Introduction
Glioma, the most common primary central nervous
system tumour, accounts for ~80% of malignant brain tumours
(1). This tumour type can be
divided into four grades (I–IV) on the basis of their degree of
malignancy (2). Despite remarkable
improvements in treatment strategies, including surgery,
chemotherapy, radiotherapy and immunotherapy, the overall survival
rate of patients with glioma remains poor, with a median survival
time of 9–12 months from diagnosis to death (3). The poor prognosis of patients with
gliomas is mainly due to its unlimited proliferation, high tumour
aggressiveness and difficulty in completing surgical excision
(4,5). Undoubtedly, investigating the
molecular mechanisms of glioma formation and progression is
essential for the development of novel and precise therapeutic
targets for patients with this disease.
MicroRNAs (miRNAs) are a large subset of
single-strand and non-coding short RNAs with a length of
approximately 18–24 nucleotides. They have been considered as
critical regulators of gene expression through interaction with
3′-untranslated regions (3′-UTRs) of their target genes, thereby
inhibiting translation or promoting degradation of the Mrna
(6). Through this mechanism,
miRNAs modulate >50% of all human protein-coding genes and
implicated in the regulation of numerous physiological and
pathological processes, including cell proliferation, cell cycle,
apoptosis, differentiation and metastasis (7–9).
However, abnormal expression of miRNAs has been observed in human
malignancies, such as glioma (10), gastric cancer (11), lung cancer (12), cervical cancer (13) and bladder cancer (14). In tumourigenesis and tumour
development, highly expressed miRNAs may play oncogenic roles by
regulating tumour suppressor genes (15), whereas downregulated miRNAs may
perform tumour-suppressing functions by directly targeting
oncogenes (16). Therefore, the
expression pattern, roles and associated mechanisms of miRNAs in
glioma should be investigated to identify novel and efficient
therapeutic methods for gliomas.
MiR-379 has been shown to be aberrantly expressed in
the progression of malignant tumours (17,18).
However, the expression, biological functions and mechanism of
miR-379 in glioma are yet to be fully understood. Hence, this study
aimed to detect miR-379 expression, investigate its functional
relevance and explore its associated molecular mechanism in
glioma.
Materials and methods
Tissue collection
A total of 26 pairs of glioma tissues and matched
adjacent normal brain tissues were collected from patients who were
undergo surgery resection at The Fourth Affiliated Hospital of
Harbin Medical University between March 2014 and October 2016. All
these participants were first diagnosed with glioma and had not
been treated with chemotherapy, radiotherapy, or immunotherapy
prior to surgery. Tissues were immediately frozen and stored in
liquid nitrogen until RNA isolation. This study was approved by the
Ethics Committee of The Fourth Affiliated Hospital of Harbin
Medical University. Written informed consent was provided by all
patients before enrollment.
Cell culture
Five human glioma cell lines (U138, U251, U343, T98
and LN229) were acquired from Chinese Academy of Sciences Cell Bank
(Shanghai, China), and cultured in Dulbecco's modified Eagle medium
(DMEM) containing 10% fetal bovine serum (FBS), 100 U/ml penicillin
and 100 mg/ml streptomycin (all from Invitrogen, Carlsbad, CA,
USA). A normal human astrocyte (NHA) cell line was purchased from
ScienCell Research Laboratories (Carlsbad, CA, USA), and was grown
in astrocyte medium supplemented with 1% astrocyte growth
supplement (both from Sciencell Research Laboratories) and 10% FBS.
All cell lines were maintained in a humidified incubator at 37°C
with 5% CO2.
Cell transfection
MiR-379 mimics and miRNA mimic negative control
(miR-NC) were obtained from GeneCopoeia™ (Guangzhou,
China). Metadherin (MTDH) overexpression vector (pcDNA3.1-MTDH) and
empty vector (pcDNA3.1) were chemically synthesised and their
sequences confirmed by Shanghai Genechem Co., Ltd. (Shanghai,
China). Cells were plated into six-well plates at a density of
5×105 cells per well and incubated at 37°C with 5%
CO2 until 50% confluence was reached. Cells were
transfected with miR-379 mimics, miR-NC, pcDNA3.1-MTDH or pcDNA3.1
using Lipofectamine 2000 transfection reagent (Invitrogen)
following the manufacturer's instructions. The medium was
subsequently replaced with fresh DMEM with 10% FBS at 8 h
posttransfection.
Bioinformatic assay
The potential targets of miR-379 were predicted
using the algorithms TargetScan (https://www.targetscan.org) and PicTar (http://pictar.mdc-berlin.de/).
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
TRIzol Reagent (Invitrogen) was used to extract
total RNA from tissues or cells. To examine miR-379 expression,
reverse transcription was conducted using a TaqMan miRNA
reverse transcription kit, followed by real-time PCR with a
TaqMan miRNA PCR kit (both from Applied Biosystems, Foster
City, CA, USA) according to the manufacturer's protocol. To
quantify the mRNA expression of MTDH, cDNA was synthesised from
total RNA by using a PrimeScript RT reagent kit (Takara Bio, Inc.,
Otsu, Japan). Quantitative PCR was also performed with
SYBR® Premix Ex Taq (Takara Bio) to measure the
mRNA expression of MTDH. The expression levels of miR-379 and mRNA
of MTDH were normalised with U6 snRNA and GAPDH, respectively. Data
were analysed using 2−ΔΔCq method (19).
Cell counting kit-8 (CCK-8) assay
CCK-8 assay (Dojindo Molecular Technologies, Inc.,
Kumamoto, Japan) was adopted to determine cell proliferative
ability. Cells were seeded onto 96-well plates with a density of
3,000 cells per well. After overnight incubation, cell transfection
or cotransfection was performed and further incubated at 37°C with
5% CO2 for 0, 1, 2 and 3 days. At each time point, CCK-8
assay was carried out according to the manufacturer's instructions.
