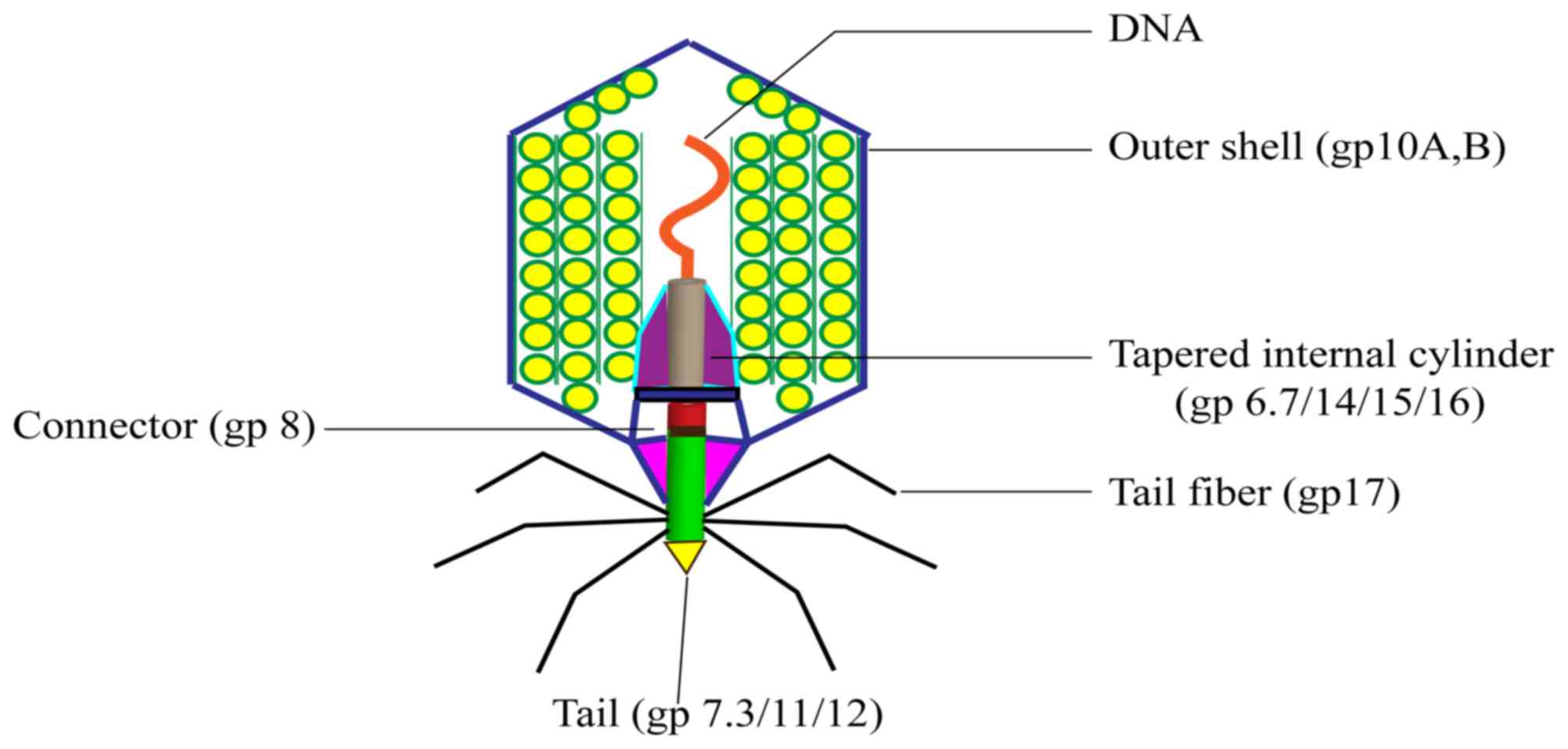
seven phages that replicate in E. coli (13). The DNA of the T7 phage is 40 kb and
is packed into the 60 nm diameter cavity formed by the capsid
protein. The capsid, or head, is the outer protective shell of the
phage, which protects the viral nucleic acid. The phage particle
consists of the following six major proteins: gp10A (primary capsid
protein); gp10B (secondary capsid protein); gp8 (connector); gp11
and gp12 (tail proteins); and gp17 (tail fiber). The majority of
the capsid protein is made up of gp10, which includes gp10A (344
amino acids) and gp10B (397 amino acids). Gp10A and gp10B are
products of gene 10 and are expressed at a ratio of 9:1 under
normal conditions. Gp10B is the result of a frame shift at the end
of the gp10A coding frame (14).
Foreign proteins or peptides are fused to the C-terminus of gp10B
(14,15). The proportion of gp10A and gp10B
may vary according to conditions; however, this does not affect the
activity of the phage (15).
