Introduction
Breast cancer is the most prevalent malignancy in
women worldwide (1). A recent
report conducted in China revealed that 272,400 new diagnoses, as
well as 70,700 mortalities, occur annually as a result of breast
cancer (2). Molecular subtyping of
breast cancer is relati(^vely well established, and tamoxifen
represents the most common drug prescribed to patients with breast
cancer. However, relapse occurs in a large proportion of patients
with estrogen-receptor positive (ER+) breast cancer treated with
tamoxifen (3), and current
clinical practice is insufficient for accurate prognosis. Previous
research has identified survival-associated genomic signatures of
breast cancer. For example, high expression of the GATA binding
protein 3 gene has been reported to be associated with prolonged
progression-free survival in patients with ER+ breast cancer
(4). Furthermore, patients with a
reduced level of Beclin 1 expression demonstrated a higher
sensitivity to tamoxifen and a prolonged survival time (5). In addition, a high protein expression
level of enhancer of zeste 2 polycomb repressive complex 2 subunit
(EZH2) has been reported to be associated with the development of
distant metastases in breast cancer (6).
However, the clinical prognostic effect of single
molecular biomarkers varies across datasets; whereas a multiple
gene expression-based staging method is robust across datasets
(7–10). In the present study, a
transcriptome-based risk score for the prediction of survival in
patients with ER+ breast cancer treated with tamoxifen was
developed using the Cox multivariate regression model. Risk scores
were developed using cyclin B2 (CCNB2), glutamyl-prolyl-tRNA
synthetase (EPRS), α-ketoglutarate dependent dioxygenase, stem-loop
binding protein (SLBP), CTD small phosphatase 1 (CTDSP1), cyclin A2
(CCNA2), bridging integrator 3 (BIN3), RNA binding protein with
multiple splicing (RBPMS), forkhead box D1 (FOXD1), gene encoding
WD repeat of SOCS box containing 2 (WSB2); and the resultant model
based upon said genes’ expression levels was revealed to
successfully predict survival time in the training and validation
datasets (GSE22219, GSE26971 and GSE58644). Median survival time of
the high-risk and the low-risk group was 3.75 and 6.5 years,
respectively. Furthermore, the associations between risk score and
clinical parameters were investigated and it was demonstrated that
age, grade and stage were significantly associated with risk score.
A 5 year survival nomogram was plotted in order to facilitate the
utilization of the risk score, which was demonstrated to be an
important clinical indicator for prognosis. In conclusion, this
study has developed a robust risk score staging system for the
prediction of survival in patients with ER+ breast cancer treated
with tamoxifen.
Materials and methods
Sample enrollment and data
pre-analysis
The following key words were searched for in the
Gene Expression Omnibus (GEO) dataset: ‘Breast cancer’,
‘tamoxifen’, ‘expression’ and ‘microarray’; and datasets with
<100 ER+ tamoxifen-treated samples, or datasets without survival
information, were then manually filtered out. Following this, four
datasets, GSE17705, GSE22219, GSE26971 and GSE56884, were then
retained for further analysis. Furthermore, samples that were not
primary tumor tissue were also excluded during this process. Raw
data were then downloaded in the CEL format from the GEO datasets.
Following this, background correction and normalization with Robust
Multiarray Averaging were carried out using the R package ‘affy’
function ‘rma’ (v1.54.0). Probe and gene names were matched
according to the manufacturer-provided annotation file. Genes with
more than one complementary probe were merged and the average
values were retained as the expression levels for the corresponding
genes.
Risk score model development
Cox univariate regression was implemented in both
GSE26971 and GSE17005 datasets via correlation of each individual
gene's expression with the survival information in both datasets
using the R package ‘survival’ Genes significantly correlated with
distant metastasis-free survival time in both GSE26971 and GSE17005
datasets were retained for further analyses as candidate genes.
Random forest variable hunting was applied for the selection of a
reasonable combination of candidate genes using R package
‘RandomForestSRC’ (v1.9.0). The parameter used was: 100 repeats and
100 iterations. Following this, multivariate Cox regression
analysis was carried out in order to develop the linear risk score
model using the selected candidate genes, and coefficients were
solved with the training dataset, GSE17005. In the validation
datasets (GSE22219, GSE26971 and GSE58644), these coefficients were
locked in order to calculate the risk score of samples in the other
datasets.
