Introduction
Globally, liver cancer has the sixth highest
incidence for cancer, and it ranks in the top three for cancer
mortality. In some Asian countries, liver cancer is the leading
cause of cancer death in men (1).
In developed countries, such as the United States and Europe, the
incidence and mortality of liver cancer is also increasing year
after year (2,3). Ninety percent of liver cancer-induced
deaths are caused by tumour metastasis; bone is a common site of
metastasis, and the spine is the most common site of bone
metastasis (BM) (4). Liver cancer
patients with BM often experience severe pain with decreased
quality of life. The prognosis for these patients is poor: The
one-year survival rate of patients with spinal metastasis is only
30 percent after surgery (5,6). So,
it is of important clinical value to explore the mechanism of liver
cancer BM, especially the mechanism of spinal metastasis, and to
develop new biomarkers to evaluate the BM risk of primary liver
cancer.
Liver cancer is a complex disease involving
epigenetic instability, chromosomal instability, and expression
abnormalities, including miRNAs. miRNAs are a type of small
non-coding RNAs with lengths of approximately 20–24 nt which can
bind to the 3′UTR of target genes through complementary base
pairing and thereby inhibit the expression of the target genes
(7). Over 1,000 miRNAs have been
found to play vital roles, and it has been estimated that over 50%
mammalian protein coding genes may be regulated by miRNAs (8). By regulating the expression level of
different target genes, miRNAs are widely involved in the
regulation of various physiological processes, including organ
development, cell differentiation, and immune response, and are
closely related to tumour development (9). Some miRNAs even act as oncogenes or
tumour suppressors and modulate tumour progression and metastasis
(10).
Recently, cumulative evidence has suggested that the
dysregulation of miRNAs plays a pivotal role in the formation and
progression of liver cancer (11).
Thus far, several miRNAs are previously reported to be dysregulated
in liver cancer, including miR-21, miR-122, miR-181b, miR-338, and
miR-491 (12–16), which are all closely associated
with the migration and metastasis of liver cancer. miR-124a
functions as a tumour suppressor in osteosarcoma, endometrial
carcinoma, prostate cancer, and glioblastoma (17–20).
It is also downregulated in liver cancer tissues (21). However, the role of miR-124a in
liver cancer and potential targets of miR-124a in hepatoma cells
have not been studied previously. In this study, we identified that
interleukin (IL)-11, which plays a critical role in tumour
progression and metastasis, is a direct target of miR-124a. Several
studies have shown that IL-11 is upregulated in BM samples from
liver cancer patients and can promote liver cancer metastasis,
thereby affecting the prognosis of patients (22–25).
Furthermore, we discovered that the overexpression of miR-124a in
HepG2 cells suppresses cell proliferation, inhibits cell migration,
and promotes cell apoptosis through the repression of IL-11. These
findings could provide new views for the clinical treatment and
prevention of the development of liver cancer.
Materials and methods
Cell culture
The human liver cancer cell line, HepG2, was
obtained from American Type Culture Collection (ATCC; Manassas, VA,
USA) and cultured in RPMI-1640 medium (HyClone; GE Healthcare Life
Sciences, Logan, UT, USA) containing 10% foetal bovine serum (FBS;
Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) at 37°C in
a humidified 5% CO2 atmosphere. miR-124a mimics and
negative controls (NCs) were purchased from Shanghai GenePharma
Co., Ltd. (Shanghai, China). The sequences are as follows: miR-124a
mimics, 5′-UAAGGCACGCGGUGAAUGCC-3′ and 5′-CAUUCACCGCGUGCCUUAUU-3′;
RNA NC, 5′-UUCUCCGAACGUGUCACGUTT-3′ and
5′-ACGUGACACGUUCGGAGAATT-3′. Small interfering RNAs against IL-11
(sc-39636) was purchased from Santa Cruz Biotechnology, Inc.
(Dallas, TX, USA). At 24 h prior to experiments, cells were
transfected with miRNA-124a mimics, RNA NC or IL-11 siRNA according
to the manufacturer's recommendations. After 24 h of incubation,
cells were trypsinized and used for experimental assays.
MTT assay
HepG2 cells and cells transfected with miR-124a
mimics, NCs or IL-11 siRNA were plated in 96-well plates at
5×103 cells/well. A volume of 20 µl sterile methyl
thiazolyl tetrazolium (MTT) was added to the cells in each group
every 24 h in triplicate. After another 4 h incubation, the
supernatant was removed, and 150 µl dimethyl sulfoxide (DMSO) was
added into each well with shaking for 15 min. Absorbance was read
at 570 nm on an automatic microplate reader to determine the
optical density. The cell proliferation rate was calculated by the
optical density of the experimental group divided by that of the
blank group. Experiments were performed in triplicate.
Transwell chamber assay
We used transwell inserts (Corning, Beijing, China)
to assess cell migration. Cells were subjected to transfection, and
a cell suspension was prepared. RPMI-1640 medium with 1% BSA was
added to the upper chambers, and the lower chambers were filled
with 500 µl RPMI-1640 medium with 5% FBS. Approximately 100 µl of a
tumour cell suspension was added to the upper chambers and cultured
for 24 h. Then, cells on the lower chamber were stained with methyl
violet for 25 min and counted under a light microscope. Ten visual
fields were selected randomly, and the assay was performed in
triplicate.
