Introduction
Cholesterol plays a critical role in normal cellular
functions, serving as a structural component within cell membranes,
a signaling modulator and a form of energy storage (1). Although cholesterol is essential for
life, its excessive accumulation or abnormal distribution leads to
significant health issues, such as hypercholesterolemia,
atherosclerosis, cardiovascular disease and nonalcoholic fatty
liver disease (NAFLD) (2).
Organisms and cells must maintain the balance between the internal
and external sources of cholesterol, while avoid either shortage or
over-accumulation of cholesterol. Both the biosynthetic and uptake
pathways for cholesterol are tightly controlled by precise
regulatory mechanisms (3).
Sterol regulatory element-binding protein 2
(SREBP-2) is considered a master regulator of cholesterol
biosynthesis (4–6). A number of gene promoter analyses
have revealed that the expression of most genes encoding
cholesterol biosynthetic enzymes, particularly
3-hydroxy-3-methylglutaryl CoA reductase (HMGCR), is regulated
largely by SREBP-2 (5). HMGCR is a
rate-limiting enzyme in cholesterol synthesis, which is also a
target of statins. The expression of SREBP-2 is regulated by many
transcription factors and microRNAs (miRs), including NF-κB and
miR-185 (5,6). When intracellular cholesterol is
depleted, SREBP-2 activates HMGCR expression to enhance cholesterol
biosynthesis (7). In contrast,
following accumulation of intracellular cholesterol, both of the
expression and activation of HMGCR by SREBP-2 are inhibited, and,
specifically, HMGCR activity is decreased by more than 90% in
cholesterol-loaded cells (5).
Previous studies have demonstrated that gerbils
promptly responded to the intake of dietary cholesterol, followed
by increased serum cholesterol, hepatic cholesterol ester and LDL-C
levels in humans (8,9). We have previously demonstrated that
simvastatin (8 mg/kg/d), an HMGCR-specific inhibitor, rapidly and
efficiently decreased serological total cholesterol (TC) level by
37.99% in hypercholesterolemic gerbils within 2 weeks (10). Moreover, lovastatin (8 mg/kg/d) has
also been shown to reduce this level by 34.03% in
hypercholesterolemic gerbils within 4 weeks (11). In comparison, simvastatin (12
mg/kg/d) has been reported to decrease the TC level by 25.63% in
hypercholesterolemic Kunming mice within 4 weeks (12). Whereas simvastatin (2 mg/kg/d) has
been shown to decrease by 9.87% in hypercholesterolemic rats within
6 weeks (13). Interestingly,
Simvastatin exhibits greater efficiency in hypercholesterolemic
gerbils than in mice or rats. These results suggested that
considerable endogenous cholesterol is synthesized in
hypercholesterolemic gerbils. In attempt to elucidate the molecular
mechanism of potent response to cholesterol regulation, we analyzed
the role of SREBP-2 during cholesterol synthesis in the gerbil
liver through regulating SREBP-2 protein expression. The results of
this study will enrich our understanding of the function of SREBP-2
in cholesterol metabolism in gerbils, which provide further
insights into the pathological mechanism of
hypercholesterolemia.
Materials and methods
Animal treatments
All experiments and animal procedures were conducted
in accordance with the Guidelines of the Capital Medical University
Animal Experiments and Experimental Animals Management Committee.
The protocol was approved by the Animal Experiments and
Experimental Animal Welfare Committee of Capital Medical University
(Permit number: 2011-X-009).
Male gerbils (Merionesunguiculatus) weighing
50–60 g (7–8 weeks of age) were obtained from a closed colony kept
at Chinese Capital Medical University. All gerbils were housed
under controlled conditions (room temperature of 23±2°C, humidity
of 55±10% and 12 h light/dark cycle) with free access to food and
water. The gerbils were allowed to acclimate to the environment for
1 week prior to experiments. Thirty gerbils were randomly divided
into three groups of ten gerbils each: a normal control group (CG),
model control group (MG) and recovery group (RG). The gerbils in
the CG were fed the normal diet for 2 weeks, and those in the MG
were fed a high-fat diet (HFD) for 2 weeks. The gerbils in the RG
were fed the HFD for 2 weeks and were then fed the normal diet for
additional 2 weeks. The HFD was composed of 80.5% normal diet, 2%
cholesterol, 7% lard, 10% yolk powder and 0.5% bile salts (Huadong
Medicine, Hangzhou, China) according to previous reports (10,11).
The nutritional differences between the normal diet and HFD are
shown in Table I. At the end of
experiment, diets were removed from cages at 12 h before the
gerbils were sacrificed, the body weights (BWs), liver weights
(LWs) were measured, and serum samples were obtained.
 | Table I.After treated by HFD, the BW, LW, and
7 serum biochemical markers value in gerbils. |
Table I.
After treated by HFD, the BW, LW, and
7 serum biochemical markers value in gerbils.
| Groups | BW (g) | LW (g) | ALT (U/l) | AST (U/l) | TC (mM) | TG (mM) | HDL-c (mM) | LDL-c (mM) | TBA (µM) |
|---|
| CG |
66.7±8.1 |
2.0±0.24 |
80.6±16.4 |
194.5±31.1 |
2.62±0.80 |
0.58±0.08 |
1.51±0.25 |
1.57±0.4 |
29.7±13.7 |
| MG |
68.5±7.6 |
2.6±0.35b |
118.2±23.7b |
278.4±21.5a |
12.74±4.5b |
0.64±0.10a |
5.15±1.4b |
11.03±2.5b |
48.2±14.7b |
| RG |
70.6±8.3 |
2.3±0.54c |
104.5±18.5c |
260.4±25.8c |
8.35±4.6d |
0.61±0.09 |
3.35±1.3c |
7.43±2.8d |
41.6±10.4 |
Biochemical analysis
Blood samples were collected from the coeliac artery
after the gerbils were anesthetized with chloralic hydras and
centrifuged at 3,500 g for 15 min to obtain sera. The levels of
serum total cholesterol (TC), triglycerides (TG), low-density
lipoprotein cholesterol (LDL-C), high-density lipoprotein
cholesterol (HDL-C), alanine aminotransferase (ALT), aspartate
aminotransferase (AST) and serum total bile acids (TBA) were
measured on an automatic chemical analyzer (Hitachi, Ltd., Tokyo,
Japan) using commercially enzymatic kits (Nanjing Jiancheng
Bioengineering Institute, Nanjing, China) according to the
manufacturer's instructions.
Hepatic histological analysis
Small pieces of gerbil liver tissue were collected
from the same location from each gerbil and fixed with 10% formalin
for 24 h. The samples were then paraffin-embedded, and 4–5 µm
sections of were prepared. The sections were subsequently stained
with hematoxylin and eosin (H&E). Histological evaluation was
performed twice by a pathologist who was blind to the animal
groups. In brief, the following criteria were used for scoring
hepatic steatosis: Grade 0 (−), no fat; grade 1 (+), fatty hepatic
cells occupying 33% of the hepatic parenchyma; grade 2 (++), fatty
hepatic cells occupying 33–66% of the hepatic parenchyma; and grade
3 (+++), fatty hepatic cells occupying >66% of the hepatic
parenchyma (14).