Statistical analysis
All statistical analyses were performed using R
software (v3.0.1; https://www.r-project.org) and R packages.
Normalizations of affymetrix raw data were performed with R package
‘affy’ using the function ‘rms’. The survival analysis and cox
probability hazard model construction were performed with R package
‘survival’. Random forest variable hunting was implemented with R
package ‘RandomForestSRC’, and receiver operating characteristic
(ROC) curves were generated with R package ‘pROC’ (11). The nomogram was plotted with the
clinical data in the training dataset using R package ‘rms’.
Results
Gene selection and model
development
The detailed workflow of gene selection and model
development is presented in Fig.
1A. The levels of association between gene expression levels
and treatment outcomes (survival data) were assessed using Cox
univariate regression. Genes associated with overall survival in
both the GSE17705 and GSE26971 datasets were identified, and a
total of 48 genes were then selected as candidates. Following this,
the random forest variable hunting was performed in order to select
for the optimal candidate genes. Following identification of 10
candidate genes (Fig. 1B), risk
scores using Cox multivariate regression and expression of 10 genes
were then calculated. The coefficients are presented in Fig. 1C, and parameters of Cox regression
are shown in Table I. The risk
scores were calculated as follows (where gene names represent their
respective expression levels): Risk score = (0.299988203)*cyclin B2
(CCNB2) + (0.640775607) *glutamyl-prolyl-tRNA synthetase (EPRS) +
(−0.756716676) *α-ketoglutarate dependent dioxygenase (FTO) +
(0.117814961) *stem-loop binding protein (SLBP) + (0.245606283)*CTD
small phosphatase 1 (CTDSP1) + (−0.161767842) *cyclin A2 (CCNA2) +
(0.196307548) *bridging integrator 3 (BIN3) + (−0.618268545) *RNA
binding protein with multiple splicing (RBPMS) + (0.580014194)
*forkhead box D1 (FOXD1) + (−0.288974361) *gene encoding WD repeat
of SOCS box containing 2 (WSB2).
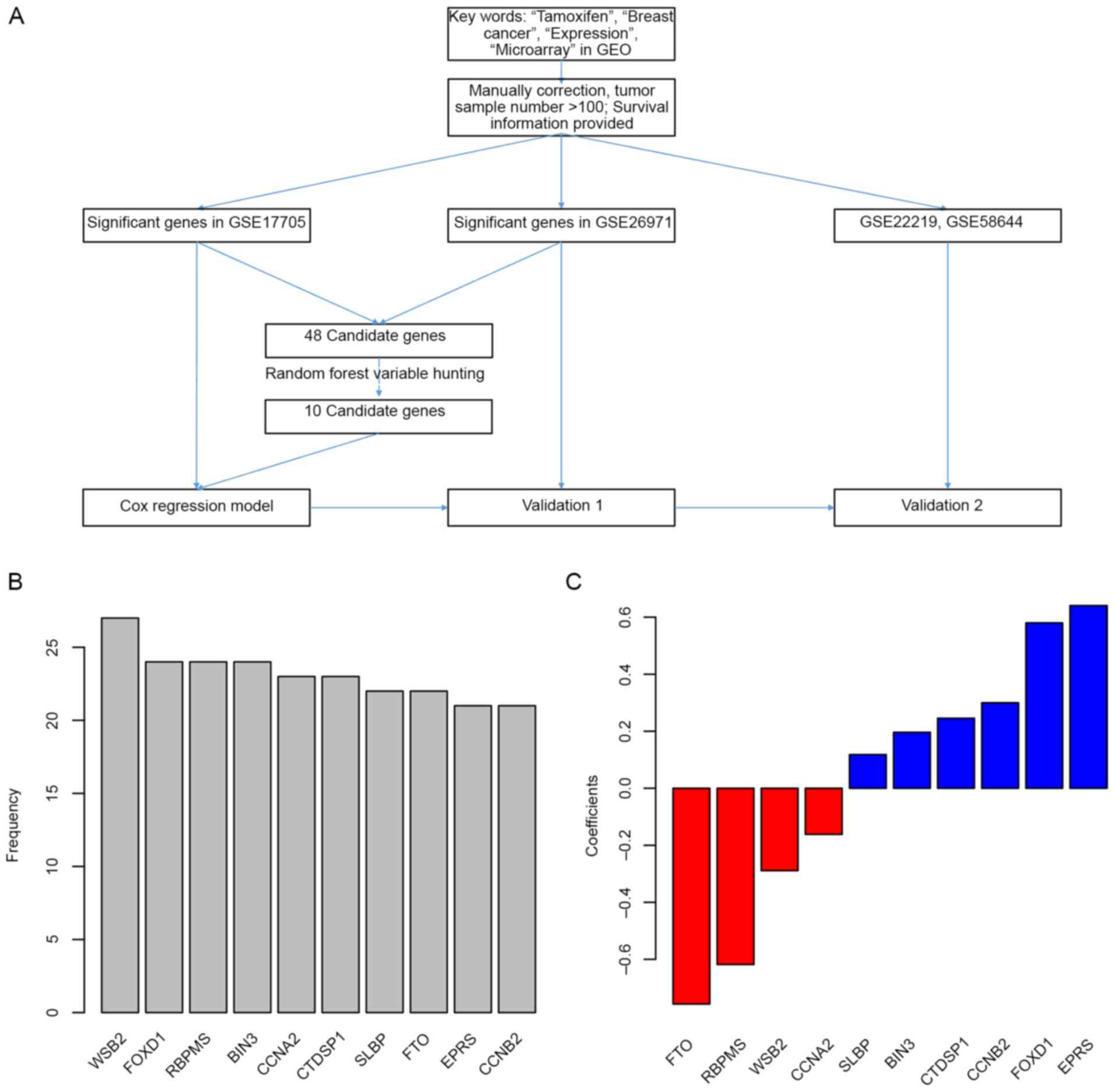 | Figure 1.Candidate gene identification. (A)