Western blotting
Cells transfected with miR-124a mimics or NCs were
lysed at an appropriate time, and total protein was extracted with
the M-PER Mammalian Protein Extraction Reagent (Thermo Fisher
Scientific, Inc.). After blocking with 5% non-fat milk in TBS with
0.05% Tween-20 (TBST) for 1 h at room temperature, nitrocellulose
membranes were incubated overnight at 4°C with primary antibody
dissolved in 5% bovine serum albumin in TBST. The primary
antibodies used in this study were as follows: anti-human-MMP2
(1:1,000, 10373–2-AP; Wuhan Sanying Biotechnology, Wuhan, China),
anti-human-MMP9 (1:1,000, 10375-2-AP; Wuhan Sanying
Biotechnologya), anti-human-tissue inhibitor of matrix
metalloproteinase-2 (TIMP-2; 1:1,000, 17353-1-AP; Wuhan Sanying
Biotechnology), anti-human-Caspase-3 (1:500, 19677-1-AP; Wuhan
Sanying Biotechnology), anti-human-B-cell lymphoma 2 (Bcl-2;
1:1,000, 12789-1-AP; Wuhan Sanying Biotechnology),
anti-human-signal transducer and activator of transcription 3
(STAT3; 1:1,000, 10253-2-AP; Wuhan Sanying Biotechnology),
anti-human-p-STAT3 (1:500, AF3293; Affinity, Cincinnati, OH, USA),
and anti-human GAPDH (1:5,000, 10494-1-AP; Wuhan Sanying
Biotechnology) as an internal control. Membranes were incubated for
1 h at room temperature with secondary anti-rabbit Ig-HRP linked
(1:3,000, GAR0072; MultiSciences, Hangzhou, China). Immunoreactive
proteins were visualized by enhanced chemiluminescence. Image J
software (National Institutes of Health, Bethesda, MD, USA) was
used to compare the density of band on a western blot. We used the
Gel Analysis method to calculate the density of the protein bands
relative to the GAPDH standard band. Then we could make statistical
analysis.
Enzyme-linked immunosorbent assay
(ELISA)
To measure the secretion of IL-11, the supernatants
from different groups were collected after transfection and assayed
by ELISA kits according to the manufacturer's recommendations
(Human IL-11 QuantiCyto ELISA kit EHC128.96; Neobio, Shenzhen,
China). The absorbance values at 450 nm were used for analysis.
Experiments were performed in triplicate.
Dual-luciferase reporter gene
assay
Using the bioinformatics tools at TargetScan
(http://www.targetcan.org/), potential
targets of miR-124a were predicted. The binding sequences for
miR-124a in the 3′untranslated regions (3′UTR) of IL-11 were
mutated at positions 150 to 156 from GUG CCU U to CGC AGCA. And
then we constructed luciferase reporter vectors for the wild-type
and mutant-type of IL-11 3′UTR to verify IL-11 is the direct target
of miR-124a. Either miR-124a mimics or NC miRNA were co-transfected
with the constructed wild-type or mutant-type luciferase reporter
vector into 293T cells (ATCC) using Lipofectamine® 2000
(Thermo Fisher Scientific, Inc.). Firefly and renilla luciferase
activities were assessed with the Dual-Luciferase Reporter Assay
(Promega Corporation, Madison, WI, USA) 48 h after cell