In brief, 10 µl of CCK8 reagent was added to each well and
incubated at 37°C for 2 h. Afterwards, absorbance at 450 nm
wavelength was detected using an automatic multi-well
spectrophotometer (Bio-Rad Laboratories, Inc., Hercules, CA,
USA).
Transwell invasion assay
Cellular invasion capacity was evaluated using
24-well polycarbonate membrane Transwell chambers (8 µm pore-size;
Corning Inc., NY, USA) pre-coated with Matrigel (BD Biosciences,
Franklin Lakes, NJ, USA). Transfected cells were collected at 48 h
post-transfection and suspended in FBS-free culture medium. A total
of 5×104 cells were plated in the upper chambers and the
lower chambers were filled with 500 µl of DMEM containing 10% FBS.
The cells were cultured at 37°C with 5% CO2 for 24 h,
and non-invasive cells remaining on the upper side of the membrane
were removed with a cotton swab. Invasive cells were fixed with
methanol, stained with 0.1% crystal violet (Sigma, St. Louis, MO,
USA) and washed with phosphate-buffered saline. Five randomly
selected fields in each chamber were photographed and counted under
an inverted microscope (Olympus Corp., Tokyo, Japan).
Luciferase reporter assay
The predicted 3′-UTR sequence of MTDH, containing
the wild-type (Wt) or mutant (Mut) putative binding sequences for
miR-379, was synthesized and inserted into the pmirGLO luciferase
reporter vector (Promega Corp., Madison, WI, USA), and be named as
pmirGLO-MTDH-3′-UTR Wt and pmirGLO-MTDH-3′-UTR Mut, respectively.
For luciferase reporter assay, cells were plated into 24-well
plates and cotransfected with miR-379 mimics or miR-NC and
pmirGLO-MTDH-3′-UTR Wt or pmirGLO-MTDH-3′-UTR Mut using
Lipofectamine 2000 transfection reagent. Cell lysates were
harvested 48 h subsequent to transfection, and luciferase
activities were detected using a Dual-Luciferase®
Reporter Assay system (Promega Corp.). Renilla luciferase
activity was employed as an internal control.
Western blot analysis
RIPA lysis buffer (Santa Cruz Biotechnology, Inc.,
Santa Cruz, CA, USA) was utilised to isolate total protein from
tissue samples or cells. The concentration of total protein was
quantified using a BCA kit (Beyotime Biotechnology, Haimen, China).
Equal amounts of total protein were separated via 10% sodium
dodecyl sulphate polyacrylamide gel electrophoresis and
electrically transferred onto PVDF membranes (Millipore, Billerica,
MA, USA). The membranes were then blocked with 5% skimmed milk at
room temperature for 2 h and then incubated with primary antibodies
recognizing MTDH (sc-517220), phosphatase and tensin homolog (PTEN)
(sc-7974), p-AKT (sc-271966), AKT (sc-81434) or GAPDH (sc-166574)
at 4°C overnight. All these primary antibodies were acquired form
Santa Cruz Biotechnology. Subsequently, the membranes were probed
with horseradish peroxidase-conjugated secondary antibodies
(sc-2005; Santa Cruz Biotechnology) and further visualised using
enhanced chemiluminescence reagents (Pierce, Rockford, IL, USA).
GAPDH was used as the loading control.
Statistical analysis
All data are expressed as the mean ± SD. Statistical
significance was analyzed with SPSS19.0 (IBM SPSS, Chicago, IL,
USA) using Student's t-tests or one-way analysis of variance
followed by the SNK multiple comparisons test. The association
between MTDH mRNA and miR-379 in glioma tissues was evaluated by
Spearman's correlation analysis. P<0.05was considered to
indicate a statistically significant difference.
Results
MiR-379 expression is decreased in
glioma tissues and cell lines
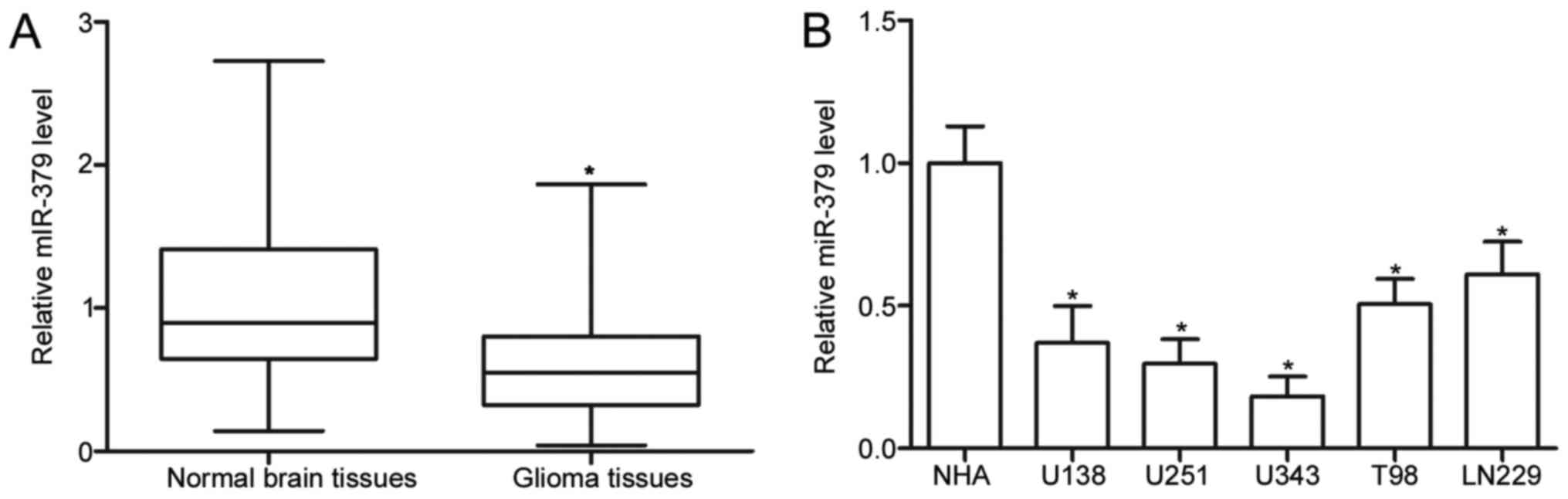
To investigate the expression pattern of miR-379 in
glioma, we initially measured the miR-379 expression in 26 pairs of
glioma tissues and matched adjacent normal brain tissues. The
results of RT-qPCR showed that miR-379 expression was downregulated
in glioma tissues compared with that in normal brain tissues
(P<0.05) (Fig. 1A). Afterwards,
we conducted RT-qPCR to determine the miR-379 expression in five
human glioma cell lines (U138, U251, U343, T98 and LN229) and a
normal NHA cell line. As shown in Fig.