A diagram of the structure of the T7 phage is
presented in Fig. 1, which
highlights the connector between the head and tail. The connector
is a ring structure that consists of multiple copies of gp8. Inside
the head, the core of the T7 phage forms a cylindrical structure
and this combines with gp8 to connect the head and tail. Gp15, gp14
and gp16 may form a pathway across the outer membrane of cells
following the initial step of infection bacteria (16).
T7 adsorption to lipopolysaccharide on the cell
surface may be similar to the T4 adsorption process (17), however, it is more difficult for
the T7 DNA to directly enter the cytoplasm of bacteria from the
adsorption position. Therefore, in order to insert T7 DNA into
bacteria, a tubular structure that traverses the outer membrane of
bacteria is required. Cooperation between gp15 and gp16 completes
the process of T7 DNA entry into the bacteria (18). The details of this process are
fully elucidated (19), and the
glycoproteins gp16 and gp15 are key factors in the transfer of T7
DNA to bacteria. The main function of the spiral ring formed by
gp15 and gp16 is assisting T7 phage DNA into host bacteria
(19).
The advantages of T7 phage display
system
T7 phage display libraries are formed of numerous
phages that carry different exogenous genes and express different
peptides on the surface of the phage shell. The amplification of
phages that carry these exogenous genes leads to the replication of
the foreign peptide in the library, which allows a large amount of
the identical exogenous peptide to be expressed on progeny phage.
In vivo and in vitro applications of the T7 phage
display system exist (20),
however, the majority of applications are currently in
vitro. The construction of T7 phage display libraries is
associated with advantages that include easy accessibility and few
equipment requirements (21).
Phage display systems usually employ filamentous
phages, including M13, fd, and f1 (22). However, these vectors are
associated with a limited cloning capacity, which in the case of
M13 is <1,500 bp, and their genome exhibits markedly reduced
stability following insertion of foreign DNA. By contrast,
recombinant T7 is very stable, even with foreign gene inserts >1
kb. Compared with M13, advantages of T7 as a phage display vector
include the following: T7 grows quickly and forms plaques within 3
h, which saves time during cloning and screening (23), while M13 grows slowly; the
construction of large display libraries is easier in T7 compared
with M13; unlike filamentous phagemids, foreign cDNA library is
directly inserted into T7 phage genome and expressed as capsid
fusion proteins; various types of cDNA libraries may be constructed
using T7 phage as a vector (23,24);
affinity elutriation, which is employed to select the proteins or
peptides of interest in the display library, is efficient and
effective with T7; and with T7, the likelihood of survival of the
recombinant phage in the environment is reduced, as the host cell
is required to produce T7 gene 10 to express proteins.
Applications of T7 phage display system
T7 phage display system for antigen
and epitope discovery
The discovery of antigen, surface antigen on
pathogenic microorganisms and cancer antigens, may be achieved
using the T7 phage display system. Various studies have
demonstrated the power of T7 phage display on the discovery of
antigens (25,26). For example, a T7 phage display cDNA
library was constructed from cell mRNA of bronchoalveolar lavage in
the granulomatous inflammatory disease, sarcoidosis, which led to
the identification of 1152 potential sarcoidosis antigens (27). The discovery of useful antigens in
epidemic diseases, such as influenza and tuberculosis, will aid the
diagnosis and treatment of these diseases. The ectodomain of
influenza A virus M2 protein, which may be expressed on the surface
of the T7 phage, provides immunoprotection against influenza A
(28). Recent studies from our
laboratory involved the construction of a Mycobacterium
tuberculosis genomic library using T7 phage as a carrier, which
was subsequently screened for the dominant M. tuberculosis
antigen recognized in the serum of patients with tuberculosis
(Yanhua Zeng et al, unpublished data).
In addition, phage peptide libraries allow the rapid
determination of the sequence of protein epitopes, and have become
a powerful tool for investigating the interaction between epitopes
and antigen receptors. At present, studies have employed phage
display to identify epitopes for human immunodeficiency virus (HIV)
(29), hepatitis C virus (30), West Nile virus (31), Mycoplasma pneumoniae
(32), Streptococcus
pneumoniae (33),
Helicobacter pylori (34)
and streptococcus disease pathogens in pigs (35). The VP1 epitope displayed by T7
phage may be used as a potential diagnostic reagent in
foot-and-mouth disease (36). In
addition, an immunodominant epitope that originated from the rat
erb-b2 receptor tyrosine kinase 2 (HER-2/neu) oncoprotein was
expressed by T7 bacteriophage nanoparticles. The induction of
robust cytotoxic T lymphocyte (CTL) responses indicated that T7
phage nanoparticles offer higher levels of immunogenicity on the
HER-2-derived minimal CTL epitope (37). Therefore, these results indicate
that phage display, particularly the T7 phage display system, may
be a powerful tool for antigen discovery and epitope
identification.