RNA extraction and cDNA cloning
Total RNA was extracted from tissues or hepatic
cells using Trizol RNA isolation reagent (Invitrogen, Carlsbad, CA,
USA) following the manufacturer's instructions. RNA quality and
concentration were assessed using a NanoDrop 1000 (Thermo Fisher
Scientific, Inc., Waltham, MA, USA). cDNA was synthesized using a
First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). Primers
(GSP1-3, 3′116-1-2) were designed for the cloning of SREBP-2 cDNA
based on the conserved regions of the human (H. sapiens),
rat (R. norvegicus) and mouse (M. musculus) mRNAs
(Table II). A 5′/3′ rapid
amplification of cDNA ends (RACE) system (18374–58 and 18373-019;
Invitrogen) was used to amplify cDNA fragment using the
inversely-transcribed cDNA as template, according to the
manufacturer's instructions, under the following conditions: 1 min
at 94°C for 1 cycle; 30 sec at 94°C, 30 sec at 66°C, and 2 min at
72°C for 5 cycles; 30 sec at 94°C, 30 sec at 64°C, and 2 min at
72°C for 5 cycles; 30 sec at 94°C, 30 sec at 62°C, and 2 min at
72°C for 30 cycles, and 10 min at 72°C as the final extension
step.
 | Table II.The homologies of SREBP-2 sequences
gerbil compared with others animals. |
Table II.
The homologies of SREBP-2 sequences
gerbil compared with others animals.
|
| cDNA sizs (bp) | ORF size (bp) | cDNA homology
(%) | Deduced amino acid
homology (%) |
|---|
| Mus
musculus | 4570 | 3393 | 89.0 | 96.5 |
| Rattus
norvegicus | 5060 | 3402 | 88.3 | 96.0 |
| Cricetulus
griseus | 4987 | 3420 | 89.4 | 95.1 |
| Homo
sapiens | 4325 | 3426 | 82.7 | 93.7 |
| Bos
taurus | 5194 | 3423 | 76.1 | 92.2 |
| Xenopus
(Silurana) tropicalis | 4167 | 3264 | 60.3 | 69.9 |
| Danio
rerio | 4496 | 3300 | 51.7 | 52.6 |
Construction of over-expression and
shRNA-expressing plasmids
Gerbil SREBP-2 cDNA was obtained by RT-PCR using
total RNA isolated from liver tissues. The PCR products were
digested with Sal I and BamH I via incorporation of
the corresponding restriction sites into the PCR primers (Table III), and were then cloned into a
vector, pIERS2-EGFP, containing the CMV promoter, thereby
generating an SREBP-2 expression construct (P55-35 and P55-39). The
entire insert in the construct was sequenced and verified by
restriction endonuclease digestion and sequencing, performed by
Ruibiotech, Inc (Beijing, China).
 | Table III.The primer sequences used for cloning
SREBP-2 and qPCR analysis. |
Table III.
The primer sequences used for cloning
SREBP-2 and qPCR analysis.
| Purpose | Name | From sequence to
sequence (5′-3′) | Product (bp) |
|---|
| Degenerate
primers | SF1 |
CGGCGGACCACCCACAATATCAT | 113 |
|
| SR1 |
CGCCGGACTTGTGCATCTTGGCGTC |
|
|
| SF2 |
CARCGGYCVTTCASCCAGGTC | 809 |
|
| SR2 |
TCAGGARGCRGCRATGGCVGTGCC |
|
|
| SF3 |
GCGTCTGTTCCCATGACCAAGT | 2,355 |
|
| SR3 |
ATGGGAACAGACGCCAAGATGC |
|
| RACE primers | GSP-1 |
CGTGCTGAATGTTGGG | 472 |
|
| GSP-2 |
GGGTTGTCCTTGGTGGGGTAG |
|
|
| GSP-3 |
AGGGCTGGAGCCTGAGGGGAG |
|
|
| 3′116-1 |
GGCTGAAGCCGCCCGCACCCTGGAGAA | 836 |
|
| 3′116-2 |
GGGTGACCGCCGTTCCTGCAATGACTG |
|
| qPCR | SREBP-2 |
CGCTCTTCAAGTACCAACCCT | 223 |
|
|
|
CATGACCAAGTCCTTCAGCTCTA |
|
|
| HMGCR |
GGGTATTGCTGGCCTGTTCA | 241 |
|
|
|
AGCATCCAGAGTGAATGTGGG |
|
|
| GAPDH |
CTGGTCGTACCACTGGCATT | 234 |
|
|
|
AGTCCAGGGCAACATAGCAC |
|
A vector-based shRNA expression system was used to
endogenously express shRNA in mammalian cells. We selected the
target regions in the SREBP-2 cDNA sequence according to Tushul's
principle (15). DNA
oligonucleotides (SL23-1, SL23-3, NC-2 and NC-4) for in
vitro transcription were designed (Table IV) with BamH I and
Hind III sites at both ends and cloned into a vector,
pRNAT-U6.1/Neo, containing the CMV promoter. A control DNA
construct (NC) was generated by insertion of a scrambled sequence
that did not show significant sequence homology to rat, mouse, or
human gene sequences. The shRNA sequences the targeting SREBP-2
gene were synthesized by Ruibiotech, Inc. All of the inserted
sequences were identified by restriction-endonuclease digestion and
sequenced by Ruibiotech, Inc.
 | Table IV.The primer sequences used for SREBP-2
shRNA and over-expression vectors constructed. |
Table IV.
The primer sequences used for SREBP-2
shRNA and over-expression vectors constructed.
| Purpose | Name | From sequence to
sequence (5′-3′) | Restriction enzymes
loci |
|---|
| shRNA vector | SL23-1 |
GATCCGGCTCTGGAGTATTTGAAATTTCAAGAGAATTT | BamH I,
HindIII |
|
|
|
CAAATACTCCAGAGCTTTTTTA |
|
|
| SL23-2 |
GATCCGCGGTCTCGAATTCTCTTATTTCAAGAGAATAA |
|
|
|
|
GAGAATTCGAGACCGTTTTTTA |
|
|
| NC-2 |
GATCCGCCGGTCTCGAATTCTCTTATTCAAGAGATAAG |
|
|
|
|
AGAATTCGAGACCGGTTTTTTA |
|
|
| NC-4 |
GATCCGTTCTCCGAACGTGTCACGTTTCAAGAGAACG |
|
|
|
|
TGACACGTTCGGAGAATTTTTTA |
|
| cDNA clone | OPF |
CGTGCCTATGACGTCAGATCCGCTAGCGCTACCGGACT | Sal I, |
|
|
|
CAGATCTCGAGCTCAAGCTTCGAATTCTGCAGTCGAC |
|
|
| OPR |
AGTCCGGAGTACGTTAGGGGGGGGGGAGGGAGAGGG | BamH I |
|
|
| GCGGATCC |
|
Cells culture and transfection
Primary hepatic cells were isolated from male
Mongolian gerbils by tissue digestion and collagenase perfusion.