1B, the miR-379 expression levels were consistently decreased
in glioma cell lines compared with NHA (P<0.05). These results
suggest that miR-379 may play important roles in glioma
progression.
Restoration of miR-379 expression
attenuates glioma cell proliferation and invasion
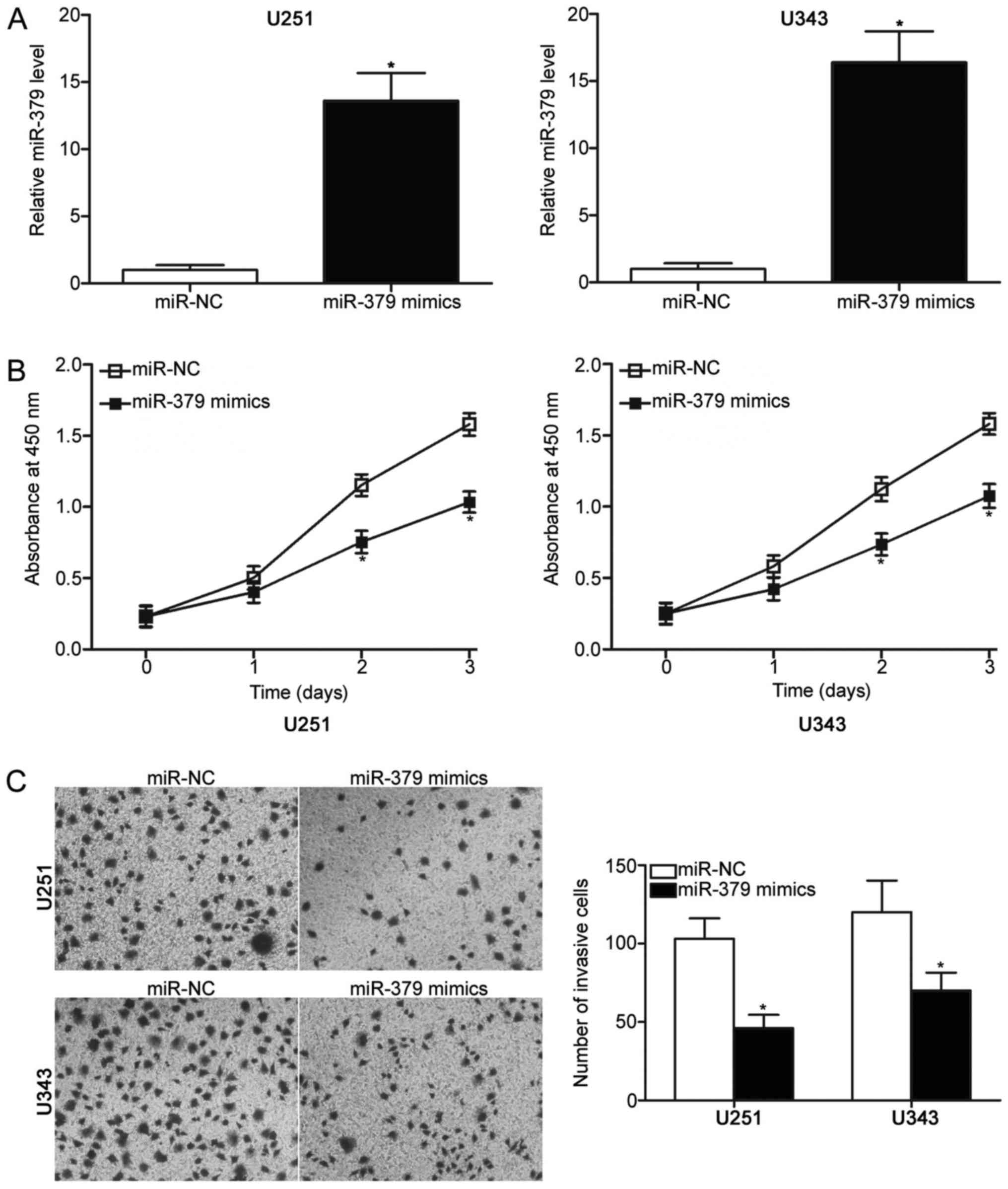
To explore the biological roles of miR-379 in
glioma, miR-379 mimics were transfected into U251 and U343 cells,
which expressed relatively lower miR-379 expression among the five
glioma cell lines. RT-qPCR analysis confirmed that miR-379 was
markedly upregulated in U251 and U343 cells after transfection with
miR-379 mimics compared with those transfected with miR-NC
(P<0.05) (Fig. 2A).
Subsequently, CCK-8 assay was conducted to detect proliferation of
U251 and U343 cells transfected with miR-379 mimics or miR-NC. The
results showed that miR-379 upregulation prohibited the
proliferation of U251 and U343 cells compared with the miR-NC group
(P<0.05) (Fig. 2B). The effect
of miR-379 overexpression on the cell invasion ability of U251 and
U343 cells was evaluated using Transwell invasion assay. Compared
with miR-NC transfected cells, U251 and U343 cells transfected with
miR-379 mimics showed significantly decreased invasion capacities
(P<0.05) (Fig. 2C). These
results suggest that miR-379 plays tumour-suppressing roles in the
progression of glioma.