transfection. The RLU value for renilla luciferase was divided by
the RLU value of firefly luciferase to obtain the final result.
Experiments were performed in triplicate.
Flow-cytometric analysis of
apoptosis
The apoptotic rates of HepG2 cells were detected
using an Annexin-V/FITC Kit (BD Biosciences, San Jose, CA, USA). In
brief, HepG2 cells were transfected with miR-124a mimics, NCs or
IL-11 siRNA and harvested 48 h after transfection by
trypsinization. The cell pellet was resuspended in 500 µl binding
buffer, 5 µl Annexin V-FITC, and 5 µl PI after washing in ice-cold
PBS. Then, it was incubated for 15 min in darkness. The ratios of
apoptotic cells were determined using a FACSCalibur flow cytometer
(BD Biosciences), and data were analysed with FlowJo software.
Experiments were performed in triplicate.
Statistical analysis
We used SPSS 19.0 software (SPSS, Inc, Chicago, IL,
USA) for statistical analysis. All data are presented as the means
± standard deviation. Comparisons between different groups were
analyzed by one-way ANOVA with a Bonferroni post hoc test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
miR-124a inhibits the proliferation
and migration of HepG2 cells
The proliferation and migration of malignant tumour
cells are important prerequisites for tumour development and
metastasis. In this study, we used MTT assays and Transwell chamber
assays to explore the biological functions of miR-124a by assessing
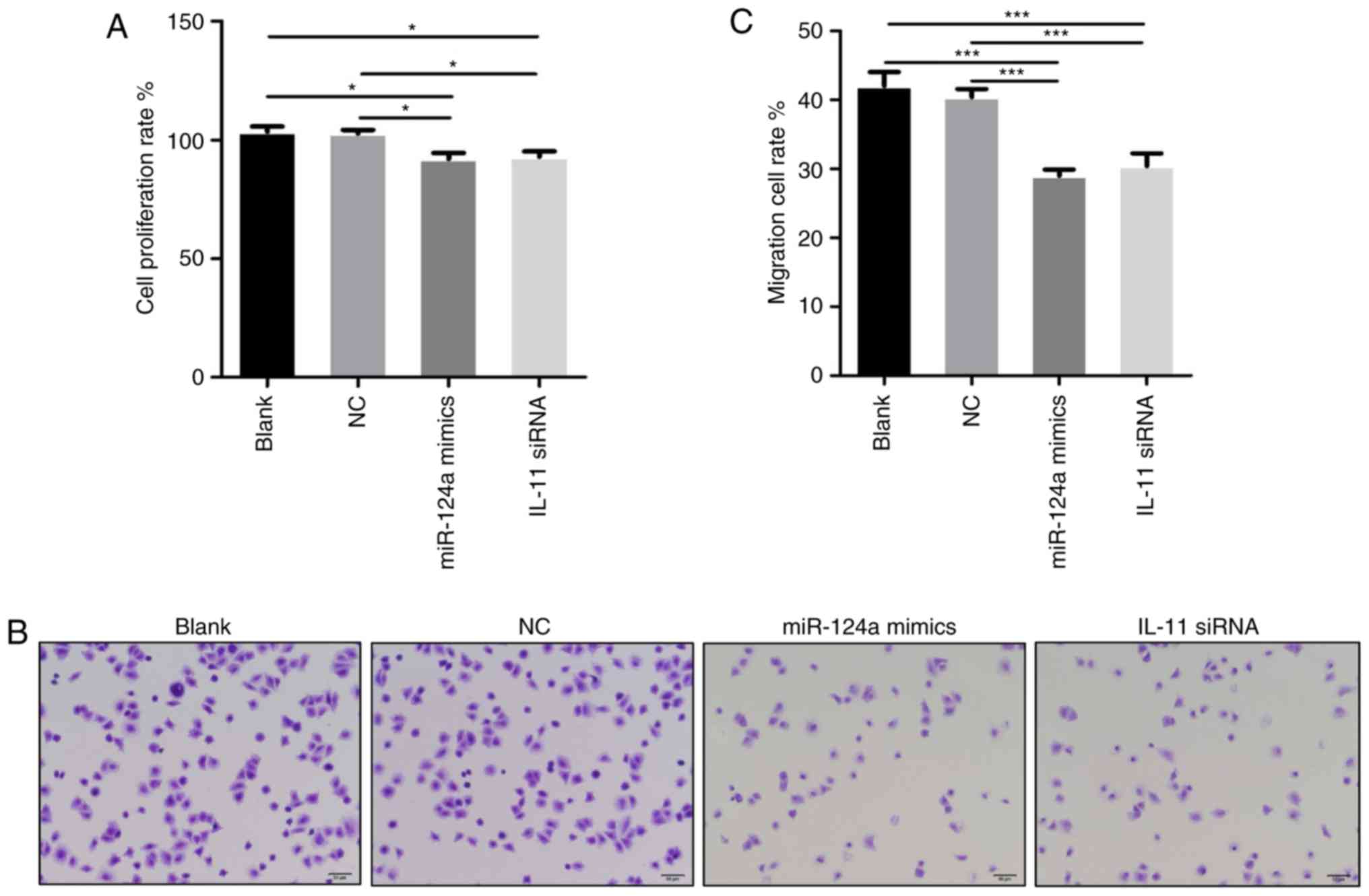
cell proliferation and migration rates. Significantly constrained
proliferation of HepG2 cells was observed when cells were
transfected with miR-124a mimics or IL-11 siRNA compared with the
blank or NC group (Fig. 1A). Cells
that migrated to the lower chamber were counted under a light
microscope to get the migration rates (Fig. 1B). There were significantly fewer
migrated cells in miR-124a mimics and IL-11 siRNA group than in the
blank and NC group (Fig. 1C). So,
these data indicate that IL-11 is involved in miR-124a-inhibited
proliferation and migration.
miR-124a promotes HepG2 cell
apoptosis
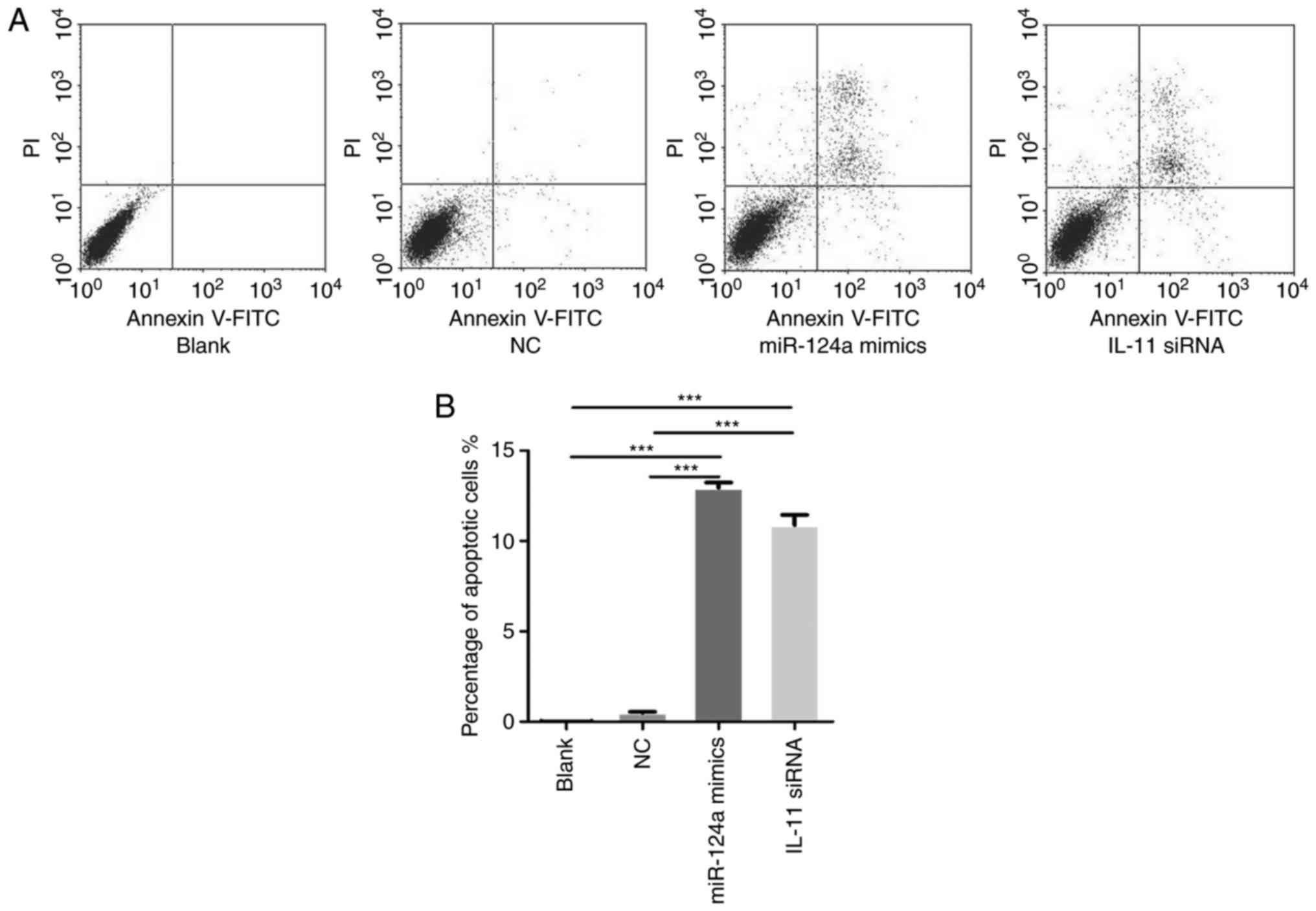
The regulation of cell apoptosis is an important
factor in tumour progression. To determine whether miR-124a and
IL-11 could affect the ratios of apoptotic cells, HepG2 cells were
divided into the following four groups: Blank, cells transfected
with NCs, cells transfected with miR-124a mimics and cells
transfected with IL-11 siRNA. Forty-eight h after transfection, the
apoptosis of HepG2 cells was assessed (Fig. 2A). Our results reveal that the cell
apoptosis rate of miR-124a mimics and IL-11 siRNA was much higher
than that of the blank and NC group (Fig. 2B). These results indicate that the
upregulation of miR-124a or downregulation of IL-11 can promote
HepG2 cell apoptosis, suggesting that IL-11 is involved in
miR-124a-regulated cell apoptosis.