T7 phage display system for the
investigation of molecule interactions
Protein interaction has a major role in numerous
biological processes. The primary process of the identification of
small peptides or proteins is by using the corresponding ligands or
receptors as molecular targets to search phage display libraries.
The proteins that interact with a novel mineralocorticoid receptor
(MR) were identified by employing T7 phage display (38). To identify proteins that interact
with MR, researchers, using MR as bait, screened T7 phage cDNA
libraries derived from either human heart or kidney RNA (38). The results identified 23
non-redundant peptides obtained from the heart library and 7 from
the kidney library. In addition, a T7 phage display cDNA library
based on N2a cells was constructed and the library was screened
with soluble porcine hemagglutinating encephalomyelitis coronavirus
(PHE-CoV) glycoproteins. The results identified a novel interaction
between neural cell adhesion molecule and PHE-CoV spike protein,
and this association proved to be critical during PHE-CoV infection
(39). Obesity leads to a variety
of consequences for a large number of individuals. An improved
screening system based on T7 phage display was reported to be an
efficient method for identifying tubby-binding proteins (40), which may lead to improvements in
the understanding of the mechanisms in obesity. Furthermore, a
novel coregulator, SH3 and SYLF domain-containing 1, which
interacts with the androgen receptor was identified using T7 phage
display (41). These studies
demonstrate the practicability of T7 phage display on the
identification of ligands or receptors. In addition, the use of T7
phage display libraries with drugs has allowed researchers to
investigate interactions between peptides expressed on the surface
of phages and drugs. Etoposide functions as an antitumor agent, and
E2F transcription factor 4 was demonstrated to be an
etoposide-binding protein through T7 phage display (42). Furthermore, T7 phage display may
also be performed to investigate the associated mechanisms of host
interaction with pathogenic microorganisms. For example, the
mechanism of the interactions between certain pathogenic
microorganisms and hosts was explored using T7 cDNA display, and it
has been hypothesized that the peptides identified in such studies
may be useful for pathogen detection and identification (43,44).
To improve the understanding of the molecular pathogenesis of
gastrointestinal anthrax, one study employed T7 phage display based
on the human stomach to identify the proteins that interact with
anthrax lethal factor (45).
Additionally, by using T7 phage displays, another study identified
the inhibitory peptides that exhibited anti-endotoxin activity
in vivo and in vitro (46). More recently, our own research
group constructed a cDNA library from human urothelium cell RNA
using T7 phage as a carrier and screened for receptors of
Mycoplasma genitalium adhesion protein (Daipei et al,
unpublished data). The results of the above studies demonstrate the
value of T7 phage display system for the investigation of the
interactions between molecules, particularly with regards to
protein interaction. With further improvements in the technology,
we anticipate that the T7 phage display system may have a central
role in proteomics research.
T7 phage display system for vaccine
development
Phage display systems, including the T7 phage
display system, have been widely used in prophylactic vaccine and
therapeutic vaccine development due to a number of advantages,
which include simplicity, high levels of safety and stability, and
ease of storage and transport (47).
Prophylactic vaccines
The T7 phage display system aids the development of
novel vaccines. A sequence encoding the immunodominant region of
small hepatitis B virus surface antigen was inserted into the DNA
of T7 phage, and the peptide (111–156 amino acids) that was
expressed on the surface of the T7 phage induced antigenicity and
immunogenicity (48). In addition,
the T7 phage display system has been employed in research
concerning foot and mouth disease (FMD), which is a highly
contagious disease during childhood. The immunodominant region of
the capsid protein VP1 of the FMD virus was displayed on the T7
phage capsid surface, which may aid research and the development of
a vaccine for FMD (36). At
present, an effective vaccine for the prevention of neosporosis is
not available. However, one study employed T7 phage display to
identify the host cell binding protein that interacts with
neosporosis, and this binding protein induced partial immunological
protection against neosporosis in mice, which indicates its
potential as a candidate vaccine (49). The results of these studies
highlight the potential of T7 phage display in the development of
preventative vaccines.
Therapeutic vaccines
Although single-chain variable fragment antibodies
(ScFvs) are extremely useful, they are often unstable and difficult
to express individually in bacteria. These limitations may avoid
through T7 phage display single antibody library technology. ScFvs
exert a positive effect on the prevention and treatment of a
variety of diseases, including those induced by hepatitis C virus
(50), avian influenza viruses
(51), rabies virus (52). In addition, the use of phage
display systems in this context facilitates screening for ScFvs.