Workflow of the study. (B) Genes identified in random forest
variable hunting. (C) Coefficients of each gene. GEO, Gene
Expression Omnibus; WSB2, gene encoding WD repeat of SOCS box
containing 2; FOXD1, forkhead box D1; RBPMS, RNA binding protein
with multiple splicing; BIN3, bridging integrator 3; CCNA2, cyclin
A2; CTDSP1, CTD small phosphatase 1; SLBP, stem-loop binding
protein; FTO, α-ketoglutarate dependent dioxygenase; EPRS,
glutamyl-prolyl-tRNA synthetase; CCNB2, cyclin B2. |
 | Table I.Parameters of candidate genes. |
Table I.
Parameters of candidate genes.
|
| Univariate | Multivariate |
|---|
|
|
|
|
|---|
| Genes | HR | 95% C.I. | P-value | HR | 95% C.I. | P-value |
|---|
| CCNB2 | 2.2 | 1.3–3.7 | 0.00252 | 0.81 | 0.38–1.71 | 0.57828 |
| CCNA2 | 0.82 | 0.7–0.95 | 0.00959 | 0.91 | 0.78–1.07 | 0.2525 |
| FOXD1 | 2.6 | 1.6–4.3 | 0.00016 | 0.78 | 0.28–2.21 | 0.64203 |
| WSB2 | 0.44 | 0.27–0.72 | 0.00119 | 0.55 | 0.3–1 | 0.05139 |
| RBPMS | 0.26 | 0.12–0.57 | 0.00077 | 0.58 | 0.21–1.62 | 0.29785 |
| CTDSP1 | 2.2 | 1.5–3.2 | 2.00E-05 | 1.74 | 1.17–2.59 | 0.00631 |
| BIN3 | 1.4 | 1.1–1.7 | 0.00815 | 1.22 | 0.96–1.54 | 0.10664 |
| SLBP | 1.3 | 1.1–1.5 | 0.00536 | 1.24 | 1.04–1.48 | 0.01777 |
| EPRS | 2.7 | 1.5–4.7 | 0.00045 | 2.88 | 1.23–6.74 | 0.0148 |
| FTO | 0.27 | 0.11–0.63 | 0.0028 | 0.68 | 0.25–1.85 | 0.45285 |
Prognostic values of the risk score in
the training dataset
Patients were divided into two groups, a high-risk
group or a low-risk group, according to their median risk score.
Following this, the difference in survival between the high-risk
and the low-risk groups was calculated, and the results revealed
that the high-risk group had a reduced relapse-free time compared
with the low-risk group, with median survival times of 3.75 vs. 6.5
years, respectively (P<0.001; Fig.
2A). The high-risk group tended to represent early metastasis,
and genes with high expression levels tended to have positive
coefficients and genes with low expression tended to have negative
coefficients (Fig. 2B). The 5-year
distant relapse-free survival rate of the high-risk group was 75%;
whereas this value was revealed as being 96% in the low-risk group.