IL-11 is a direct target of
miR-124a
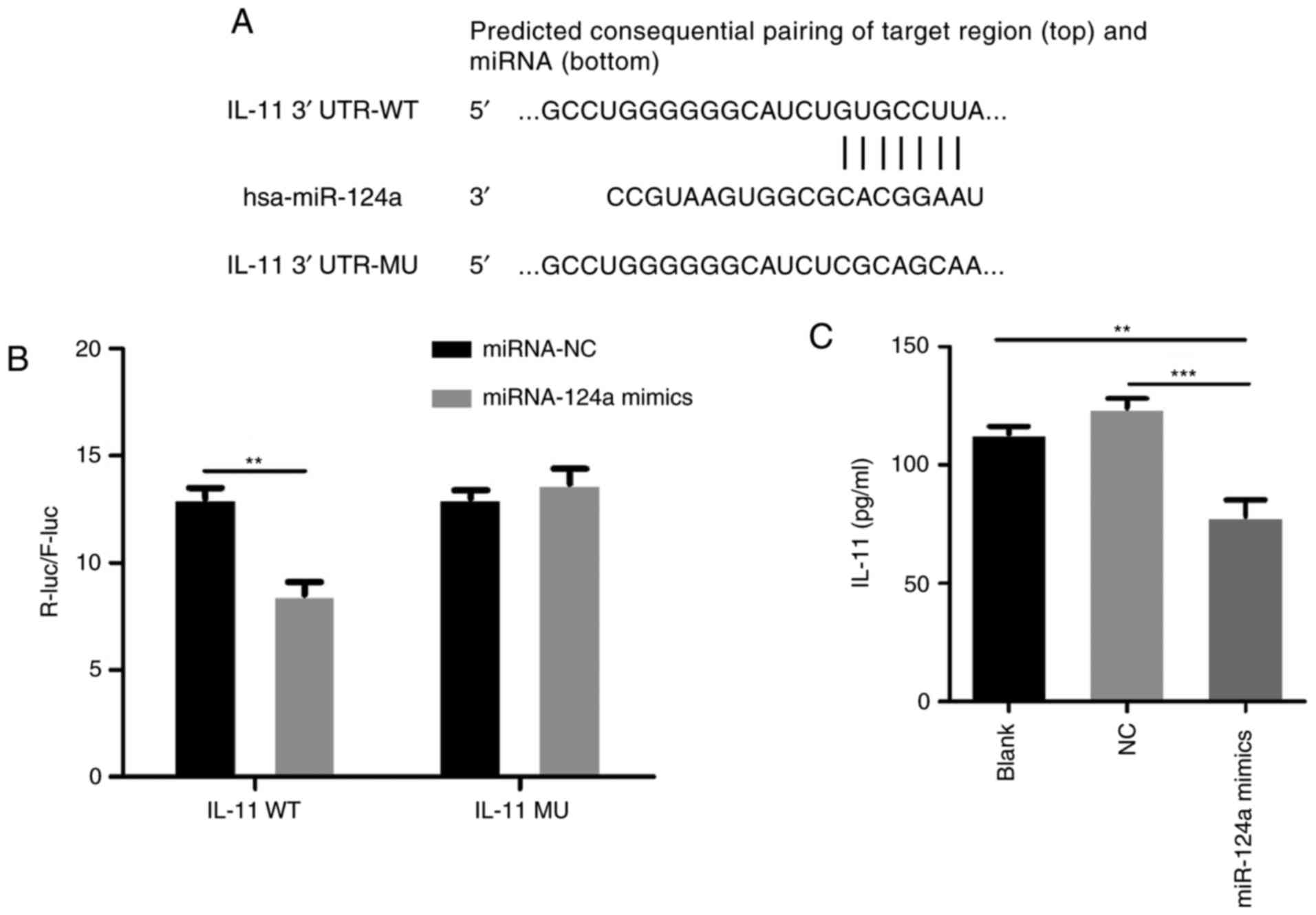
miRNAs can bind to the 3′UTR of the target gene
transcripts to inhibit gene expression. Using the bioinformatics
tools at TargetScan (http://www.targetcan.org/), potential targets of
miR-124a were predicted. Several results were subsequently analysed
by the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways
(www.genome.jp/kegg/), and IL-11 was
evaluated as an effective target of miR-124a. IL-11 contains highly
conserved binding sites in the 3′UTR (Fig. 3A). To further confirm that IL-11 is
a direct target of miR-124a, DNA fragments containing the wild and
mutant IL-11 3′UTR were magnified by PCR and cloned into the
luciferase vector psiCHECK-2 to construct the luciferase reporter
gene vectors. In order to have higher transfection efficiency to
eliminate individual differences in low transfection efficiency, we
used 293T cells to conduct dual-luciferase reporter gene assay.
When we transfected either miR-124a mimics or control with
wild-type or mutant-type into 293T cells for 48 h, the luciferase
activity of IL-11 wild-type with miR-124a mimics was decreased by
approximately 40% relative to the miR-124a NC group. However,
miR-124a did not significantly affect the luciferase activity of
IL-11 with mutant-type 3′UTR (Fig.
3B). ELISA revealed that the secretion of IL-11 was decreased
after transfection with miR-124a mimics compared with the blank and
NC group (Fig. 3C). These results
therefore suggest that miR-124a can bind directly to IL-11 and that
IL-11 is a direct target of miR-124a.