HIV gp120-specific ScFvs were selected for use as therapeutic
vaccines, and subsequent tests demonstrated excellent efficiency
against infection in vitro (53). With regards to the application of
T7 phage display to therapeutic vaccines, a T7 phage cDNA library
of the infective larval stage of Brugia malayi, the
causative agent of lymphatic filariasis, was established. Upon
screening of this library with sera from patients, the researchers
were able to identify potential vaccine candidates (54). Trichinella spiralis nudix
hydrolase protein, which was identified by a T7 phage display cDNA
library, led to the generation of local protective immunity in mice
(55). The results of these
studies indicate that T7 phage display is promising for the
identification of candidate vaccine antigens of infectious agents,
and for therapeutic vaccine development more generally.
T7 phage display system for cancer
diagnosis and treatment
The traditional approaches to cancer diagnosis and
treatment are inefficient, expensive and have low precision. Over
the last decade, however, the diagnosis and treatment of cancer has
progressed greatly, including the application of novel molecular
imaging probes and therapeutic agents (56–58).
Furthermore, advances in phage display technology and the
development of phage particles have provided further opportunities
for the detection and treatment of cancer (59,60).
A trackable nanoplatform for positron emission tomography that is
based on T7 phage particles was developed as a cancer imaging
method (61). This method,
therefore, provides the foundations for the construction of
cancer-specific theranostic agents and may be applicable to other
receptor/ligand systems for theranostic agent construction. A few
of peptides that may inhibit tumor growth were generated by
utilizing T7 phage display technology (62–64).
Using multimodal biopanning of T7 phage-displayed peptides, the
site that roxithromycin binds to was identified as the
extracellular domain of angiomotin, which is an anti-angiogenic
inhibitor angiostatin, leading to the restriction of
angiogenesis-dependent tumor growth and metastasis (65). Furthermore, the proteins that
interact with sulfoquynovosylacylpropanediol, a novel anticancer
agent, were identified using T7 phage display based on HeLa, lung
tumor and human umbilical vein endothelial cells (66). Prostate cancer is an important
health risk to men, and a novel photothermal therapy employed gold
nanoparticles that were formed from genetically modified T7 phages
(67). The aforementioned studies
highlight the potential of the T7 phage particle and T7 phage
display in cancer diagnosis and treatment.
Additionally, human monoclonal antibodies that are
isolated by T7 phage display libraries may directly detect tumor
markers. In breast cancer, research has previously focused on
tumor-associated antigens that may be used for detection of cancer.
Cancer antigen 15-3 is a traditional biomarker in breast cancer,
and its levels are increased in 75% of advanced-stage patients,
while levels are raised in <10% of early stage patients
(68). Certain results have
indicated that the diagnostic accuracy of breast cancer using these
serologic biomarkers may be improved by the use of T7 phage display
in combination with autoantibodies (68). Autoantibodies, which exhibit
specificity and stability in sera, against tumor-associated
antigens may be used for the early detection of cancer in
noninvasive serological tests (69). A series of representative antigens
that led to humoral responses in patients with gastric cancer (GC)
were identified by using T7 phage display-based serological
analysis of recombinant cDNA expression libraries. Subsequently,
phage-antigen microarrays were produced and used to research the
autoantibodies produced in patients with GC (69). At present, the early diagnosis of
head and neck squamous cell carcinoma (HNSCC) is difficult.
However, 5,133 selectively cloned tumor antigens were identified by
phage display libraries based on three kinds of diverse HNSCC
tissues (70). In addition, to
identify valid lung cancer-associated antigens, early screening of
patients with lung cancer was performed by diagnostic protein chips
derived from a lung cancer T7 phage cDNA library (71,72).
As an anticancer drug, methotrexate (MTX) is applied in tumor
therapy, and the identification of MTX-binding proteins was
performed using T7 phage display (73). Generally, it is seemingly
impossible by the naked eyes to count the cancer biomarker miRNAs,
but it may be quantified by utilizing the method based on T7 phage
display system (74). These
studies indicate that the T7 phage display system may aid the
development of anticancer drugs and improvement the accuracy of
diagnosis.
Concluding remarks and further
perspectives
Phage display technology is a fast, highly efficient
and relatively cheap method in bioscience research. The diversity
among phage species underlies the differences among the various
phage display systems, with commonly used vectors including the M13
and T7 phages. The present review has summarized the advantages of
the T7 phage display system over the M13 phage display system. The
T7 phage display system has various practical applications in
biomedical research, including the investigation of mechanisms of
disease, improving the accuracy of cancer diagnosis and the design
of potential therapeutic agents and vaccines. A diagram of the
principle, process and application of the T7 phage display system
is presented in Fig. 2.