The isolated hepatic cells were identified according to morphology
by light microscopy and periodic acid-Schiff reaction (PAS)
(16). They were then seeded at a
density of 5×105 cells/well in a 6-well plate and
cultured for 24 h to achieve a 60–80% confluence. To obtain stable
transfectants, the hepatic cells were transfected with the
over-expression and shRNA-plasmid constructs using Lipofect 2000
Plus reagent (Thermo Fisher Scientific) by incubation in serum-free
medium for 4 h at 37°C, according to the manufacturer's
recommendation. Subsequently, the transfection medium was removed
and fresh complete growth medium was added. After 48 h
post-transfection, GFP protein expression was assessed in the
hepatic cells.
Quantitative PCR (qPCR)
SREBP-2 and HMGCR gene expression levels were
detected in the gerbil livers and in primary hepatic cells by qPCR.
GAPDH gene expression was used as an internal control. The primer
sequences are shown in Table
III. Each qPCR mixture (total volume of 25 µl) contained 12.5
µl SYBR Premix ExTaq (Takara Biotechnology Co., Ltd., Dalian,
China), 2 µl normalized template cDNA from each tissue or from the
hepatic cells, 1 µl qPCR forward primer, 1 µl reverse primer and
8.5 µl RNase-free H2O (Tiangen, Beijing, China). The
qPCR amplification program was as follows: 95°C for 10 min,
followed by 40 cycles of 95°C for 5 sec and, 60°C for 30 sec, and a
final temperature increment of 0.5°C/sec from 65°C to 95°C. The
2−ΔΔCt method was used to analyze the relative
expression levels of the SREBP-2 and HMGCR genes.
Western blotting analysis
The gerbil livers or primary hepatic cells were
weighed, homogenized, and centrifuged in RIPA buffer containing
Protease and Phosphatase Inhibitor Cocktails (Thermo Fisher
Scientific, Inc.). Protein concentrations were determined using a
BCA assay kit (Boster Bioengineering Institute, Wuhan, China).
Then, 40 µg of each total protein extract were separated on 10%
SDS-PAGE gels and transferred to nitrocellulose membranes (EMD
Millipore, Billerica, MA, USA). The membranes were blocked with 5%
milk powder/TBS containing 5% Tween-20 at room temperature (RT) for
60 min, washed with TBST for 5 times, and then incubated with a
polyclonal anti-SREBP-2 (cat. no. ab28482; Abcam, Cambridge, MA,
USA) or anti-HMGCR (cat no. ab180615; Abcam) antibody or a control
GAPDH antibody (cat no. 2118; Cell Signaling Technology, Inc., St.
Louis, MO, USA) overnight at 4°C. After washed in TBST for 5 times,
the membranes were incubated with relevant secondary antibodies
conjugated with horseradish peroxidase (Merck KGaA, Darmstadt,
Germany) at RT for 1 h, and followed by another round of TBST
washes, and developed using Supersignal Chemiluminescence Substrate
(Thermo Fisher Scientific, Inc.). Protein signals were imaged and
analyzed using ChemiDoc XRS+ (Bio-Rad Laboratories,
Inc., Hercules, CA, USA). The semi-quantitative results were
normalized to expression of the housekeeping protein GAPDH after
gray scale scanning.
Cholesterol measurement
A total of 106 cells was washed 3 times
with cold PBS prior to lysis and extracted with 200 µl of a mixture
of chloroform: isopropanol: NP-40 (7:11:0.1) in a
micro-homogenizer. The extract was centrifuged for 10 min at 15,000
g. The organic phase was transferred to a new tube and dried
in a vacuum for 30 min. Then, the dried lipids were dissolved in
200 µl of 1× Assay Diluent and 1–50 µl of each sample were
quantified using commercially available kits (Cell Biolabs, Inc,
San Diego, CA, USA).
Statistical analysis
The results are presented as the means ± standard
error. Differences among the diet groups were detected by one-way
ANOVA with Duncan's post-hoc test with SAS version 9.0
statistical packages. P<0.01 or P<0.05 were considered to
indicate a statistically significant difference.
Results
Serum lipid level was increased by
HFD, but rapidly recovered when switched to normal diet in the
hypercholesterolemic gerbils
High calorie and fat intake is an important risk
factor for hypercholesterolemia, fatty liver, atherosclerosis and
coronary heart disease. As shown in Table I, HFD treatment significantly
increased the serum ALT (P<0.01), AST (P<0.05), TC
(P<0.01), TG (P<0.05), LDL-C (P<0.01), HDL-C (P<0.01),
and TBA level (P<0.01) in the MG compared with the CG. The serum
LDL-C level in the MG was increased by 7.0-fold compared with that
in the CG, whereas the serum TC level increased by 4.8-fold.
However, the normal diet significantly decreased the serum ALT
(P<0.05), AST (P<0.05), TC (P<0.01), LDL-C (P<0.01),
and HDL-C levels (P<0.05) in the RG compared with the MG. Serum
TG and TBA levels were also reduced in the RG compared with the MG;
however, these differences were not statistical significant. These
results indicated that HFD effectively increased the serum ALT,
AST, TC, LDL-C and HDL-C levels, whereas such effect was recovered
in RG group.