Effects of miR-124a on proteins
associated with cell proliferation, migration, and apoptosis
We have shown that miR-124a inhibits the
proliferation and migration of HepG2 cells and promotes their
apoptosis. However, the underlying molecular mechanism remains
indefinite. In our study, the expression levels of MMP2, MMP9,
TIMP-2, Caspase-3, Bcl-2, STAT3, and p-STAT3 at the protein level
were discovered by Western blotting (Fig. 4A). With the upregulation of
miR-124a, the expression of MMP2 and MMP9, that are associated with
cancer cell metastasis, decreased significantly. In contrast, the
expression of TIMP-2, which is an inhibitor of MMP2, increased. The
expression of Caspase-3, an important indicator of apoptosis, was
significantly upregulated, whereas the expression of Bcl-2, a
recognized cell survival factor, was decreased. The apoptotic rate
of HepG2 cells increased as a consequence. These results suggest
that miR-124a mimics can inhibit metastasis and promote the
apoptosis of HepG2 cells. The protein levels of STAT3 and
phosphorylated STAT3 (p-STAT3) both significantly decreased,
indicating that STAT3 pathway activity was decreased (Fig. 4B).
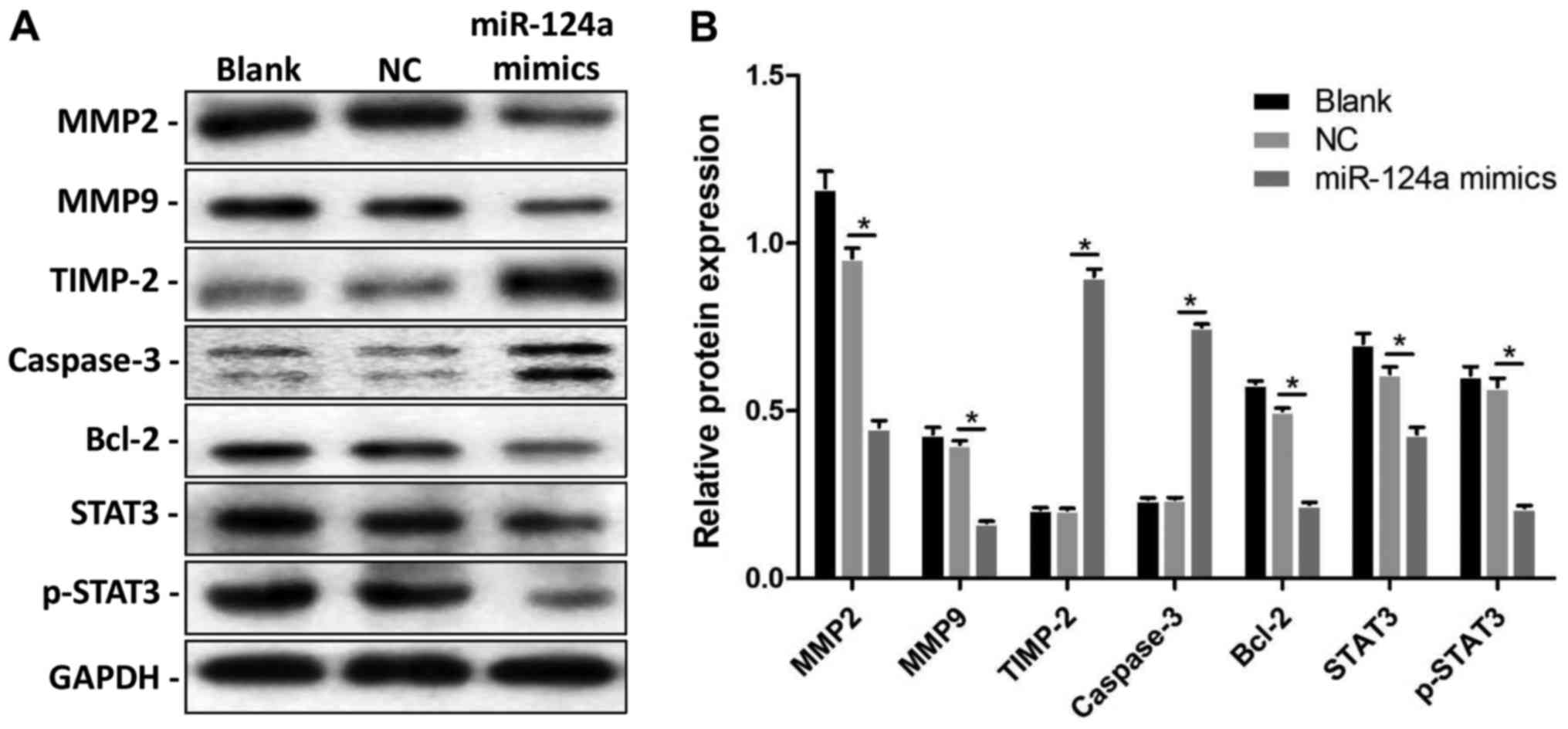 | Figure 4.Effects of miR-124a on proteins
associated with cell proliferation, migration, and apoptosis. (A)
MMP2, MMP9, TIMP-2, Caspase-3, Bcl-2, STAT3, and p-STAT3 protein
levels were detected by Western blotting, and GAPDH as an internal
control. (B) Relative protein expressions of MMP2, MMP9, TIMP-2,
Caspase-3, Bcl-2, STAT3, and p-STAT3 in the miR-124a mimic group
and respective negative groups. *P<0.05 vs. NC group. MMP,
matrix metalloproteinase; TIMP-2, tissue inhibitor of matrix
metalloproteinase-2; Bcl-2, B-cell lymphoma 2; STAT3, signal
transducer and activator of transcription 3; NC, negative
control. |
Discussion
Liver cancer is the third most common cause of
cancer-related death due to its high mortality. With continuous
improvements in medicine, surgical techniques, and antineoplastic
drugs, the five-year survival rate for liver cancer continues to
increase. However, because of the high frequency of recurrence and
high migratory capacity, the liver cancer patients have a poor
prognosis with only a postoperative 5-year survival rate of 30–40%,
and the patient's quality of life is often unsatisfactory due to
tumour metastasis. Although a variety of tumour suppressor genes
and oncogenes have been identified, our understanding of the
potential molecular mechanisms in liver cancer metastasis and
progression is insufficient. It is necessary to identify and
characterize molecular markers that are effective in predicting
liver cancer metastasis to establish the prognosis for patients and
to determine the most effective therapeutic methods.