The current review has outlined the various
advantages of the T7 phage display system; however, improvements in
the technology are still required. The current limitations of T7
phage display include the following: There are limits to the
complexity of peptide display libraries in E. coli, with
conversion efficiencies in the region of
107−108, and a peptide library may only have
a capacity of 109 different sequences; as phage display
technology, including T7 phage display, is dependent on the
expression of genes within live host cells, it is difficult to
effectively express and display toxic molecules; the genes of
encoded peptides have certain preferences, whereby the same amino
acid for degenerate codon use frequency is not the same, which
limits the diversity of the peptide library; amino acid
modifications, such as phosphorylation, are restricted by the
biology of the host bacterium. Therefore, future studies should
focus on improving these limitations to further improve T7 phage
display systems.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 31370207), Hunan
Provincial Key Laboratory for Special Pathogens Prevention and
Control (grant no. 2014-5), Construct Program of the Key Discipline
in Hunan Province (grant no. 2011-75) and the Hunan Province
Cooperative Innovation Center for Molecular Target New Drug Study
(grant no. 2015-351).
References
|
1
|
Smith GP: Filamentous fusion phage: Novel
expression vectors that display cloned antigens on the virion
surface. Science. 228:1315–1317. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
McCafferty J, Griffiths AD, Winter G and
Chiswell DJ: Phage antibodies: Filamentous phage displaying
antibody variable domains. Nature. 348:552–554. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Barbas CF III, Kang AS, Lerner RA and
Benkovic SJ: Assembly of combinatorial antibody libraries on phage
surfaces: The gene III site. Proc Natl Acad Sci USA. 88:7978–7982.
1991. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Smith GP: Surface presentation of protein
epitopes using bacteriophage expression systems. Curr Opin
Biotechnol. 2:668–673. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bass S, Greene R and Wells JA: Hormone
phage: An enrichment method for variant proteins with altered
binding properties. Proteins. 8:309–314. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Smith GP and Scott JK: Libraries of
peptides and proteins displayed on filamentous phage. Methods
Enzymol. 217:228–257. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gao C, Mao S, Kaufmann G, Wirsching P,
Lerner RA and Janda KD: A method for the generation of
combinatorial antibody libraries using pIX phage display. Proc Natl
Acad Sci USA. 99:12612–12616. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gao C, Mao S, Lo CH, Wirsching P, Lerner
RA and Janda KD: Making artificial antibodies: A format for phage
display of combinatorial heterodimeric arrays. Proc Natl Acad Sci
USA. 96:6025–6030. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jespers LS, Messens JH, De Keyser A,
Eeckhout D, Van den Brande I, Gansemans YG, Lauwereys MJ, Vlasuk GP
and Stanssens PE: Surface expression and ligand-based selection of
cDNAs fused to filamentous phage gene VI. Biotechnology (N Y).
13:378–382. 1995.PubMed/NCBI
|
|
10
|
Hufton SE, Moerkerk PT, Meulemans EV, de
Bruïne A, Arends JW and Hoogenboom HR: Phage display of cDNA
repertoires: The pVI display system and its applications for the
selection of immunogenic ligands. J Immunol Methods. 231:39–51.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zheng Z, Jiang H, Huang Y, Wang J, Qiu L,
Hu Z, Ma X and Lu Y: Screening of an anti-inflammatory peptide from
Hydrophis cyanocinctus and analysis of its activities and mechanism
in DSS-induced acute colitis. Sci Rep. 6:256722016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang Z, Wang D, Chen J, Sela DA and Nugen
SR: Development of a novel bacteriophage based biomagnetic
separation method as an aid for sensitive detection of viable
Escherichia coli. Analyst. 141:1009–1016. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Demerec M and Fano U:
Bacteriophage-resistant mutants in escherichia coli. Genetics.
30:119–136. 1945.PubMed/NCBI
|
|
14
|
Sipley J, Stassi D, Dunn J and Goldman E:
Analysis of bacteriophage T7 gene 10A and frameshifted 10B
proteins. Gene Expr. 1:127–136. 1991.PubMed/NCBI
|
|
15
|
Condron BG, Atkins JF and Gesteland RF:
Frameshifting in gene 10 of bacteriophage T7. J Bacteriol.
173:6998–7003. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Molineux IJ: No syringes please, ejection
of phage T7 DNA from the virion is enzyme driven. Mol Microbiol.