HFD increased accumulation of lipids
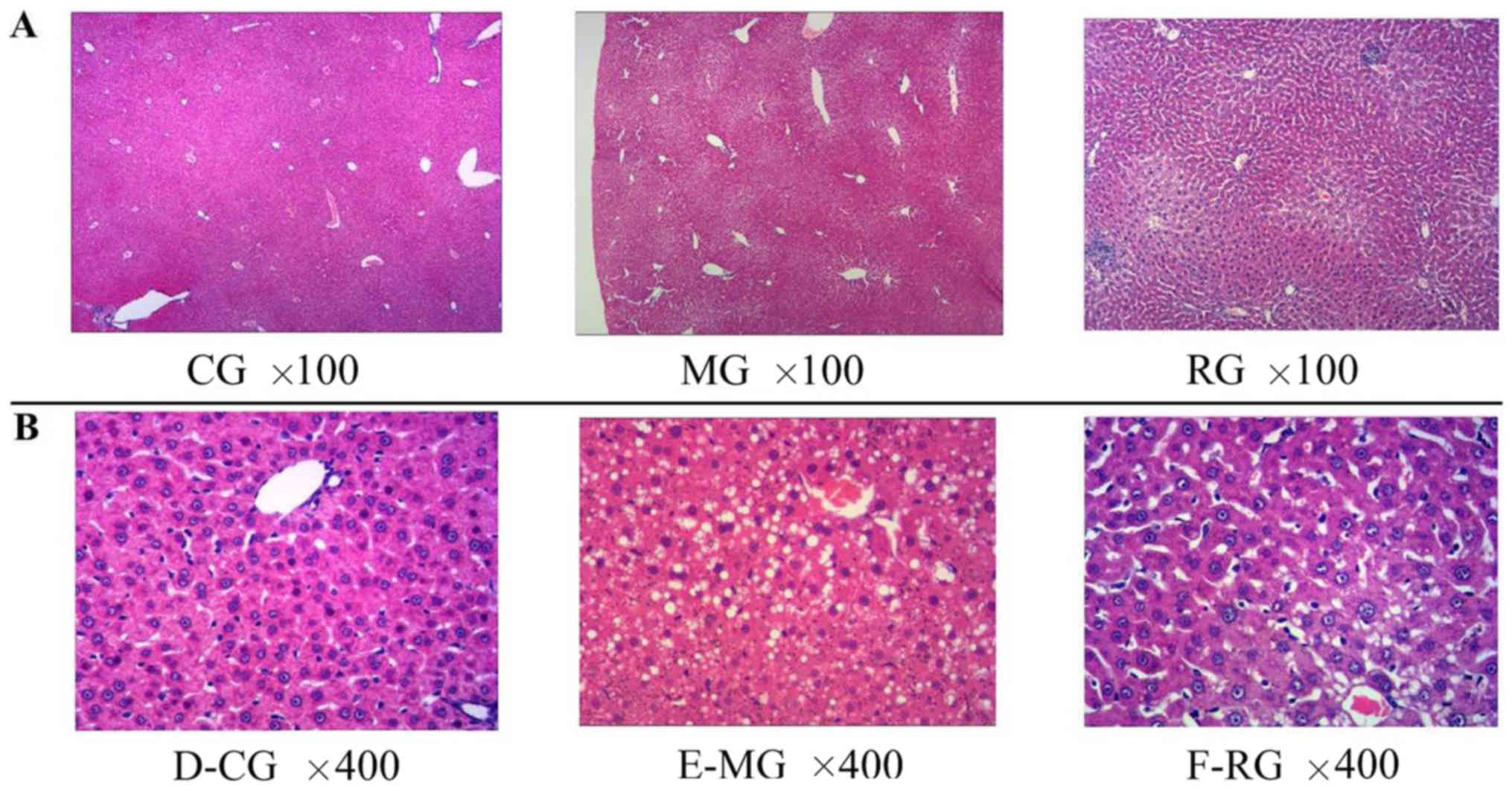
in gerbil liver
Body weight, and liver weight and histology were
used as indication for fatty liver (Table I). The results showed that HFD
significantly increased LW (P<0.01) in the MC group compared
with the CG group but that it did not significantly alter the BWs
(P>0.05). However, the normal diet significantly decreased LWs
(P<0.05) in RG group in contrast to MG group. Significant
histological differences were revealed in the animals in the MG
compared to those in the CG (Fig.
1). Strikingly, microvesicular fatty infiltration were found in
both MG and RG groups, whereas it was nearly absent in CG group.
The fatty liver scores were significantly higher in the MG
(1.7±0.9) (P<0.01) and RG (1.2±0.9) (P<0.05) than in the CG
(0.4±0.6). Furthermore, significant histological differences were
observed in RG group compared to CG group (Fig. 1A and B).
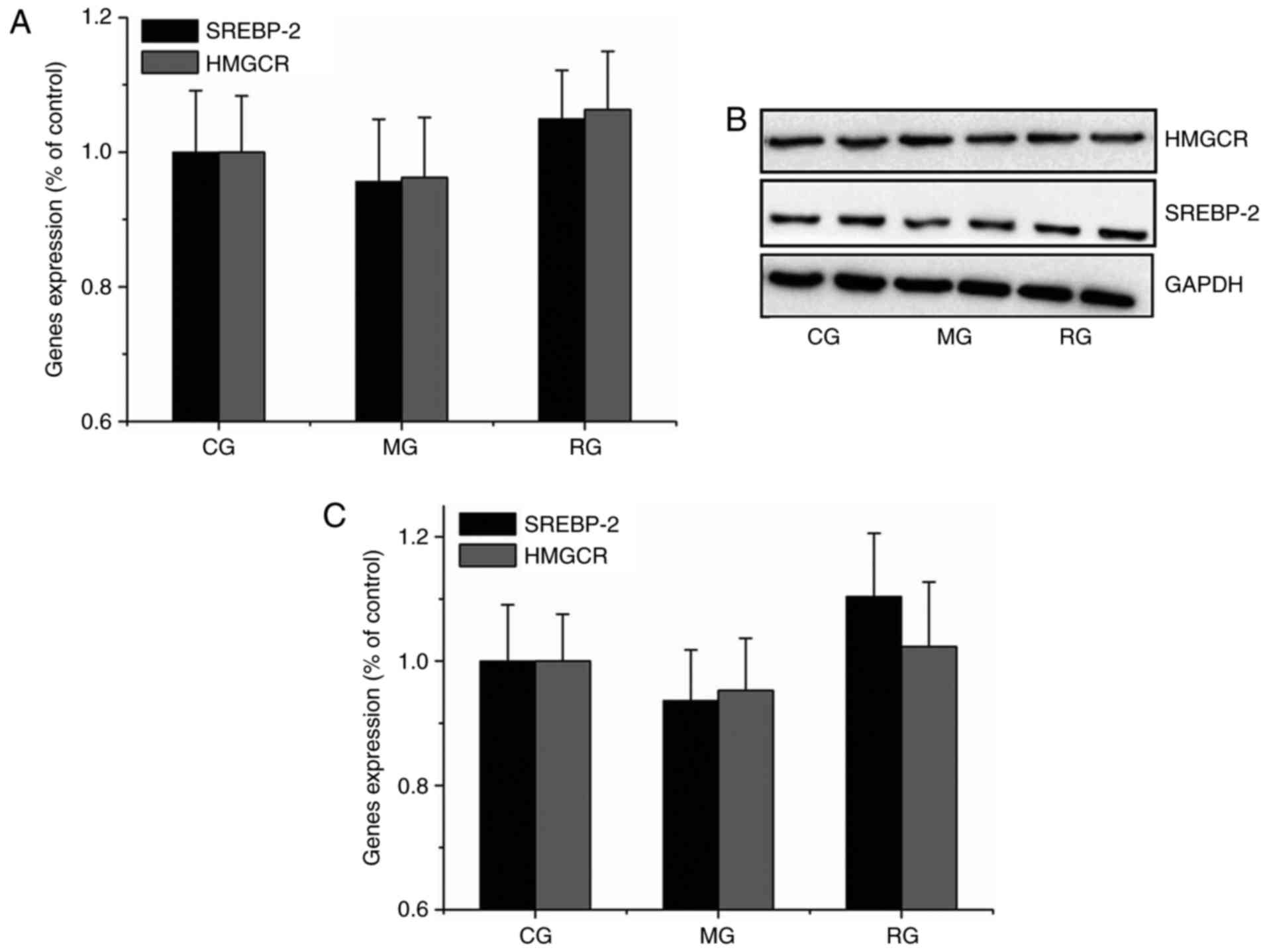
Neither SREBP-2 nor HMGCR expression
was inhibited in hypercholesterolemic gerbil livers
To assess whether susceptibility to dietary
cholesterol is related to SREBP-2 and HMGCR expression, their RNA
expression level was analyzed in gerbils. Similar results were
obtained in qPCR and Western blot analyses. Compared with the CG,
neither SREBP-2 nor HMGCR expression was significantly differed
(P>0.05) from MG and RG groups (Fig. 2), indicating that HFD treatment did
not sensitively or effectively decrease SREBP-2 and HMGCR RNA
levels, despite of increased fat deposit shown in the gerbil
livers.