Cumulative evidence suggests that the abnormal
regulation of microRNAs can contribute to tumourigenesis (26). Changes in miRNA expression are
involved in tumour cell progression and metastasis (17,27).
Thus, miRNAs are increasingly considered as potential diagnostic
and therapeutic targets (28). In
the present study, we reviewed the literature and found that
miR-124a is downregulated in liver cancer tissues, including
hepatocellular carcinoma, cholangiocarcinoma and hepatoblastoma
(21,26,27).
Meanwhile, miR-124a has been reported to inhibit tumour metastasis
in several cancers, including lung cancer (29) and nasopharyngeal carcinoma
(30). However, the effects of
miR-124a in liver cancer metastasis and its biological mechanism
are not fully understood. First, to illuminate the role of miR-124a
in hepatoma cells, HepG2 cells were transfected with miR-124a
mimics. As is well known, the migration, proliferation, and
apoptosis evasion of tumour cells are essential prerequisites for
tumour development and metastasis. In our study, miR-124a was
upregulated through the successful implementation of miR-124a
mimics in HepG2 cells. miR-124a mimics promoted cell apoptosis and
inhibited cancer cell proliferation and migration. Knockdown of
IL-11 by siRNA suppressed the proliferation, migration and promoted
apoptosis of HepG2 cells. These results suggest that IL-11 is
involved with miR-124a-regulated HepG2 cell metastasis.
Furthermore, we would like to explore the underlying biological
mechanism in the miR-124a-mediated inhibition of tumour
metastasis.
A key part of tumour migration and metastasis is the
degradation of extracellular matrix (ECM) components. MMPs can
degrade ECM, and the MMP activity is regulated by TIMPs. Many
studies have confirmed that MMP expression is higher in human
primary liver cancer and liver metastases compared to normal liver
tissue, and some studies have shown relationships between MMP
expressions and tumour progression, migration, and metastasis,
indicating an important role of MMPs in tumour malignancy (31–33).
In this study, the upregulation of miR-124a increased TIMP-2
expression and reduced MMP2 and MMP9 expression in HepG2 cells.
Thus, it can be speculated that miR-124a could inhibit the
migration of tumour cells by targeting the TIMP/MMP pathway.
To study the correlations of the apoptotic features
with relevant signalling molecules, the classical apoptotic genes
Caspase-3 and Bcl-2 were analysed by Western blotting. Cell
apoptosis is related to the activation, expression, and regulation
of many apoptotic genes, among which the family of Bcl-2 and
Caspases are widely recognized. Bcl-2 is a member of regulator
proteins and acts as an anti-apoptotic protein by inhibiting cell
death (34,35). Caspase-3 make great contributions
to the implementation stage of cell apoptosis and is widely
involved in many pathological processes, including tumourigenesis,
infection, trauma, and autoimmune disease (36,37).
HepG2 cell line with wildtype p53 was used in our study. The p53
protein could induce apoptosis by activating the expression of
pro-apoptotic genes as well as inhibiting the expression of
anti-apoptotic genes. It is reported that miR-124 could directly
target inhibitor of apoptosis-stimulating protein of p53 (iASPP)
and subsequently induce an upregulation of p53 in some tumours,
leading to the apoptosis promotion. In the present study, miR-124a
mimics promoted Caspase-3 expression and inhibited Bcl-2 expression
in HepG2 cells. These results imply that miR-124a can promote
apoptosis by promoting Caspase-3 expression and blocking Bcl-2
expression.
IL-11 is located on chromosome 9 and belongs to the
IL-6 cytokine family. It has been confirmed that IL-11 plays a
significant role in the progression of pancreatic (38), gastric (39), and renal cancers (40) and promotes the BM of liver cancer
(41). STAT signalling plays a
significant role in the transfer of extracellular signals into the
nucleus, resulting in the regulation of transcription, and is
essential in the uncontrolled growth, angiogenesis, and metastasis
of cancer cells (42,43). STAT3 is a direct target of miR-124
and is overexpressed in most hepatocellular carcinomas with poorer
prognosis, however it is not overexpressed in normal liver tissues
or in surrounding non-tumour tissues (44,45).
Without IL-11/STAT3 signalling, tumour onset is delayed in mice
(46). In the present study, the
dual-luciferase reporter assay verified that IL-11 is a direct
target of miR-124a, and we observed decreased secretion of IL-11 in
miR-124a mimic-transfected HepG2 cells. Knockdown of IL-11 by siRNA
suppressed the proliferation of HepG2 cells. Moreover, the activity
of the STAT3 pathway was decreased, indicating that miR-124a can
repress liver cancer proliferation and metastasis by disrupting
IL-11/STAT3 signalling.
The organ colonization of diffuse tumour cells can
be promoted by IL-11 mRNA, and activating STAT3 signalling
(47). Meanwhile, the upregulation
of intratumoural IL-11 has been associated with BM after
hepatectomy and is an independent prognostic factor for the
progress of BM in liver cancer patients (41,48).
These studies suggest that IL-11 is closely related to liver cancer
BM. According to the inhibitory effects of miR-124a on IL-11,
miR-124a may not only inhibit hepatoma cell proliferation,
migration, and invasion but also act as a target to repress the BM
of liver cancer. However, cumulative evidence has suggested that
miR-124 could inhibit metastatic potential of liver cancer, by
directly targeting integrin αV, ROCK2 and EZH2 (49,50).
So, further researches are required to investigate the potential
functional targets of miR-124a, and identifying the actual
mechanisms by which miR-124a affects the progression and metastasis
of liver cancer requires further clarification.
In summary, we determined that miR-124a can inhibit
cell proliferation and migration by directly targeting IL-11,
indicating that IL-11 is an important functional mediator of
miR-124a in hepatoma cells. These findings may help in
investigating potential mechanisms and provide new insights for the
clinical treatment to prevent the development and metastasis of
liver cancer.
Acknowledgements
The present study was supported by grants from the
Zhejiang Provincial Science and Technology Department Public
Service Technology Research Project of China (grant no.
2015C33203).