40:1–8. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Leiman M: Research of effectiveness is a
challenge in psychotherapy. Duodecim. 120:2645–2653. 2004.(In
Finnish). PubMed/NCBI
|
|
18
|
Chang CY, Kemp P and Molineux IJ: Gp15 and
gp16 cooperate in translocating bacteriophage T7 DNA into the
infected cell. Virology. 398:176–186. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lupo D, Leptihn S, Nagler G, Haase MJ,
Molineux I and Kuhn A: The T7 ejection nanomachine components
gp15-gp16 form a spiral ring complex that binds DNA and a lipid
membrane. Virology. 486:263–271. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Johns M, George AJ and Ritter MA: In vivo
selection of sFv from phage display libraries. J Immunol Methods.
239:137–151. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hu CF, Peng XJ, Zhou YY, Tan YP, Li SQ and
Zhu YG: Construction of T7 phage display library from the anther of
Honglian hybrid line of rice. Yi Chuan. 30:771–775. 2008.(In
Chinese). View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Paschke M: Phage display systems and their
applications. Appl Microbiol Biotechnol. 70:2–11. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li W and Caberoy NB: New perspective for
phage display as an efficient and versatile technology of
functional proteomics. Appl Microbiol Biotechnol. 85:909–919. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Danner S and Belasco JG: T7 phage display:
A novel genetic selection system for cloning RNA-binding proteins
from cDNA libraries. Proc Natl Acad Sci USA. 98:12954–12959. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hansen MH, Ostenstad B and Sioud M:
Identification of immunogenic antigens using a phage-displayed cDNA
library from an invasive ductal breast carcinoma tumour. Int J
Oncol. 19:1303–1309. 2001.PubMed/NCBI
|
|
26
|
Larman HB, Zhao Z, Laserson U, Li MZ,
Ciccia A, Gakidis MA, Church GM, Kesari S, Leproust EM, Solimini NL
and Elledge SJ: Autoantigen discovery with a synthetic human
peptidome. Nat Biotechnol. 29:535–541. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Talwar H, Rosati R, Li J, Kissner D, Ghosh
S, -Madrid FF and Samavati L: Development of a T7 phage display
library to detect sarcoidosis and tuberculosis by a panel of novel
antigens. EBioMedicine. 2:341–350. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hashemi H, Pouyanfard S, Bandehpour M,
Noroozbabaei Z, Kazemi B, Saelens X and Mokhtari-Azad T:
Immunization with M2e-displaying T7 bacteriophage nanoparticles
protects against influenza A virus challenge. PLoS One.
7:e457652012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gazarian KG, Palacios-Rodríguez Y,
Gazarian TG and Huerta L: HIV-1 V3 loop crown epitope-focused
mimotope selection by patient serum from random phage display
libraries: Implications for the epitope structural features. Mol
Immunol. 54:148–156. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Rechkina EA, Denisova GF, Masalova OV,
Lideman LF, Denisov DA, Lesnova EI, Ataullakhanov RI, Gur'ianova SV
and Kushch AA: Epitope mapping of antigenic determinants of
hepatitis C virus proteins by phage display. Mol Biol (Mosk).
40:357–368. 2006.(In Russian). View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sun EC, Zhao J, Yang T, Liu NH, Geng HW,
Qin YL, Wang LF, Bu ZG, Yang YH, Lunt RA, et al: Identification of
a conserved JEV serocomplex B-cell epitope by screening a
phage-display peptide library with a mAb generated against West
Nile virus capsid protein. Virol J. 8:1002011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Beghetto E, De Paolis F, Montagnani F,
Cellesi C and Gargano N: Discovery of new Mycoplasma pneumoniae
antigens by use of a whole-genome lambda display library. Microbes
Infect. 11:66–73. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Beghetto E, Gargano N, Ricci S, Garufi G,
Peppoloni S, Montagnani F, Oggioni M, Pozzi G and Felici F:
Discovery of novel streptococcus pneumoniae antigens by screening a
whole-genome lambda-display library. FEMS Microbiol Lett.
262:14–21. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li Y, Ning YS, Wang YD, Hong YH, Luo J,
Dong WQ and Li M: Production of mouse monoclonal antibodies against
Helicobacter pylori Lpp20 and mapping the antigenic epitope by
phage display library. J Immunol Methods. 325:1–8. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Fan H, Wang Y, Tang F and Lu C:
Determination of the mimic epitope of the M-like protein adhesin in
swine Streptococcus equi subsp. Zooepidemicus. BMC
Microbiol. 8:1702008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wong CL, Sieo CC and Tan WS: Display of
the VP1 epitope of foot-and-mouth disease virus on bacteriophage T7
and its application in diagnosis. J Virol Methods. 193:611–619.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Pouyanfard S, Bamdad T, Hashemi H,
Bandehpour M and Kazemi B: Induction of protective anti-CTL epitope
responses against HER-2-positive breast cancer based on multivalent
T7 phage nanoparticles. PLoS One. 7:e495392012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yang J, Fuller PJ, Morgan J, Shibata H,
McDonnell DP, Clyne CD and Young MJ: Use of phage display to
identify novel mineralocorticoid receptor-interacting proteins. Mol
Endocrinol. 28:1571–1584. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Gao W, He W, Zhao K, Lu H, Ren W, Du C,
Chen K, Lan Y, Song D and Gao F: Identification of NCAM that
interacts with the PHE-CoV spike protein. Virol J. 7:2542010.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Caberoy NB, Zhou Y, Jiang X, Alvarado G
and Li W: Efficient identification of tubby-binding proteins by an
improved system of T7 phage display. J Mol Recognit. 23:74–83.