SREBP-2 cDNA sequence showed that
gerbils were closely related to cricetidae
To study the function of SREBP-2 in lipid
metabolism, we cloned gerbil SREBP-2 cDNA for first time (Genbank
accession no. KR081464), and showed that full-length SREBP-2 cDNA
sequence is 3,949 bp long, containing a 142 bp of 5′-untranslated
region (UTR), a 759 bp of 3′UTR and an open reading frame (ORF) of
3,048 bp encoding a peptide with 1,135 amino acids. Bioinformatics
searches using Target Scan 6.1 and miRBase programs revealed one
response element for miR-185 within the 3′-UTR of gerbil SREBP-2
mRNA. The predicted molecular mass of SREBP-2 protein is 123.1 kDa
with a theoretical isoelectric point is 8.34. A characteristic
helix-loop-helix (HLH) domain span form the
326th-378th amino acids was identified, a key
domain through which SREBP-2 regulates the expression of target
genes.
Basic Local Alignment Search Tool (BLAST) analysis
of the amino acid sequences showed that the gerbil (M.
unguiculatus) sequences had high similarities to mouse (M.
musculus, Genbank accession no. NM_033218, 96.5% identity), rat
(R. norvegicus, Genbank accession no. NM_001033694, 96.0%
identity) and hamster (C. griseus, Genbank accession
no.U12330, 95.1% identity) sequences (Table II). In addition, BLAST analysis of
the nucleotide sequences also revealed that the gerbil sequences
had high similarities to hamster (92.9% identity), rat (90.0%
identity) and mouse sequences (89.0% identity) (Table II).
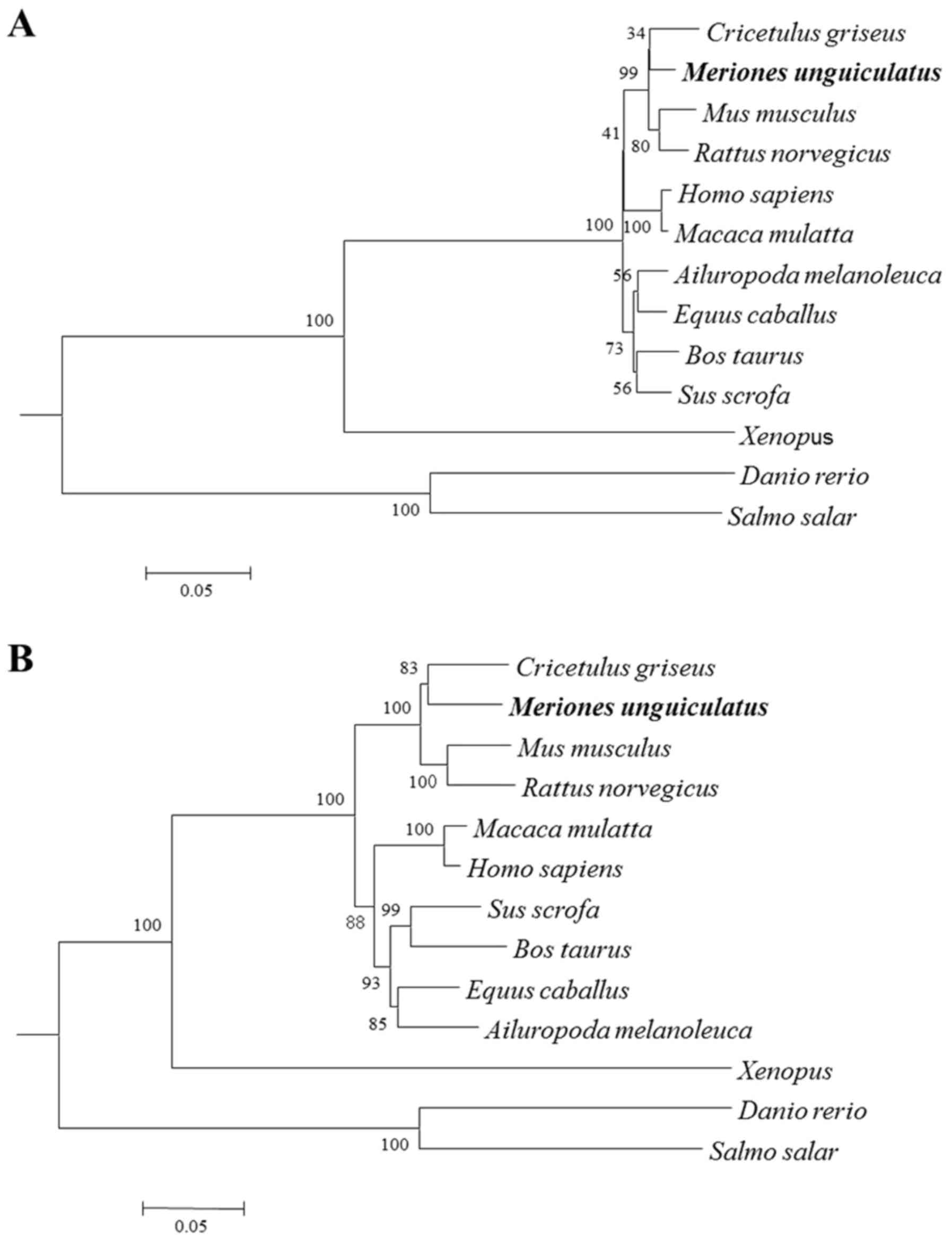
Phylogenetic trees were constructed by analyzing the
nucleotide and deduced amino acid sequences of gerbil SREBP-2
compared with those of other species. These phylogenetic trees,
generated based on the neighbor-joining (NJ) method, had similar
topologies. Gerbil SREBP-2 was found to be more closely related to
the hamster sequence, with high bootstrap support, and these
sequences fell into distinct clades with sequences from
Muridae (Fig. 3),
suggesting that the gerbils are more closely related to
Cricetidae.