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Deuffic S, Poynard T, Buffat L and
Valleron AJ: Trends in primary liver cancer. Lancet. 351:214–215.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Altekruse SF, Henley SJ, Cucinelli JE and
McGlynn KA: Changing hepatocellular carcinoma incidence and liver
cancer mortality rates in the United States. Am J Gastroenterol.
109:542–553. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Natsuizaka M, Omura T, Akaike T, Kuwata Y,
Yamazaki K, Sato T, Karino Y, Toyota J, Suga T and Asaka M:
Clinical features of hepatocellular carcinoma with extrahepatic
metastases. J Gastroenterol Hepatol. 20:1781–1787. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Roodman GD: Mechanisms of bone metastasis.
N Engl J Med. 350:1655–1664. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
He J, Zeng ZC, Tang ZY, Fan J, Zhou J,
Zeng MS, Wang JH, Sun J, Chen B, Yang P and Pan BS: Clinical
features and prognostic factors in patients with bone metastases
from hepatocellular carcinoma receiving external beam radiotherapy.
Cancer. 115:2710–2720. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Simonson B and Das S: MicroRNA
therapeutics: The next magic bullet? Mini Rev Med Chem. 6:467–474.
2015. View Article : Google Scholar
|
|
9
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Garzon R, Calin GA and Croce CM: MicroRNAs
in cancer. Annu Rev Med. 60:167–179. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Huang S and He X: The role of microRNAs in
liver cancer progression. Br J Cancer. 104:235–240. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tsai WC, Hsu PW, Lai TC, Chau GY, Lin CW,
Chen CM, Lin CD, Liao YL, Wang JL, Chau YP, et al: MicroRNA-122, a
tumor suppressor microRNA that regulates intrahepatic metastasis of
hepatocellular carcinoma. Hepatology. 49:1571–1582. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhu Q, Wang Z, Hu Y, Li J, Li X, Zhou L
and Huang Y: miR-21 promotes migration and invasion by the
miR-21-PDCD4-AP-1 feedback loop in human hepatocellular carcinoma.
Oncol Rep. 27:1660–1668. 2012.PubMed/NCBI
|
|
14
|
Wang B, Hsu SH, Majumder S, Kutay H, Huang
W, Jacob ST and Ghoshal K: TGFbeta-mediated upregulation of hepatic
miR-181b promotes hepatocarcinogenesis by targeting TIMP3.
Oncogene. 29:1787–1797. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huang XH, Chen JS, Wang Q, Chen XL, Wen L,
Chen LZ, Bi J, Zhang LJ, Su Q and Zeng WT: miR-338-3p suppresses
invasion of liver cancer cell by targeting smoothened. J Pathol.
225:463–472. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhou Y, Li Y, Ye J, Jiang R, Yan H, Yang
X, Liu Q and Zhang J: MicroRNA-491 is involved in metastasis of
hepatocellular carcinoma by inhibitions of matrix metalloproteinase
and epithelial to mesenchymal transition. Liver Int. 33:1271–1280.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Geng S, Zhang X, Chen J, Liu X, Zhang H,
Xu X, Ma Y, Li B, Zhang Y, Bi Z and Yang C: The tumor suppressor
role of miR-124 in osteosarcoma. PLoS One. 9:e915662014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li Y, Zhang Z, Liu X, Huang T, He W, Shen
Y, Liu X, Hong K and Cao Q: miR-124 functions as a tumor suppressor
in the endometrial carcinoma cell line HEC-1B partly by suppressing
STAT3. Mol Cell Biochem. 388:219–231. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chu M, Chang Y, Guo Y, Wang N, Cui J and
Gao WQ: Regulation and methylation of tumor suppressor miR-124 by
androgen receptor in prostate cancer cells. PLoS One.
10:e01161972015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li W, Huang H, Su J, Ji X, Zhang X, Zhang
Z and Wang H: miR-124 Acts as a tumor suppressor in glioblastoma
via the inhibition of signal transducer and activator of
transcription 3. Mol Neurobiol. 54:2555–2561. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Budhu A, Jia HL, Forgues M, Liu CG,
Goldstein D, Lam A, Zanetti KA, Ye QH, Qin LX, Croce CM, et al:
Identification of metastasis-related microRNAs in hepatocellular
carcinoma. Hepatology. 47:897–907. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xiang ZL, Zeng ZC, Fan J, Tang ZY and Zeng
HY: Expression of connective tissue growth factor and
interleukin-11 in intratumoral tissue is associated with poor
survival after curative resection of hepatocellular carcinoma. Mol
Biol Rep. 39:6001–6006. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sommer J, Effenberger T, Volpi E, Waetzig
GH, Bernhardt M, Suthaus J, Garbers C, Rose-John S, Floss DM and
Scheller J: Constitutively active mutant gp130 receptor protein
from inflammatory hepatocellular adenoma is inhibited by an
anti-gp130 antibody that specifically neutralizes interleukin 11
signaling. J Biol Chem. 287:13743–13751. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jia XQ, Cheng HQ, Li H, Zhu Y, Li YH, Feng
ZQ and Zhang JP: Inhibition of connective tissue growth factor
overexpression decreases growth of hepatocellular carcinoma cells
in vitro and in vivo. Chin Med J (Engl). 124:3794–3799.
2011.PubMed/NCBI
|
|
25
|
Gao YB, Xiang ZL, Zhou LY, Wu ZF, Fan J,
Zeng HY and Zeng ZC: Enhanced production of CTGF and IL-11 from
highly metastatic hepatoma cells under hypoxic conditions: An
implication of hepatocellular carcinoma metastasis to bone. J
Cancer Res Clin Oncol. 139:669–679. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Murray MJ, Raby KL, Saini HK, Bailey S,
Wool SV, Tunnacliffe JM, Enright AJ, Nicholson JC and Coleman N:
Solid tumors of childhood display specific serum microRNA profiles.