2010.PubMed/NCBI
|
|
41
|
Blessing AM, Ganesan S, Rajapakshe K, Ying
Sung Y, Bollu Reddy L, Shi Y, Cheung E, Coarfa C, Chang JT,
McDonnell DP and Frigo DE: Identification of a Novel Coregulator,
SH3YL1, that interacts with the androgen receptor n-terminus. Mol
Endocrinol. 29:1426–1439. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Takami M, Takakusagi Y, Kuramochi K,
Tsukuda S, Aoki S, Morohashi K, Ohta K, Kobayashi S, Sakaguchi K
and Sugawara F: A screening of a library of T7 phage-displayed
peptide identifies E2F-4 as an etoposide-binding protein.
Molecules. 16:4278–4294. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ren HJ, Liu RD, Wang ZQ and Cui J:
Construction and use of a Trichinella spiralis phage display
library to identify the interactions between parasite and host
enterocytes. Parasitol Res. 112:1857–1863. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lauterbach SB, Lanzillotti R and Coetzer
TL: Construction and use of Plasmodium falciparum phage display
libraries to identify host parasite interactions. Malar J.
2:472003. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Cardona-Correa A and Rios-Velazquez C:
Profiling lethal factor interacting proteins from human stomach
using T7 phage display screening. Mol Med Rep. 13:3797–3804. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Fang L, Xu Z, Wang GS, Ji FY, Mei CX, Liu
J and Wu GM: Directed evolution of an LBP/CD14 inhibitory peptide
and its anti-endotoxin activity. PLoS One. 9:e1014062014.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Silman NJ: World influenza congress Europe
2009. Expert Rev Vaccines. 9:273–275. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Tan GH, Yusoff K, Seow HF and Tan WS:
Antigenicity and immunogenicity of the immunodominant region of
hepatitis B surface antigen displayed on bacteriophage T7. J Med
Virol. 77:475–480. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Lv Q, Xing S, Gong P, Chang L, Bian Z,
Wang L, Zhang X and Li J: A 78 kDa host cell invasion protein of
Neospora caninum as a potential vaccine candidate. Exp
Parasitol. 148:56–65. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Lun YZ, Cheng J, Zhong YW, Yu ZG, Wang Q,
Wang F and Feng J: Cloning, expression and identification by
immunohistochemistry of humanized single-chain variable fragment
antibody against hepatitis C virus core protein. Pol J Microbiol.
60:13–17. 2011.PubMed/NCBI
|
|
51
|
Gabbard J, Velappan N, Di Niro R, Schmidt
J, Jones CA, Tompkins SM and Bradbury AR: A humanized anti-M2 scFv
shows protective in vitro activity against influenza. Protein Eng
Des Sel. 22:189–198. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Aavula SM, Nimmagadda SV, Biradhar N, Sula
S, Chandran D, Lingala R and Villuppanoor SA: Generation and
characterization of an scFv directed against Site II of rabies
glycoprotein. Biotechnol Res Int. 2011:6521472011. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Abdel-Motal UM, Sarkis PT, Han T, Pudney
J, Anderson DJ, Zhu Q and Marasco WA: Anti-gp120 minibody gene
transfer to female genital epithelial cells protects against HIV-1
virus challenge in vitro. PLoS One. 6:e264732011. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Gnanasekar M, Rao KV, He YX, Mishra PK,
Nutman TB, Kaliraj P and Ramaswamy K: Novel phage display-based
subtractive screening to identify vaccine candidates of Brugia
malayi. Infect Immun. 72:4707–4715. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Long SR, Wang ZQ, Jiang P, Liu RD, Qi X,
Liu P, Ren HJ, Shi HN and Cui J: Characterization and functional
analysis of Trichinella spiralis Nudix hydrolase. Exp
Parasitol. 159:264–273. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Conde J, Doria G and Baptista P: Noble
metal nanoparticles applications in cancer. J Drug Deliv.
2012:7510752012. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Mitsunaga M, Ogawa M, Kosaka N, Rosenblum
LT, Choyke PL and Kobayashi H: Cancer cell-selective in vivo near
infrared photoimmunotherapy targeting specific membrane molecules.