These results indicated that the developed risk score was an
effective predictive indicator for the distant relapse survival
time period of patients with ER+ breast cancer treated with
tamoxifen.
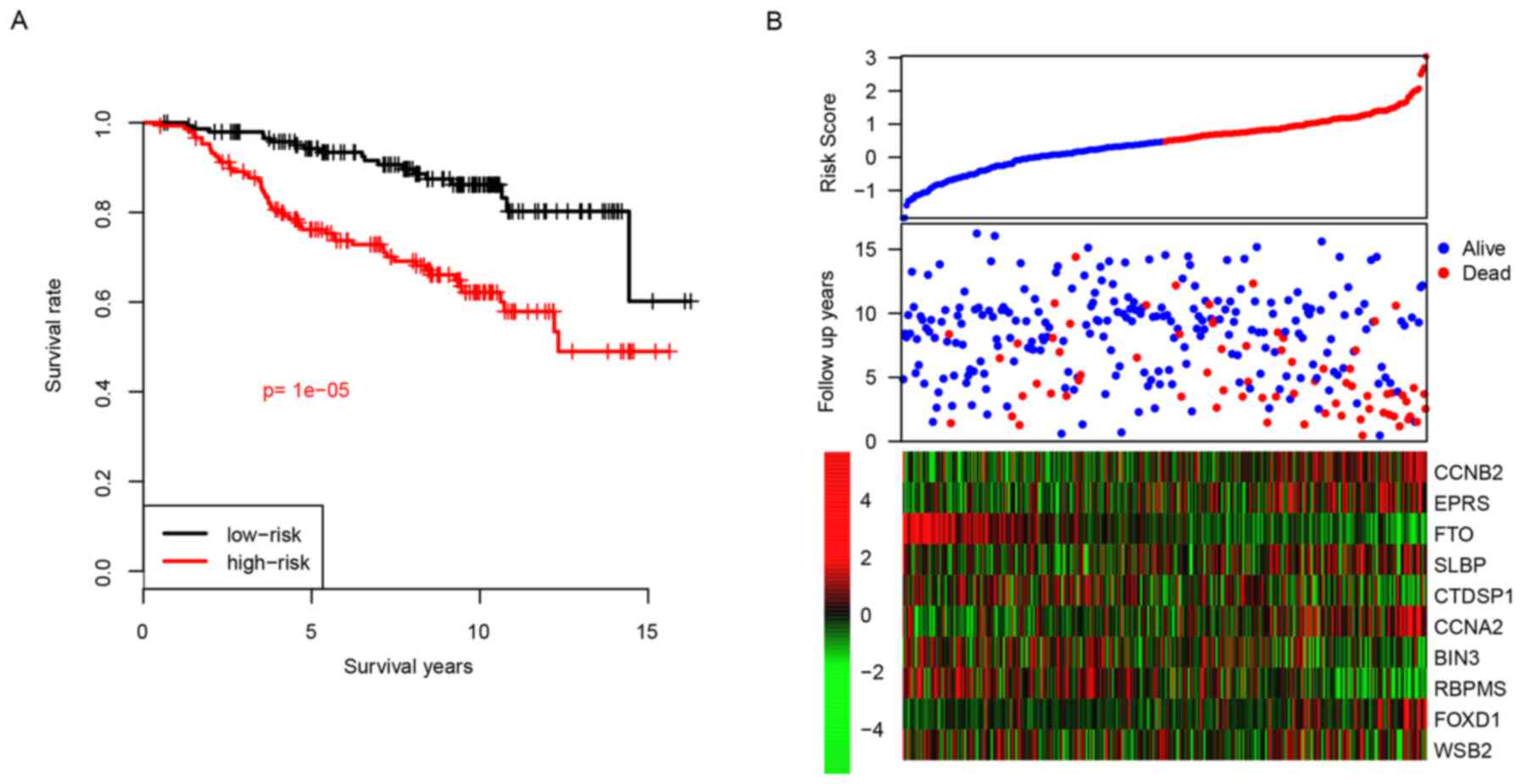 | Figure 2.Performance of risk score in the
training dataset. (A) Survival difference between high-risk and
low-risk group and (B) detailed survival information and expression
of candidate genes. CCNB2, cyclin B2; EPRS, glutamyl-prolyl-tRNA
synthetase; FTO, α-ketoglutarate dependent dioxygenase; SLBP,
stem-loop binding protein; CTDSP1, CTD small phosphatase 1; CCNA2,
cyclin A2; BIN3, bridging integrator 3; RBPMS, RNA binding protein
with multiple splicing; FOXD1, forkhead box D1; WSB2, gene encoding
WD repeat of SOCS box containing 2. |
Risk score performance validation
Considering that the risk score staging system was
developed based upon gene expression data in the GSE17705 dataset,
there was a potential risk that the model would over-fit to the
dataset. In order to assess the robustness of the developed risk
score model, three independent datasets (GSE22219, GSE26971 and
GSE58644) were used for further validation. Following the locking
of the coefficients for the 10 genes, a risk score for each patient
was calculated. In addition to patients belonging to the training
dataset, the patients belonging to each of the three independent
datasets were artificially divided into high-risk and low-risk
groups using median risk score values as cutoff values. The
patients with high-risk scores tended to have early relapse, as was
similarly demonstrated in patients belonging to the training
dataset (Fig. 3A). Furthermore,
the gene expression profiles for the 10 genes in the both the
low-risk and the high-risk groups resemble those demonstrated by
the training dataset (Fig. 3B).