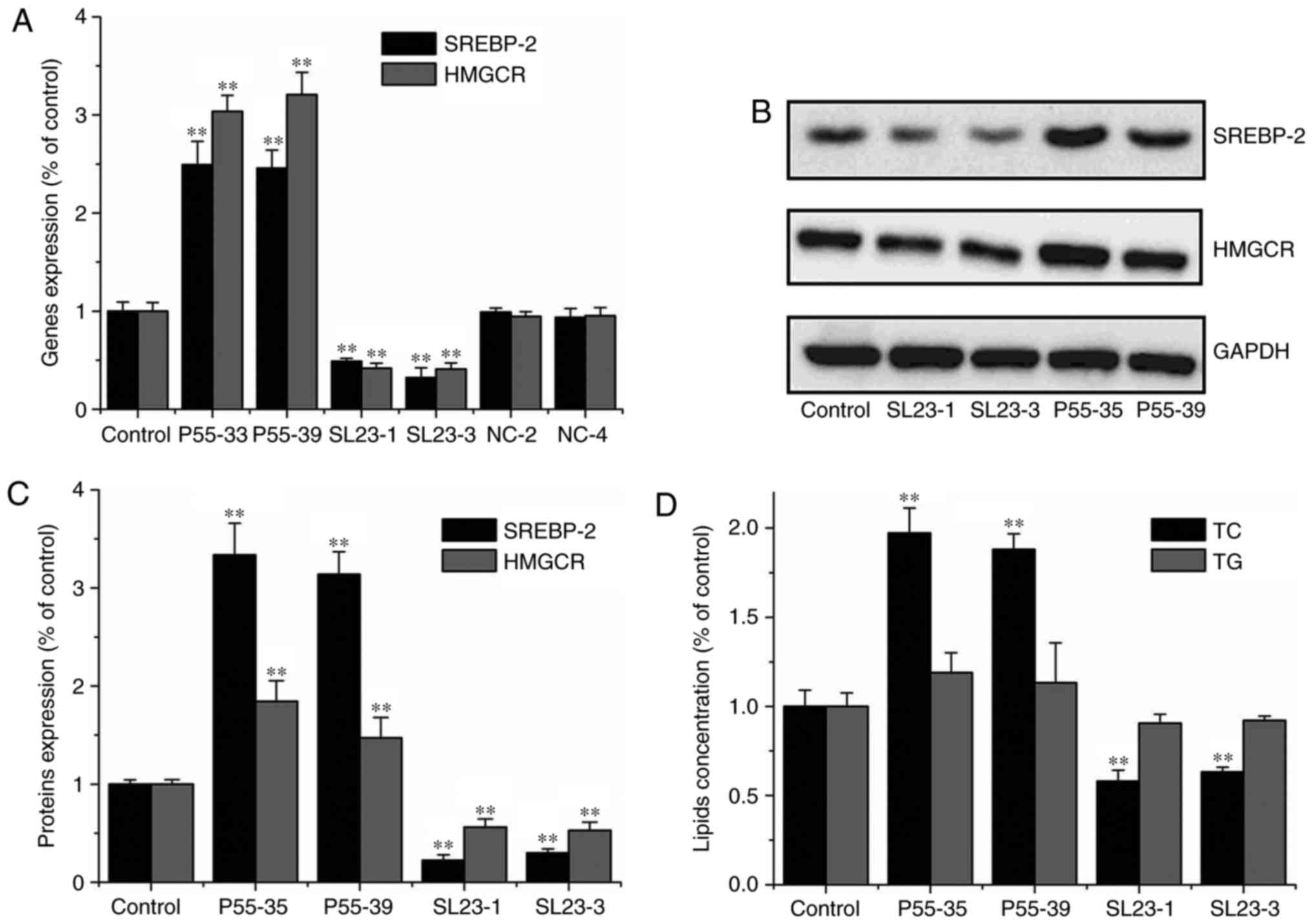
Over-expression of SREBP-2 promoted
HMGCR expression and increased TC level in hepatic cells
To assess whether increased SREBP-2 promoted the
expression and activation of HMGCR in gerbil hepatic cells, an
SREBP-2 over-expression plasmid (P55-33 and P55-39) was transfected
into gerbil primary hepatic cells. SREBP-2 and HMGCR gene
expression levels were significantly increased compared with those
in cells transfect with the control plasmid (P<0.01) (Fig. 4A-C). Consequently, intracellular
cholesterol level was also significantly increased (P<0.01);
however, intracellular triglyceride level was not increased
(Fig. 4D). These results showed
that over-expression of SREBP-2 by plasmid (P55-33 and P55-39)
significantly increased SREBP-2 and HMGCR expression in primary
hepatic cells. These results suggested that HMGCR expression and
cholesterol biosynthesis were effectively promoted by SREBP-2
expression in primary hepatic cells.
Inhibition of SREBP-2 expression and
decreased TC in hepatic cells
The shRNA plasmids (SL23-1 and SL23-3) were
constructed to explore SREBP-2-mediated regulation of HMGCR
expression and cholesterol biosynthesis in gerbil hepatic cells.
RNA and protein level of SREBP-2 in the primary hepatic cells
transfected with both knock-down plasmids, SL23-1 and SL23-3,
respectively, were significantly decreased, compared with cells
transfected with control plasmids (NC-2 and NC-4) (P<0.01)
(Fig. 4A-C). Moreover, knock-down
of SREBP-2 reduced HMGCR expression and intracellular cholesterol
levels in the hepatic cells (Fig.
4D). The results showed that inhibition of SREBP-2 expression
reduced HMGCR activation and decreased cholesterol biosynthesis in
the gerbil hepatic cells. However, inhibition of SREBP-2 expression
did not change intracellular triglyceride levels in the primary
hepatic cells (Fig. 4D).
Discussion
Although several animal models have been used to
explore the mechanism of cholesterol metabolism, there are
remarkable differences in mechanism between animals and humans. Rat
is a less satisfactory animal model, as the serum cholesterol
concentrations exhibit a relatively small elevation in response to
dietary cholesterol (17).
Moreover, rats metabolize excessive dietary cholesterol, result in
increased levels of the cholic acids and their secondary
derivatives, which are less well reabsorbed than cholic acid and
the derived bile acids (BAs) (17). Mice do not possess
cholesterol-ester transport protein (CETP) and exhibit up to
40-fold higher LDL clearance by the liver compared to humans
(18,19). Furthermore, most serum cholesterol
in mice is carried in HDL particles (18,19).
Currently, genetically modified models are also used in cholesterol
metabolism research, such as ApoE−/−
hypercholesterolemic mice. However, simvastatin caused a
paradoxical increasement in serum TC and cholesterol biosynthesis
in ApoE−/− mice (12).