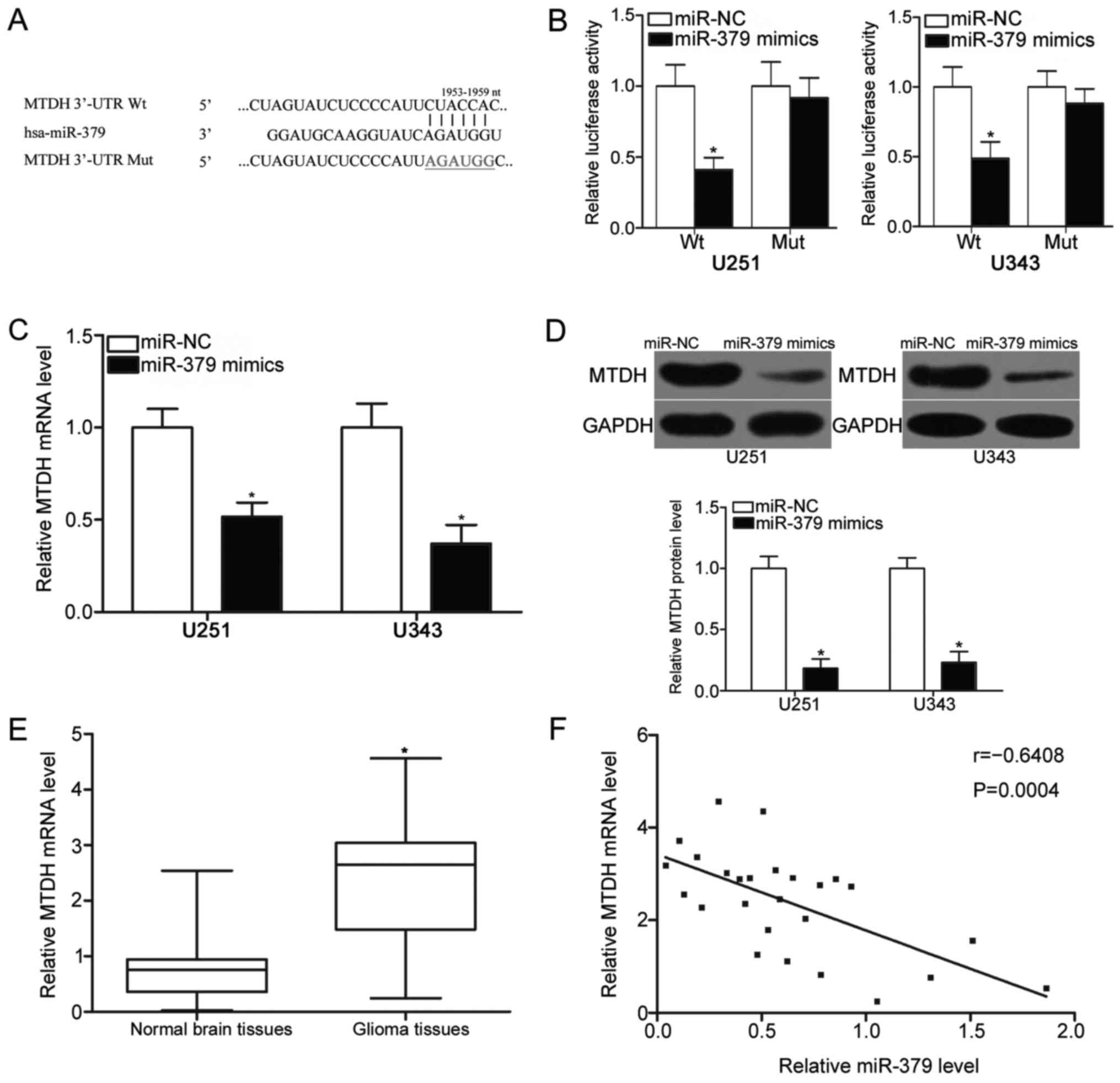
MTDH is a direct target of miR-379 in
glioma cells
To delineate the molecular mechanisms by which
miR-379 inhibits glioma cell proliferation and invasion, we
identified the targets of miR-379 in glioma. Bioinformatics
analysis indicated that MTDH, which has been reported to contribute
to the initiation and formation of glioma (20–22),
was predicted as a candidate target of miR-379 (Fig. 3A). Luciferase reporter assay was
further performed to confirm whether the 3′-UTR of MTDH could be
directly targeted by miR-379. MiR-379 mimics or miR-NC was
transfected into U251 and U343 cells along with luciferase reporter
plasmids containing Wt or Mut putative binding sequences for
miR-379. As shown in Fig. 3B,
miR-379 overexpression significantly decreased the luciferase
activities of pmirGLO-MTDH-3′-UTR Wt (P<0.05), whereas no such
inhibitory effect was observed when miR-379 mimics was
cotransfected with pmirGLO-MTDH-3′-UTR Mut (P>0.05).
RT-qPCR and western blot analysis were adopted to
identify how MTDH expression was altered at mRNA and protein levels
upon miR-379 overexpression and to understand the regulatory
effects of miR-379 on endogenous MTDH expression. The results
revealed that the ectopic expression of miR-379 reduced the MTDH
expression in U251 and U343 cells at mRNA (P<0.05) (Fig. 3C) and protein (P<0.05) (Fig. 3D) levels. We further measured the
mRNA expression of MTDH in 26 pairs of glioma tissues and matched
adjacent normal brain tissues and determined the association
between the miR-379 and MTDH mRNA expression levels in glioma
tissues. The RT-qPCR data indicated that the mRNA expression level
of MTDH was higher in glioma tissues than in adjacent normal brain
tissues (P<0.05) (Fig. 3E).