These results demonstrate that the risk score model is robust
across datasets for the prediction of distant relapse in patients
with ER+ breast cancer treated with tamoxifen.
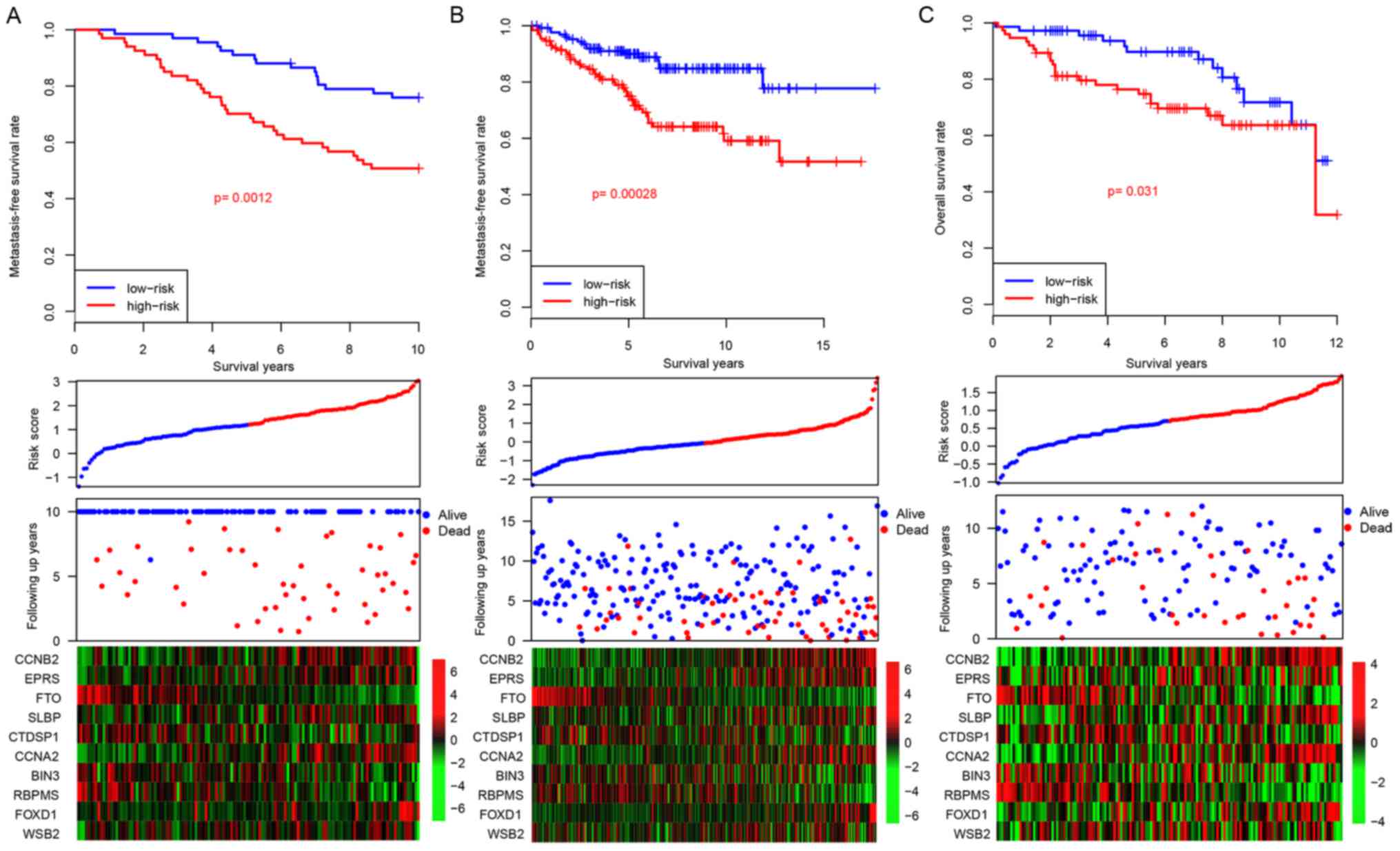 | Figure 3.Risk score in the test datasets. The
performance of risk score in three independent datasets: (A)
GSE22219, (B) GSE26971 and (C) GSE58644. CCNB2, cyclin B2; EPRS,
glutamyl-prolyl-tRNA synthetase; FTO, α-ketoglutarate dependent
dioxygenase; SLBP, stem-loop binding protein; CTDSP1, CTD small
phosphatase 1; CCNA2, cyclin A2; BIN3, bridging integrator 3;
RBPMS, RNA binding protein with multiple splicing; FOXD1, forkhead
box D1; WSB2, gene encoding WD repeat of SOCS box containing 2. |
Risk score and clinical
information
Subsequently, the associations between clinical
parameters (stage, age, grade, lymph node invasion and primary
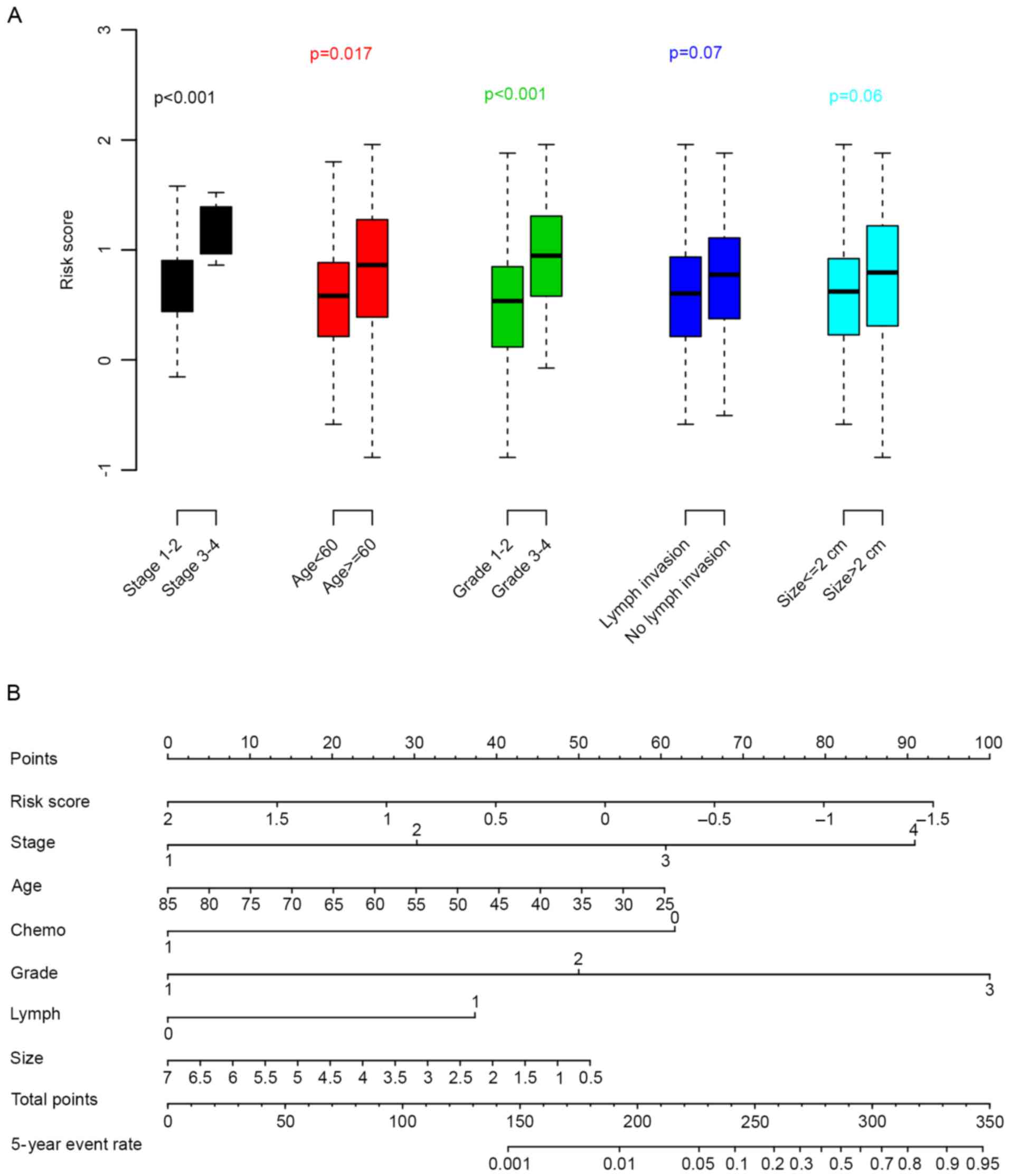
tumor size) with the risk score were evaluated. As revealed in
Fig. 4A, age, tumor stage and
grade were significantly associated with the risk score; whereas
the other clinical parameters were not (P>0.05). To facilitate
the utilization of the risk score, a 5-year distant relapse
nomogram was plotted (Fig. 4B).
According to this nomogram, risk score was one of the most
important metastatic indicators.
Discussion
Tamoxifen is the most frequently used drug for the
treatment of patients with ER+ breast cancer. However, tamoxifen
drug resistance has previously been observed (2). The underlying mechanism of how