Mongolian gerbil is an appropriate animal model to
study cholesterol metabolism. The levels of serum cholesterol in
the gerbils fed the different oils with no or very low levels of
dietary cholesterol were consistent with those seen in human fed
the same oils (8). Moreover,
gerbils and humans have similar BA patterns (17). In this study, HFD treatment rapidly
increased the concentration of serum TC (P<0.01), LDL-C
(P<0.01), TBA level (P<0.01) and the hepatic storage of
cholesterol, consistent with previous researches (12,16).
However, our results were inconsistent with a previous study
conducted by r Mercer & Holub (20). HFD used in the previous study was
deficient in bile salts and supplemental plant sterols (0.05%)
(20). Bile acid promoted the
intestinal absorption and hepatic storage of cholesterol (21). The diet with cholesterol and bile
acid increased cholesterol levels in the serum and liver, but
neither SREBP-2 nor HMGCR expression the liver was affected, which
agreed with a previous report (22). In contrast, the plant sterols
inhibited intestinal absorption of cholesterol (23). The consumption of 3.8–4.0 g/day of
plant sterol esters enhanced the whole-body cholesterol synthesis
and increased HMGCR and LDL receptor mRNA levels in human
mononuclear blood cells (23).
To maintain cholesterol homeostasis, biosynthesis,
intake and efflux during cholesterol metabolism are precisely
regulated (11). The conversion of
cholesterol into bile acids and its subsequent fecal excretion is
the most important approach to eliminate the cholesterol from the
body (19). In this study, we also
found that the serum TC, LDL-C and HDL-C levels were significant
decreased in RG group compared with MG group; however, the serum
TBA level was not significantly decreased. These results showed
that bile acids were synthesized by the liver in large amounts for
the efflux of excess cholesterol in the RG group. In addition,
histological analysis revealed that fat accumulation in the liver
was decreased in RG group, suggesting that cholesterol homeostasis
was effectively restoring by cholesterol excretion in the RG.
HMGCR is not only the rate-limiting enzyme in
cholesterol synthesis, but it is also a key target of certain
cholesterol-lowering drugs. It is well known that HMGCR is
ubiquitinated and rapidly degraded in cholesterol-loaded cells
(24). In such cells, HMGCR
expression was inhibited and HMGCR activity was decreased by over
90% (25). In this study, we found
that high intake of cholesterol effectively increased the serum TC
level but that the expression and activation of HMGCR in the liver
did not significantly differ between the MG and CG. We also found
that SREBP-2 expression in gerbil liver was not significantly
difference between MG and CG group. These results showed that
activation of HMGCR by SREBP-2 despite of a high cholesterol
intake. Further analysis revealed that altered SREBP-2 expression
resulted in the effective regulation of HMGCR and intracellular
cholesterol levels in hepatic cells. Our data also indicated that
stable expression of SREBP-2 maintained the expression and
activation of HMGCR, resulting in the synthesis of endogenous
cholesterol and accumulation lipids in livers of the gerbils fed
with HFD. Therefore, SREBP-2 expression contributes to the
susceptibility of gerbils to hypercholesterolemia. Consistent with
our conclusion, a previous study also revealed that SREBP-2
expression is not suppressed by cholesterol intake in insulin
receptor knockout (LIRKO) mice (26). Further, in the presence of a high
glucose concentration, cholesterol uptake and SREBP-2 expression
are simultaneously elevated in enterocyte (27). Along with previous findings, our
results further supported that the SREBP-2 expression pattern may
be associated with glycometabolism.
MicroRNAs are able to post-transcriptional regulate
gene expression by imperfect pairing with mRNAs in the 3′UTRs of
(28). HFD intake has been shown
to result in elevated miR-185 and miR-185 expression and effective
suppress SREBP-2 gene and protein through binding to four microRNA
response elements (MREs) in the 3′UTR mRNA (29). We found that the 3′UTR of the
gerbil SREBP-2 mRNA is only 759 bp, which is shorter than those in
humans, hamsters, and mice (1.6, 1.4 and 1.0 kb, respectively).
Moreover, there is only one predicted MRE of miR-185 in the gerbil
SREBP-2 mRNA. This may be contributed to altered SREBP-2 expression
pattern we observed in gerbil.
In the present study, we have cloned the SREBP-2
gene in gerbils. We identified high sequence homology among gerbils
and mice, rats and hamsters based on the nucleotide and deduced
amino acids sequences. Interestingly, phylogenetic analyses based
on the nucleotide and deduced amino acid sequences showed that
gerbils are more closely related to Cricetidae, which
differed from previous reports (29,30)
and from the information on the NCBI website (Taxonomy: 10047).
Elucidation of the SREBP-2 gene sequence will enhance understanding
of the susceptibility to hypercholesterolemia in Mongolian
gerbil.
We showed that HFD consumption resulted in rapid
elevation in the serum TC and LDL-C levels, but not altered HMGCR
or SREBP-2 expression; therefore, HMGCR activation was not
inhibited in the livers of hypercholesterolemic gerbils. Moreover,
HMGCR expression and activation were effectively regulated by
expression of SREBP-2 expression, as demonstrated by
over-expression and knock-down analyses using cultured hepatic
cells. Therefore, the failure to reduction in SREBP-2 expression
could be major reason for the susceptibility of gerbils to
hypercholesterolemia; it may be due to the shorter 3′UTR and lack
of MRE of miR-185 in the gerbil SREBP-2 mRNA.
Acknowledgements
The present study was supported by the National
Science Foundation of China (nos. 31000985, 31572348 and 31272393),
Key Projects in the National Science and Technology Pillar Program
(no. 2015BAI09B01) and Beijing Natural Science Foundation (no.
7141002).
Glossary
Abbreviations
Abbreviations:
|
TC
|
total cholesterol
|
|
TG
|
triglyceride
|
|
HDL-C
|
high density lipoprotein
cholesterol
|
|
LDL-C
|
low density lipoprotein
cholesterol
|
|
BWs
|
body weight
|
|
LWs
|
liver weight
|
|
ALT
|
alanine amino transferase
|
|
AST
|
aspartate amino transferase
|
|
TBA
|
serum total bile acids
|
|
SREBP-2
|
sterol regulatory element binding
protein 2
|
|
HMGCR
|
3-hydroxy-3-methylglutaryl CoA
reductase
|
|
miRs
|
micro RNA
|
|
UTR
|
untranslated region
|
|
MREs
|
microRNA response elements
|
References
|
1
|
Maxfield FR and Tabas I: Role of
cholesterol and lipid organization in disease. Nature. 438:612–621.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Liu J, Sempos CT, Donahue RP, Dorn R,
Trevisan J and Grundy SM: Nonhigh-density lipoprotein and
very-low-density lipoprotein cholesterol and their risk predictive
values in coronary heart disease. Am J Cardiol. 98:1363–1368. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sato R, Yang J, Wang X, Evans MJ, Ho YK,
Goldstein JL and Brown MS: Assignment of the membrane attachment,
DNA binding and transcriptional activation domains of sterol
regulatory element-binding protein-1 (SREBP-1). J Biol Chem.