Cancer Epidemiol Biomarkers Prev. 24:350–360. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zeng B, Li Z, Chen R, Guo N, Zhou J, Zhou
Q, Lin Q, Cheng D, Liao Q, Zheng L and Gong Y: Epigenetic
regulation of miR-124 by hepatitis C virus core protein promotes
migration and invasion of intrahepatic cholangiocarcinoma cells by
targeting SMYD3. FEBS Lett. 586:3271–3278. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Rossi JJ: New hope for a microRNA therapy
for liver cancer. Cell. 137:990–992. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang Y, Li H, Han J and Zhang Y:
Down-regulation of microRNA-124 is correlated with tumor metastasis
and poor prognosis in patients with lung cancer. Int J Clin Exp
Pathol. 8:1967–1972. 2015.PubMed/NCBI
|
|
30
|
Peng XH, Huang HR, Lu J, Liu X, Zhao FP,
Zhang B, Lin SX, Wang L, Chen HH, Xu X, et al: miR-124 suppresses
tumor growth and metastasis by targeting Foxq1 in nasopharyngeal
carcinoma. Mol Cancer. 13:1862014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kessenbrock K, Plaks V and Werb Z: Matrix
metalloproteinases: Regulators of the tumor microenvironment. Cell.
141:52–67. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Musso O, Théret N, Campion JP, Turlin B,
Milani S, Grappone C and Clément B: In situ detection of matrix
metalloproteinase-2 (MMP2) and the metalloproteinase inhibitor
TIMP2 transcripts in human primary hepatocellular carcinoma and in
liver metastasis. J Hepatol. 26:593–605. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jiang F, Chen L, Yang YC, Wang XM, Wang
RY, Li L, Wen W, Chang YX, Chen CY, Tang J, et al: CYP3A5 functions
as a tumor suppressor in hepatocellular carcinoma by regulating
mTORC2/Akt signaling. Cancer Res. 75:1470–1481. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yuan Z, Liu S, Yao J, Zeng Q, Tan S and
Liu Z: Expression of Bcl-2 genes in channel catfish after bacterial
infection and hypoxia stress. Dev Comp Immunol. 65:79–90. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lasithiotaki I, Antoniou KM, Derdas SP,
Sarchianaki E, Symvoulakis EK, Psaraki A, Spandidos DA,
Stathopoulos EN, Siafakas NM and Sourvinos G: The presence of
Merkel cell polyomavirus is associated with deregulated expression
of BRAF and Bcl-2 genes in non-small cell lung cancer. Int J
Cancer. 133:604–611. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cohen GM: Caspases: The executioners of
apoptosis. Biochem J. 326:1–16. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Korsmeyer SJ: BCL-2 gene family and the
regulation of programmed cell death. Cancer Res. 59 7
Suppl:S1693–S1700. 1999.
|
|
38
|
Ren C, Chen Y, Han C, Fu D and Chen H:
Plasma interleukin-11 (IL-11) levels have diagnostic and prognostic
roles in patients with pancreatic cancer. Tumour Biol.
35:11467–11472. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhou C, Ji J, Cai Q, Shi M, Chen X, Yu Y,
Zhu Z and Zhang J: MTA2 enhances colony formation and tumor growth
of gastric cancer cells through IL-11. BMC Cancer. 15:343–352.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Pan D, Xu L, Liu H, Zhang W, Liu W, Liu Y,
Fu Q and Xu J: High expression of interleukin-11 is an independent
indicator of poor prognosis in clear-cell renal cell carcinoma.
Cancer Sci. 106:592–597. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Xiang ZL, Zeng ZC, Tang ZY, Fan J, He J,
Zeng HY and Zhu XD: Potential prognostic biomarkers for bone
metastasis from hepatocellular carcinoma. Oncologist. 16:1028–1039.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang A, Duan G, Zhao C, Gao Y, Liu X, Wang
Z, Li W, Wang K and Wang W: Reduced RKIP expression levels are
associated with frequent non-small cell lung cancermetastasis and
STAT3 phosphorylation and activation. Oncol Lett. 13:3039–3045.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yang S, Luo C, Gu Q, Xu Q, Wang G, Sun H,
Qian Z, Tan Y, Qin Y, Shen Y, et al: Activating JAK1 mutation may
predict the sensitivity of JAK-STAT inhibition in hepatocellular
carcinoma. Oncotarget. 7:5461–5469. 2016.PubMed/NCBI
|
|
44
|
Liu S, Hu C, Wang Y, Shi G, Li Y and Wu H:
miR-124 inhibits proliferation and invasion of human retinoblastoma
cells by targeting STAT3. Oncol Rep. 36:2398–2404. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Calvisi DF, Ladu S, Gorden A, Farina M,
Conner EA, Lee JS, Factor VM and Thorgeirsson SS: Ubiquitous
activation of Ras and Jak/Stat pathways in human HCC.
Gastroenterology. 130:1117–1128. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wu J, Wang Y, Xu X, Cao H, Sahengbieke S,
Sheng H, Huang Q and Lai M: Transcriptional activation of FN1 and
IL11 by HMGA2 promotes the malignant behavior of colorectal cancer.
Carcinogenesis. 37:511–521. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ,
Tao QF, Liu F, Pan W, Wang TT, Zhou CC, et al: A long noncoding RNA
activated by TGF-β promotes the invasion-metastasis cascade in
hepatocellular carcinoma. Cancer Cell. 25:666–681. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lamarca A, Mendiola M, Bernal E, Heredia
V, Díaz E, Miguel M, Pastrian LG, Burgos E, Feliu J and Barriuso J:
Tumoural expression of connective tissue growth factor (CTGF)
impacts on survival in patients diagnosed with hepatocellular
carcinoma (HCC). Curr Cancer Drug Targets. 15:435–444. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Cai QQ, Dong YW, Wang R, Qi B, Guo JX, Pan
J, Liu YY, Zhang CY and Wu XZ: miR-124 inhibits the migration and
invasion of human hepatocellular carcinoma cells by suppressing
integrin αV expression. Sci Rep. 7:407332017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zheng F, Liao YJ, Cai MY, Liu YH, Liu TH,
Chen SP, Bian XW, Guan XY, Lin MC, Zeng YX, et al: The putative
tumour suppressor microRNA-124 modulates hepatocellular carcinoma
cell aggressiveness by repressing ROCK2 and EZH2. Gut. 61:278–289.
2012. View Article : Google Scholar : PubMed/NCBI
|