Nat Med. 17:1685–1691. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Luo S, Zhang E, Su Y, Cheng T and Shi C: A
review of NIR dyes in cancer targeting and imaging. Biomaterials.
32:7127–7138. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Allegra A, Penna G, Alonci A, Rizzo V,
Russo S and Musolino C: Nanoparticles in oncology: The new
theragnostic molecules. Anticancer Agents Med Chem. 11:669–686.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Shubayev VI, Pisanic TR II and Jin S:
Magnetic nanoparticles for theragnostics. Adv Drug Deliv Rev.
61:467–477. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Li Z, Jin Q, Huang C, Dasa S, Chen L, Yap
LP, Liu S, Cai H, Park R and Conti PS: Trackable and targeted phage
as positron emission tomography (PET) agent for cancer imaging.
Theranostics. 1:371–380. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Jemal A, Siegel R, Ward E, Murray T, Xu J
and Thun MJ: Cancer statistics, 2007. CA Cancer J Clin. 57:43–66.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Koolpe M, Dail M and Pasquale EB: An
ephrin mimetic peptide that selectively targets the EphA2 receptor.
J Biol Chem. 277:46974–46979. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Sakamoto K, Kamada Y, Sameshima T, Yaguchi
M, Niida A, Sasaki S, Miwa M, Ohkubo S, Sakamoto JI, Kamaura M, et
al: K-Ras(G12D)-selective inhibitory peptides generated by random
peptide T7 phage display technology. Biochem Biophys Res Commun.
484:605–611. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Takakusagi K, Takakusagi Y, Suzuki T,
Toizaki A, Suzuki A, Kawakatsu Y, Watanabe M, Saito Y, Fukuda R,
Nakazaki A, et al: Multimodal biopanning of T7 phage-displayed
peptides reveals angiomotin as a potential receptor of the
anti-angiogenic macrolide Roxithromycin. Eur J Med Chem.
90:809–821. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Izaguirre-Carbonell J, Kawakubo H, Murata
H, Tanabe A, Takeuchi T, Kusayanagi T, Tsukuda S, Hirakawa T,
Iwabata K, Kanai Y, et al: Novel anticancer agent, SQAP, binds to
focal adhesion kinase and modulates its activity. Sci Rep.
5:151362015. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Oh MH, Yu JH, Kim I and Nam YS:
Genetically programmed clusters of gold nanoparticles for cancer
cell-targeted photothermal therapy. ACS Appl Mater Interfaces.
7:22578–22586. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Dong X, Yang M, Sun H, Lü J, Zheng Z, Li Z
and Zhong L: Combined measurement of CA 15-3 with novel
autoantibodies improves diagnostic accuracy for breast cancer. Onco
Targets Ther. 6:273–279. 2013.PubMed/NCBI
|
|
69
|
Zayakin P, Ancāns G, Siliņa K, Meistere I,
Kalniņa Z, Andrejeva D, Endzeliņš E, Ivanova L, Pismennaja A,
Ruskule A, et al: Tumor-associated autoantibody signature for the
early detection of gastric cancer. Int J Cancer. 132:137–147. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Lin HS, Talwar HS, Tarca AL, Ionan A,
Chatterjee M, Ye B, Wojciechowski J, Mohapatra S, Basson MD, Yoo
GH, et al: Autoantibody approach for serum-based detection of head
and neck cancer. Cancer Epidemiol Biomarkers Prev. 16:2396–2405.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Li HM, Guo K, Yu Z, Feng R and Xu P:
Diagnostic value of protein chips constructed by
lung-cancer-associated markers selected by the T7 phage display
library. Thorac Cancer. 6:469–474. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Yuan N, Xin GH, Zuo XX, Huang SK, Wang Y,
Hou L, Qin TJ and Zhao XH: Combination of phage display and SEREX
for screening early lung cancer associated antigens. Zhejiang Da
Xue Xue Bao Yi Xue Ban. 43:388–396. 2014.(In Chinese). PubMed/NCBI
|
|
73
|
Kuroiwa Y, Takakusagi Y, Kusayanagi T,
Kuramochi K, Imai T, Hirayama T, Ito I, Yoshida M, Sakaguchi K and
Sugawara F: Identification and characterization of the direct
interaction between methotrexate (MTX) and high-mobility group box
1 (HMGB1) protein. PLoS One. 8:e630732013. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Zhou X, Cao P, Zhu Y, Lu W, Gu N and Mao
C: Phage-mediated counting by the naked eye of miRNA molecules at
attomolar concentrations in a Petri dish. Nat Mater. 14:1058–1064.
2015. View Article : Google Scholar : PubMed/NCBI
|