269:17267–17273. 1994.PubMed/NCBI
|
|
4
|
Yang M, Liu W, Pellicane C, Sahyoun C,
Joseph BK, Gallo-Ebert C, Donigan M, Pandya D, Giordano C, Bata A
and Nickels JT Jr: Identification of miR-185 as a regulator of de
novo cholesterol biosynthesis and low density lipoprotein uptake. J
Lipid Res. 55:226–238. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Brown MS and Goldstein JL: The SREBP
pathway: Regulation of cholesterol metabolism by proteolysis of a
membrane-bound transcription factor. Cell. 89:331–340. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhao GJ, Tang SL, Lv YC, Ouyang XP, He PP,
Yao F, Tang YY, Zhang M, Tang YL, Tang DP, et al: NF-Κβ suppresses
the expression of ATP-bingding cassette transporter A1/G1 by
regulating SREBP-2 and miR-33a in mice. Int J Cardiol. 171:e93–e95.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
van der Wulp MY, Verkade HJ and Groen AK:
Regulation of cholesterol homeostasis. Mol Cell Endocrinol.
368:1–16. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hegsted DM and Gallagher A: Dietary fat
and cholesterol and serum cholesterol in the gerbil. J Lipid Res.
8:210–214. 1967.PubMed/NCBI
|
|
9
|
Forsythe WA III: Comparison of dietary
casein or soy protein effects on plasma lipids and hormone
concentrations in the gerbil (Meriones unguiculatus). J Nutr.
116:1165–1171. 1986.PubMed/NCBI
|
|
10
|
Li CL, Guo HG, Shi QJ, Lou Q and Sa XY:
Establishment of a Mongolian gerbil model of hypercholesterolemia
and the effect of simvastatin. Chin J Comp Med. 20:47–52. 2010.
|
|
11
|
Choudhury RP, Carrelli AL, Stern JD,
Chereshnev I, Soccio R, Elmalem VI, Fallon JT, Fisher EA and Reis
ED: Effects of simvastatin on plasma lipoproteins and response to
arterial injury in wild-type and apolipoprotein-E-deficient mice. J
Vasc Res. 41:75–83. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ying HZ, Liu YH, Yu B, Wang ZY, Zang JN
and Yu CH: Dietary quercetin ameliorates nonalcoholic
steatohepatitis induced by a high-fat diet in gerbils. Food Chem
Toxicol. 52:53–60. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jia YJ, Xu RX, Sun J, Tang Y and Li JJ:
Enhanced circulating PCSK9 concentration by berberine through
SREBP-2 pathway in high fat diet-fed rats. J Transl Med.
12:1032014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bonkovsky HL, Jawaid Q, Tortorelli K,
LeClair P, Cobb J, Lambrecht RW and Banner BF: Non-alcoholic
steatohepatitis and iron: Increased prevalence of mutations of the
HFE gene in non-alcoholic steatohepatitis. J Hepatol. 31:421–429.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Khanna A, Morelli AE, Zhong C, Takayama T,
Lu L and Thomson AW: Effects of liver-derived dendritic cell
progenitors on Th1- and Th2-like cytokine responses in vitro and in
vivo. J Immunol. 164:1346–1354. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen LQ, Li CL, Guo HG, Sa XY and Chen ZW:
Establishment of a primary culture protocol of Mongolian gerbil
hepatocytes. Acta Lab Anim Sci Sin. 21:15–18. 2013.
|
|
17
|
Bartizal KF Jr, Beaver MH and Wostmann BS:
Cholesterol metabolism in gnotobiotic gerbils. Lipids. 17:791–797.
1982. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dietschy JM, Turley SD and Spady DK: Role
of liver in the maintenance of cholesterol and low density
lipoprotein homeostasis in different animal species, including
humans. J Lipid Res. 34:1637–1659. 1993.PubMed/NCBI
|
|
19
|
Xie C, Turley SD and Dietschy JM:
Cholesterol accumulation in tissues of the Niemann-pick type C
mouse is determined by the rate of lipoprotein-cholesterol uptake
through the coated-pit pathway in each organ, Proc. Natl Acad Sci
USA. 96:11992–11997. 1999. View Article : Google Scholar
|
|
20
|
Mercer NJ and Holub BJ: Measurement of
hepatic sterol synthesis in the Mongolian gerbil in vivo using
[3H]water: Diurnal variation and effect of type of dietary fat. J
Lipid Res. 22:792–799. 1981.PubMed/NCBI
|
|
21
|
Murphy C, Parini P, Wang J, Björkhem I,
Eggertsen G and Gåfvels M: Cholic acid as key regulator of
cholesterol synthesis, intestinal absorption and hepatic storage in
mice. Biochim Biophys Acta. 1735:167–175. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hosomi R, Fukunaga K, Arai H, Kanda S,
Nishiyama T and Yoshida M: Fish protein hydrolysates affect
cholesterol metabolism in rats fed non-cholesterol and
high-cholesterol diets. J Med Food. 15:299–306. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Plat J and Mensink RP: Plant stanol and
sterol esters in the control of blood cholesterol levels: Mechanism
and safety aspects. Am J Cardiol. 96:15D–22D. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sever N, Song BL, Yabe D, Goldstein JL,
Brown MS and DeBose-Boyd RA: Insig-dependent ubiquitination and
degradation of mammalian 3-hydroxy-3-methylglutaryl-CoA reductase
stimulated by sterols and geranylgeraniol. J Biol Chem.
278:52479–52490. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Goldstein JL and Brown MS: Regulation of
the mevalonate pathway. Nature. 343:425–430. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Miao J, Haas JT, Manthena P, Wang Y, Zhao
E, Vaitheesvaran B, Kurland IJ and Biddinger SB: Hepatic insulin
receptor deficiency impairs the SREBP-2 response to feeding and
statins. J Lipid Res. 55:659–667. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Grenier E, Mailhot G, Dion D, Ravid Z,
Spahis S, Bendayan M and Levy E: Role of the apical and basolateral
domains of the enterocyte in the regulation of cholesterol
transport by a high glucose concentration. Biochem Cell Biol.
91:476–486. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Filipowicz W, Bhattacharyya SN and
Sonenberg N: Mechanisms of post-transcriptional regulation by
microRNAs: Are the answers in sight? Nat Rev Genet. 9:102–114.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chevret P and Dobigny G: Systematics and
evolution of the subfamily Gerbillinae (Mammalia, Rodentia,
Muridae). Mol Phylogenet Evol. 35:674–688. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Michaux J and Catzeflis F: The bush like
radiation of muroid rodents is exemplified by the molecular
phylogeny of the LCAT nuclear gene. Mol Phylogenet Evol.
17:280–293. 2000. View Article : Google Scholar : PubMed/NCBI
|