Furthermore, a significant inverse association between miR-379
expression and MTDH mRNA expression was found in glioma tissues
(r=−0.6408, P=0.0004) (Fig. 3F).
In conclusion, MTDH may be a novel target of miR-379 in glioma.
MTDH upregulation partially rescues
the suppressive effects of miR-379 on glioma cell progression and
invasion
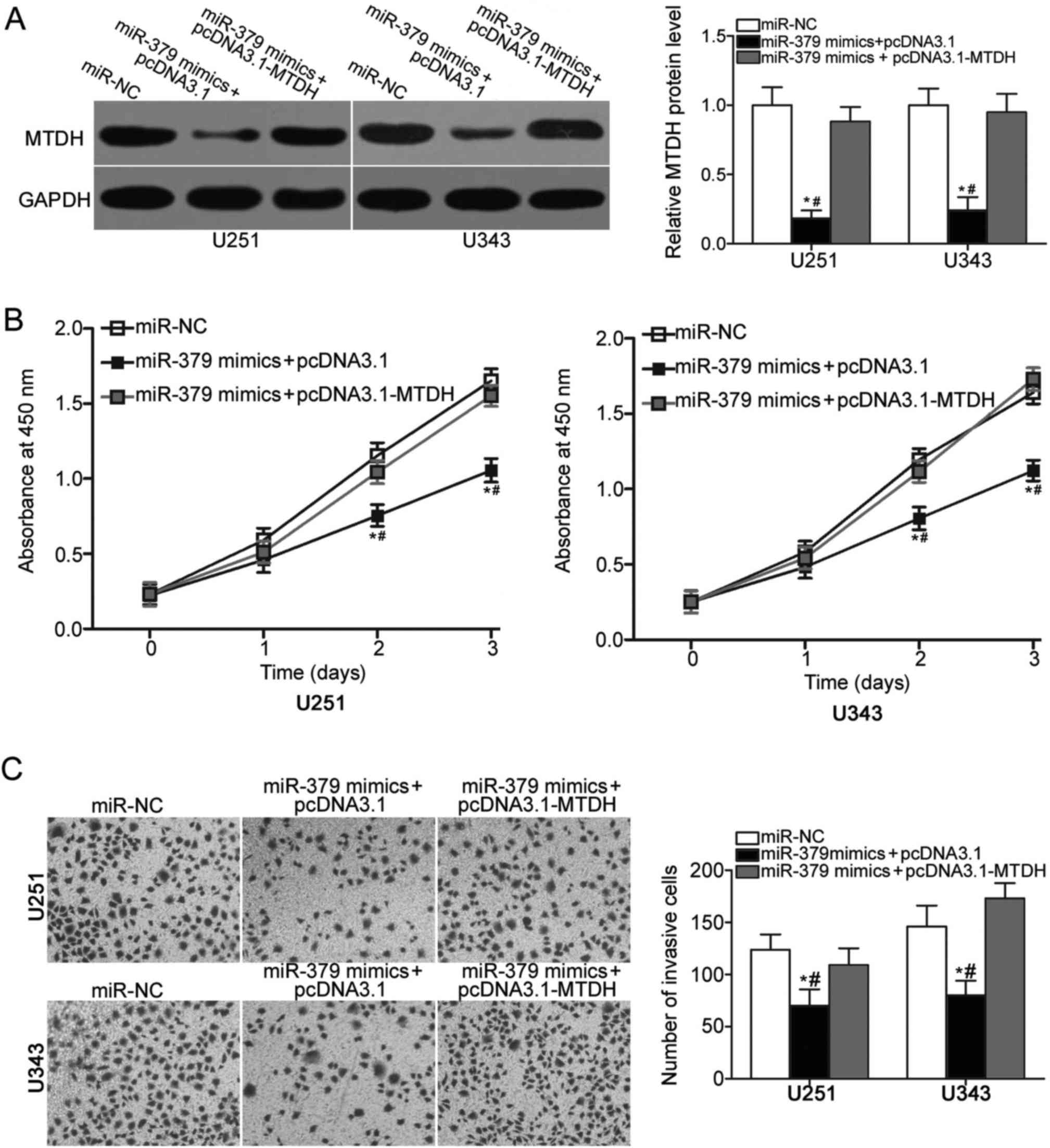
To further verify that the inhibitory effects of
miR-379 on glioma cell proliferation and invasion were mediated by
MTDH, we carried out a series of rescue experiments. U251 and U343
cells were cotransfected with miR-379 mimics and the MTDH
overexpression vector pcDNA3.1-MTDH or the empty vector pcDNA3.1.
After transfection, western blot analysis showed that
cotransfection of pcDNA3.1-MTDH partially restored the decreased
MTDH protein level in U251 and U343 cells that was induced by
miR-379 mimics (P<0.05) (Fig.
4A). In addition, functional assays demonstrated that the
restoration of MTDH expression rescued the inhibition of cell
proliferation (P<0.05) (Fig.
4B) and invasion (P<0.05) (Fig.
4C) caused by miR-379 overexpression in U251 and U343 cells.
These results suggest that miR-379 may serve tumour suppressive
roles in glioma, at least in part, by inhibiting MTDH
expression.