tamoxifen drug resistance develops remains unclear. In order to
predict the survival time of patients treated with tamoxifen, this
study has developed a predictive risk score staging system based
upon gene expression levels. According to the developed model, the
risk score successfully predicted the survival time of patients
across both training and test datasets. In addition, associations
between risk score and pathological parameters were assessed. The
proposed nomogram demonstrated that the risk score was one of the
most important indicators for prognosis.
Among the included genes, FOXD1 has previously been
reported to promote migration and to be associated with drug
resistance in glioma (12). CCNA2
was revealed to correlate closely with distant metastasis-free,
recurrence-free and overall survival in breast cancer; in addition,
it also contributes to tamoxifen resistance in patients with ER+
breast cancer (13). CCNB2 has
previously been demonstrated to serve as an independent biomarker
for invasive breast cancer, and elevated CCNB2 has previously been
revealed to be associated with poor patient survival (14). Although little is known about FTO
expression and breast cancer, gene polymorphism of FTO has been
revealed to be associated with carcinogenesis and survival of
patients with breast cancer (15,16).
Another gene, CTDSP1, inhibits cancer cell migration and invasion
(17). According to recent
findings, EPRS is a regulator of cell proliferation in ER+ breast
cancer, and reduced EPRS expression has been demonstrated to be
associated with decreased distant relapse-free survival in patients
treated with tamoxifen for 5 years (18). Enhanced RBPMS expression has been
revealed to significantly repress activator protein 1 signaling
activity, and thus regulate the proliferation and migration of
breast cancer cells (19). The
aforementioned candidate genes were either associated with survival
of breast cancer patients or tamoxifen resistance/sensitivity, thus
explaining why a risk score based upon the expression levels of
said genes has proved to be effective for the survival prediction
time period of patients with ER+ breast cancer. However, it was
revealed that none of the candidate genes were significantly
associated with survival across all of the included datasets (data
not shown), thus indicating that the expression level of a single
gene as a predictive measure for the survival time period of
patients with ER+ breast cancer is not as robust as a cumulative
risk score.
In conclusion, the current model developed in this
study is robust across datasets in the prediction of the survival
time of patients with ER+ breast cancer treated with tamoxifen.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2012. View Article : Google Scholar
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zembutsu H: Pharmacogenomics toward
personalized tamoxifen therapy for breast cancer. Pharmacogenomics.