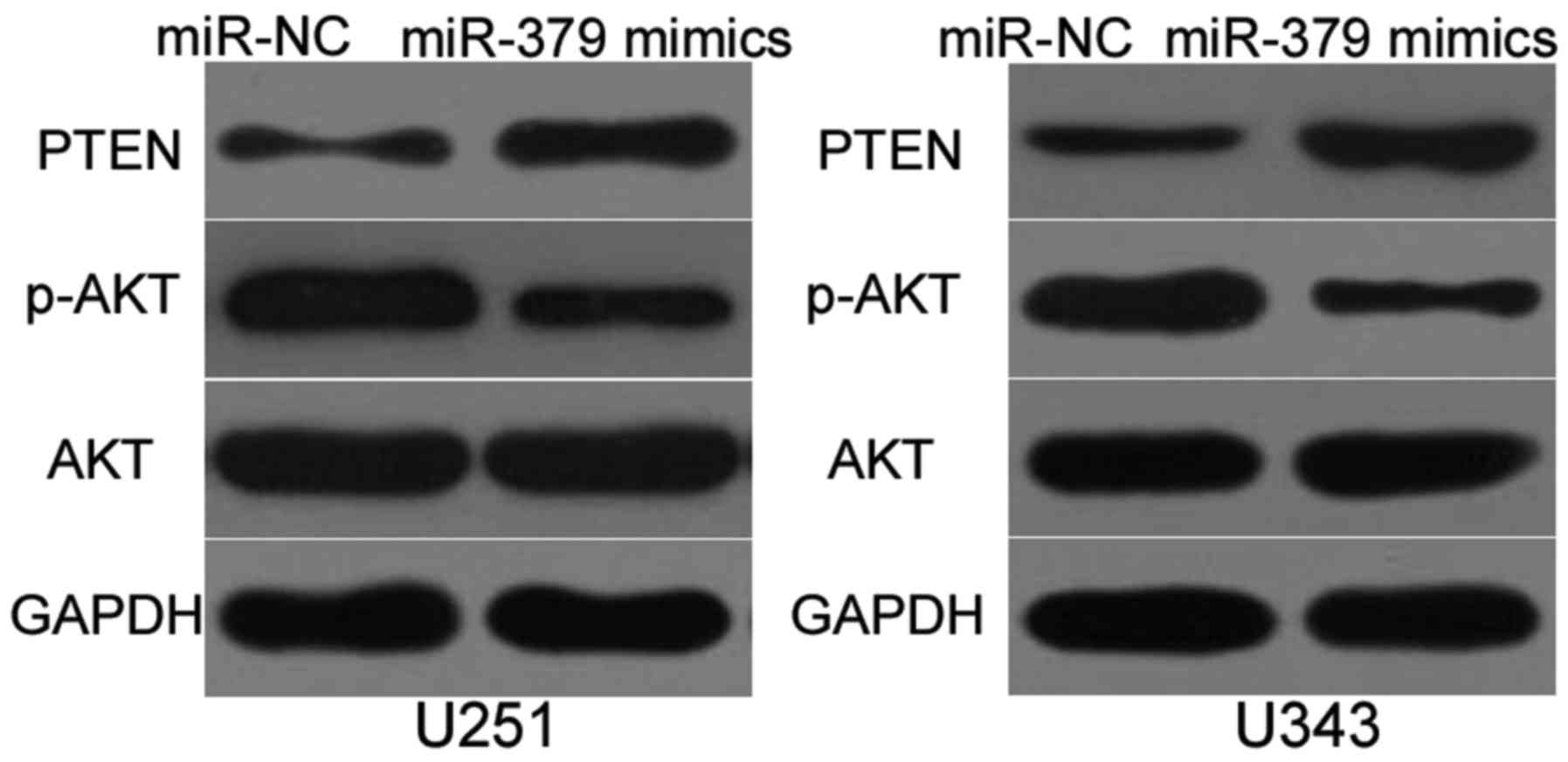
MiR-379 inhibits PTEN/AKT pathway in
glioma
MTDH contributes to the activation of the PTEN/AKT
signaling pathway (20,23,24).
Hence, we investigated whether miR-379 could inhibit PTEN/AKT
pathway in glioma. Western blot analysis was utilised to detect
PTEN, p-AKT and AKT expression in U251 and U343 cells transfected
with miR-379 mimics or miR-NC. The results demonstrated that
enforced expression of miR-379 increased the expression of PTEN and
reduced p-AKT, whereas the expression of total AKT was unaltered
compared with the miR-NC group (Fig.
5). These results suggest that miR-379 inhibits PTEN/AKT
signaling pathway in glioma.
Discussion
Numerous miRNAs are aberrantly expressed in glioma
and are implicated in glioma occurrence and development (25–27).
Therefore, the development of miRNAs as potential therapeutic
targets for the treatment of patients with glioma has been
proposed. In this research, miR-379 expression was downregulated in
glioma tissues and cell lines. In addition, the restoration of
miR-379 expression inhibited cell proliferation and invasion of
glioma. MTDH was validated as a novel target of miR-379 in glioma,
and its upregulation partially rescued the suppressive effects of
miR-379 on glioma cell progression and invasion. Moreover, miR-379
inhibited the activation of the PTEN/AKT pathway in glioma. These
results suggested that miR-379 attenuates glioma progression by
directly targeting MTDH and indirectly regulating the PTEN/AKT
pathway.
MiR-379 has been reported to be downregulated in
multiple types of human malignancies. For example, miR-379 is
decreased in gastric cancer tissues and cell lines, and its low
expression is associated with lymph node metastasis and TNM stage.
Additionally, miR-379 is identified as an independent prognostic
marker for the prediction of the 5-year survival of patients with
gastric cancer (17). In
hepatocellular carcinoma, miR-379 expression levels are low in
tumour tissues and are correlated with advanced TNM stage and
metastasis (18). In breast
cancer, miR-379 is downregulated in tumour tissues compared with
normal breast tissues. A decreased miR-379 expression is
significantly correlated with tumour stage of breast cancer
(28). Downregulation of miR-379
was also observed in bladder cancer (29), osteosarcoma (30), lung cancer (31), medulloblastomas (32) and pleural mesothelioma (33). These findings suggest that miR-379
may be used as a prognostic marker in these specific types of
cancer.
Deregulated miR-379 expression contributes to the
initiation and progression of several types of cancer. For
instance, manipulation of miR-379 levels suppresses cell metastasis
and epithelial-mesenchymal transition (EMT) in gastric cancer
through the regulation of FAK/AKT signaling pathway (17). Chen et al reported that
ectopic expression of miR-379 decreases cell migration, invasion
and EMT of hepatocellular carcinoma by directly targeting FAK and
regulating AKT signaling pathway (18). Khan et al found that
enforced miR-379 expression inhibits breast cancer cell
proliferation via the blockade of cyclin B1 (28). Wu et al showed that miR-379
upregulation attenuates cell growth and metastasis by directly
targeting MDM2 (29). Li et
al revealed that miR-379 targets PDK1 to reduce osteosarcoma
cell proliferation, in vitro invasion and in vivo
tumour growth (30). Hao et
al demonstrated that miR-379 overexpression increases cisplatin
chemosensitivity in non-small cell lung cancer by downregulating
EIF4G2 (31). These findings
suggest that miR-379 is worth investigating as a novel therapeutic
target for patients with these specific types of cancer.
The direct targets of miR-379 should be identified
to understand its role in tumourigenesis and tumour development. In
this study, MTDH, also known as astrocyte elevated gene-1, was
identified as a direct target of miR-379 in glioma. MTDH is located
at chromosome 8q22 and has been reported to be regulated in various
human cancers, such as hepatocellular carcinoma (34), colorectal cancer (35), gastric cancer (36), bladder cancer (37) and breast cancer (38). In glioma, MTDH was overexpressed
and significantly correlated with histological grade (39). The survival time of glioma patients
and with high MTDH expression was shorter than that of patients
with low MTDH expression. High MTDH expression is also identified
as an independent prognostic indicator of the survival of patients
with gliomas (40). Previous
studies demonstrated that MTDH is involved in the initiation and
progression of glioma through the regulation of cell proliferation,
colony formation, metastasis, EMT in vitro and decreased
tumour growth in vivo (20–22).
Therefore, these findings suggested that MTDH could represent a
possible target for the therapy of patients with this deadly
disease.
In summary, miR-379 is downregulated in glioma
tissues and cell lines. The restoration of miR-379 expression
attenuates the proliferation and invasion of glioma cells by
directly targeting MTDH and indirectly regulating the PTEN/AKT
signaling pathways. These results suggest that the miR-379/MTDH
signaling pathway is a potential target to treat patients with
glioma.
References
|
1
|
Ohgaki H: Epidemiology of brain tumors.
Methods Mol Biol. 472:323–342. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegal T: Clinical impact of molecular
biomarkers in gliomas. J Clin Neurosci. 22:437–444. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Helseth R, Helseth E, Johannesen TB,
Langberg CW, Lote K, Rønning P, Scheie D, Vik A and Meling TR:
Overall survival, prognostic factors, and repeated surgery in a
consecutive series of 516 patients with glioblastoma multiforme.
Acta Neurol Scand. 122:159–167. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lefranc F, Brotchi J and Kiss R: Possible
future issues in the treatment of glioblastomas: Special emphasis
on cell migration and the resistance of migrating glioblastoma
cells to apoptosis. J Clin Oncol. 23:2411–2422. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Onishi M, Ichikawa T, Kurozumi K and Date
I: Angiogenesis and invasion in glioma. Brain Tumor Pathol.
28:13–24. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Winter J, Jung S, Keller S, Gregory RI and
Diederichs S: Many roads to maturity: microRNA biogenesis pathways
and their regulation. Nat Cell Biol. 11:228–234. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Macfarlane LA and Murphy PR: MicroRNA:
Biogenesis, function and role in cancer. Curr Genomics. 11:537–561.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Imbar T and Eisenberg I: Regulatory role
of microRNAs in ovarian function. Fertil Steril. 101:1524–1530.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dogini DB, Pascoal VD, Avansini SH, Vieira
AS, Pereira TC and Lopes-Cendes I: The new world of RNAs. Genet Mol
Biol. 37 1 Suppl:S285–S293. 2014. View Article : Google Scholar
|
|
10
|
Zhu Y, Zhao H, Rao M and Xu S:
MicroRNA-365 inhibits proliferation, migration and invasion of
glioma by targeting PIK3R3. Oncol Rep. 37:2185–2192. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang Y, Guan DH, Bi RX, Xie J, Yang CH
and Jiang YH: Prognostic value of microRNAs in gastric cancer: A
meta-analysis. Oncotarget. 8:55489–55510. 2017.PubMed/NCBI
|
|
12
|
Fan N, Zhang J, Cheng C, Zhang X, Feng J
and Kong R: MicroRNA-384 represses the growth and invasion of
non-small-cell lung cancer by targeting astrocyte elevated
gene-1/Wnt signaling. Biomed Pharmacother. 95:1331–1337. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wen F, Xu JZ and Wang XR: Increased
expression of miR-15b is associated with clinicopathological
features and poor prognosis in cervical carcinoma. Arch Gynecol
Obstet. 295:743–749. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ganji SM, Saidijam M, Amini R,
Mousavi-Bahar SH, Shabab N, Seyedabadi S and Mahdavinezhad A:
Evaluation of MicroRNA-99a and MicroRNA-205 expression levels in
bladder cancer. Int J Mol Cell Med. 6:87–95. 2017.PubMed/NCBI
|
|
15
|
Yang T, Thakur A, Chen T, Yang L, Lei G,
Liang Y, Zhang S, Ren H and Chen M: MicroRNA-15a induces cell
apoptosis and inhibits metastasis by targeting BCL2L2 in non-small
cell lung cancer. Tumour Biol. 36:4357–4365. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wu G, Liu J, Wu Z, Wu X and Yao X:
MicroRNA-184 inhibits cell proliferation and metastasis in human
colorectal cancer by directly targeting IGF-1R. Oncol Lett.