16:287–296. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liu J, Prager-van der Smissen WJ, Look MP,
Sieuwerts AM, Smid M, Meijer-van Gelder ME, Foekens JA, Hollestelle
A and Martens JW: GATA3 mRNA expression, but not mutation,
associates with longer progression-free survival in ER-positive
breast cancer patients treated with first-line tamoxifen for
recurrent disease. Cancer Lett. 376:104–109. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gu Y, Chen T, Li G, Xu C, Xu Z, Zhang J,
He K, Zheng L, Guan Z, Su X, et al: Lower Beclin 1 downregulates
HER2 expression to enhance tamoxifen sensitivity and predicts a
favorable outcome for ER positive breast cancer. Oncotarget.
8:52156–52177. 2016.PubMed/NCBI
|
|
6
|
Reijm EA, Timmermans AM, Look MP,
Meijer-van Gelder ME, Stobbe CK, van Deurzen CH, Martens JW,
Sleijfer S, Foekens JA, Berns PM and Jansen MP: High protein
expression of EZH2 is related to unfavorable outcome to tamoxifen
in metastatic breast cancer. Ann Oncol. 25:2185–2190. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bou Samra E, Klein B, Commes T and Moreaux
J: Development of gene expression-based risk score in
cytogenetically normal acute myeloid leukemia patients. Oncotarget.
3:1–832. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Salazar R, Roepman P, Capella G, Moreno V,
Simon I, Dreezen C, Lopez-Doriga A, Santos C, Marijnen C, Westerga
J, et al: Gene expression signature to improve prognosis prediction
of stage II and III colorectal cancer. J Clin Oncol. 29:17–24.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bou Samra E, Klein B, Commes T and Moreaux
J: Identification of a 20-gene expression-based risk score as a
predictor of clinical outcome in chronic lymphocytic leukemia
patients. Biomed Res Int. 2014:4231742014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kim SK, Kim SY, Kim JH, Roh SA, Cho DH,
Kim YS and Kim JC: A nineteen gene-based risk score classifier
predicts prognosis of colorectal cancer patients. Mol Oncol.
8:1653–1666. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Robin X, Turck N, Hainard A, Tiberti N,
Lisacek F, Sanchez JC and Müller M: pROC: An open-source package
for R and S+ to analyze and compare ROC curves. BMC Bioinformatics.
12:772011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gao YF, Zhu T, Mao XY, Mao CX, Li L, Yin
JY, Zhou HH and Liu ZQ: Silencing of Forkhead box D1 inhibits
proliferation and migration in glioma cells. Oncol Rep.
37:1196–1202. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gao T, Han Y, Yu L, Ao S, Li Z and Ji J:
CCNA2 is a prognostic biomarker for ER+ breast cancer and tamoxifen
resistance. PLoS One. 9:e917712014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shubbar E, Kovács A, Hajizadeh S, Parris
TZ, Nemes S, Gunnarsdóttir K, Einbeigi Z, Karlsson P and Helou K:
Elevated cyclin B2 expression in invasive breast carcinoma is
associated with unfavorable clinical outcome. BMC Cancer. 13:12013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tan A, Dang Y, Chen G and Mo Z:
Overexpression of the fat mass and obesity associated gene (FTO) in
breast cancer and its clinical implications. Int J Clin Exp Pathol.
8:13405–13410. 2015.PubMed/NCBI
|
|
16
|
Zeng X, Ban Z, Cao J, Zhang W, Chu T, Lei
D and Du Y: Association of FTO mutations with risk and survival of
breast cancer in a Chinese population. Dis Markers.
2015:1010322015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sun T, Fu J, Shen T, Lin X, Liao L, Feng
XH and Xu J: The small c-terminal domain phosphatase 1 inhibits
cancer cell migration and invasion by dephosphorylating
ser(p)68-twist1 to accelerate twist1 protein degradation. J Biol
Chem. 291:11518–11528. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Katsyv I, Wang M, Song WM, Zhou X, Zhao Y,
Park S, Zhu J, Zhang B and Irie HY: EPRS is a critical regulator of
cell proliferation and estrogen signaling in ER+ breast cancer.
Oncotarget. 7:69592–69605. 2016.PubMed/NCBI
|
|
19
|
Fu J, Cheng L, Wang Y, Yuan P, Xu X, Ding
L, Zhang H, Jiang K, Song H, Chen Z and Ye Q: The RNA-binding
protein RBPMS1 represses AP-1 signaling and regulates breast cancer
cell proliferation and migration. Biochim Biophys Acta. 1853:1–13.
2015. View Article : Google Scholar : PubMed/NCBI
|