14:3215–3222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xu M, Qin S, Cao F, Ding S and Li M:
MicroRNA-379 inhibits metastasis and epithelial-mesenchymal
transition via targeting FAK/AKT signaling in gastric cancer. Int J
Oncol. 51:867–876. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen JS, Li HS, Huang JQ, Dong SH, Huang
ZJ, Yi W, Zhan GF, Feng JT, Sun JC and Huang XH: MicroRNA-379-5p
inhibits tumor invasion and metastasis by targeting FAK/AKT
signaling in hepatocellular carcinoma. Cancer Lett. 375:73–83.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yang J, Fan B, Zhao Y and Fang J:
MicroRNA-202 inhibits cell proliferation, migration and invasion of
glioma by directly targeting metadherin. Oncol Rep. 38:1670–1678.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tong L, Chu M, Yan B, Zhao W, Liu S, Wei
W, Lou H, Zhang S, Ma S, Xu J and Wei L: MTDH promotes glioma
invasion through regulating miR-130b-ceRNAs. Oncotarget.
8:17738–17749. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Park SY, Choi M, Park D, Jeong M, Ahn KS,
Lee J, Fisher PB, Yun M and Lee SG: AEG-1 promotes mesenchymal
transition through the activation of Rho GTPases in human
glioblastoma cells. Oncol Rep. 36:2641–2646. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li WF, Dai H, Ou Q, Zuo GQ and Liu CA:
Overexpression of microRNA-30a-5p inhibits liver cancer cell
proliferation and induces apoptosis by targeting MTDH/PTEN/AKT
pathway. Tumour Biol. 37:5885–5895. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xu C, Kong X, Wang H, Zhang N, Kong X,
Ding X, Li X and Yang Q: MTDH mediates estrogen-independent growth
and tamoxifen resistance by down-regulating PTEN in MCF-7 breast
cancer cells. Cell Physiol Biochem. 33:1557–1567. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhi T, Jiang K, Zhang C, Xu X, Wu W, Nie
E, Yu T, Zhou X, Bao Z, Jin X, et al: MicroRNA-1301 inhibits
proliferation of human glioma cells by directly targeting N-Ras. Am
J Cancer Res. 7:982–998. 2017.PubMed/NCBI
|
|
26
|
Jiang K, Zhi T, Xu W, Xu X, Wu W, Yu T,
Nie E, Zhou X, Bao Z, Jin X, et al: MicroRNA-1468-5p inhibits
glioma cell proliferation and induces cell cycle arrest by
targeting RRM1. Am J Cancer Res. 7:784–800. 2017.PubMed/NCBI
|
|
27
|
Li P, Wang X, Shan Q, Wu Y and Wang Z:
MicroRNA-130b promotes cell migration and invasion by inhibiting
peroxisome proliferator-activated receptor-γ in human glioma. Oncol
Lett. 13:2615–2622. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Khan S, Brougham CL, Ryan J, Sahrudin A,
O'Neill G, Wall D, Curran C, Newell J, Kerin MJ and Dwyer RM:
miR-379 regulates cyclin B1 expression and is decreased in breast
cancer. PLoS One. 8:e687532013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wu D, Niu X, Tao J, Li P, Lu Q, Xu A, Chen
W and Wang Z: MicroRNA-379-5p plays a tumor-suppressive role in
human bladder cancer growth and metastasis by directly targeting
MDM2. Oncol Rep. 37:3502–3508. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li Z, Shen J, Chan MT and Wu WK:
MicroRNA-379 suppresses osteosarcoma progression by targeting PDK1.
J Cell Mol Med. 21:315–323. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hao GJ, Hao HJ, Ding YH, Wen H, Li XF,
Wang QR and Zhang BB: Suppression of EIF4G2 by miR-379 potentiates
the cisplatin chemosensitivity in nonsmall cell lung cancer cells.
FEBS Lett. 591:636–645. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kaur K, Kakkar A, Kumar A, Purkait S,
Mallick S, Suri V, Sharma MC, Julka PK, Gupta D, Suri A and Sarkar
C: Clinicopathological characteristics, molecular subgrouping, and
expression of miR-379/miR-656 cluster (C14MC) in adult
medulloblastomas. J Neurooncol. 130:423–430. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yamamoto K, Seike M, Takeuchi S, Soeno C,
Miyanaga A, Noro R, Minegishi Y, Kubota K and Gemma A: MiR-379/411
cluster regulates IL-18 and contributes to drug resistance in
malignant pleural mesothelioma. Oncol Rep. 32:2365–2372. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Poon TC, Wong N, Lai PB, Rattray M,
Johnson PJ and Sung JJ: A tumor progression model for
hepatocellular carcinoma: Bioinformatic analysis of genomic data.
Gastroenterology. 131:1262–1270. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Song H, Li C, Li R and Geng J: Prognostic
significance of AEG-1 expression in colorectal carcinoma. Int J
Colorectal Dis. 25:1201–1209. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Dong L, Qin S, Li Y, Zhao L, Dong S, Wang
Y, Zhang C and Han S: High expression of astrocyte elevated gene-1
is associated with clinical staging, metastasis, and unfavorable
prognosis in gastric carcinoma. Tumour Biol. 36:2169–2178. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Nikpour M, Emadi-Baygi M, Fischer U,
Niegisch G, Schulz WA and Nikpour P: MTDH/AEG-1 contributes to
central features of the neoplastic phenotype in bladder cancer.
Urol Oncol. 32:670–677. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang Y, Klijn JG, Zhang Y, Sieuwerts AM,
Look MP, Yang F, Talantov D, Timmermans M, Meijer-van Gelder ME, Yu
J, et al: Gene-expression profiles to predict distant metastasis of
lymph-node-negative primary breast cancer. Lancet. 365:671–679.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
He Z, He M, Wang C, Xu B, Tong L, He J,
Sun B, Wei L and Chu M: Prognostic significance of astrocyte
elevated gene-1 in human astrocytomas. Int J Clin Exp Pathol.
7:5038–5044. 2014.PubMed/NCBI
|
|
40
|
Xia Z, Zhang N, Jin H, Yu Z, Xu G and
Huang Z: Clinical significance of astrocyte elevated gene-1
expression in human oligodendrogliomas. Clin Neurol Neurosurg.
112:413–419. 2010. View Article : Google Scholar : PubMed/NCBI
|