Introduction
Gastric cancer is one of the top three leading
causes of cancer death in the United States and China (1). It is a multifactor disease with high
prevalence risk which can be presence for several years before its
symptoms develop and its clinical outcome is difficult to predict.
The factors of both genetics and epigenetic are usual causes of
gastric cancer progression and development (2,3),
however the underlying molecular mechanism is still unclear.
Macroautophagy (hereafter referred to as autophagy)
is a homeostatic process involved in turnover of long-lived
proteins and whole organelles by lysosomal activity that eliminates
supernumerary or damaged organelle (4–6). It
has been widely accepted that appropriate autophagic flux had
beneficial effect on reducing the risk for diseases, especially in
cancer (7). In general, autophagy
is a survival mechanism induced in adverse environment. However,
autophagy deficiency also lead to the tumors malignancy degree
increased, gastric cancer included (8). Moreover, gastric carcinogenesis could
be inhibited through autophagy induction (9,10).
Due to the pivotal role of autophagy in gastric cancer progression,
the key player autophagic adaptation has been considered as drug
target for gastric cancer prevention and management.
SP1 is a zinc finger transcription factor belongs to
the SP1 multigene family (SP2, SP3 and SP4), which plays an
important role in development but also play a role in cancers
(11). SP1 regulates target gene
transcription by binding to their promoter contain GC boxes
(12,13). Unique SP1 expression has been
observed in malignant gastric tissues, and proven to have a close
correlation with insulin-like growth factor I receptor (IGF1R)
expression in gastric cancer (14–16).
Besides that, dysregulated SP1 expression may contributed to the
growth and metastasis of gastric cancer and can be a potential
therapeutic target. However, little was known about how SP1
function in autophagy of gastric cancer.
In our study, we found that overexpression of SP1
decreased the autophagic flux in gastric cancer cells, whereas SP1
deficiency induces autophagy of cancer cells. Intriguingly, we
observed SP1 could enhance the expression level of p62, by directly
binding to the promoter of p62. Additionally, we found that p62
expression levels were upregulated in gastric tissues specimens
together with SP1 expression. Together, we provided evidence for a
novel mechanism regulating autophagy in gastric cancer cells.
Materials and methods
Ethics statement
The human gastric specimens were obtained during
surgery from patients at the Department of General Surgery, The
Second Hospital of Wenzhou Medical University. All the procedures
were reviewed and approved by the Ethics Committee of The Second
Hospital of Wenzhou Medical University (L-2016-27), and informed
consent was obtained from all patients.
Cell culture and transfection
Human gastric cancer cells AGS and N87 (ATCC,
Manassas, VA, USA) were culture in Dulbecco's modified Eagle's
medium (DMEM) supplemented with 10% FBS, 1 mM glutamine, 100 U/ml
penicillin, and 100 mg/ml streptomycin. Transfection was performed
using Lipofectamine 2000 according to the instructions.
Plasmid and luciferase assay
For construction luciferase reporter plasmids, the
promoter fragment of p62 was amplified from human genome cDNA and
inserted into PGL3-basic vector. SP1, control plasmids, and p62
siRNA oligo were all purchased from Genechem (Shanghai, China).
AGS cells were planted in a 48-well plate and
co-transfected with SP1 plasmid or control, pRL-TK and
pGL3-p62-promoter. Forty-eight hours after transfection, cells were
lysed and relative luciferase activity was analyzed with the Dual
Luciferase Reporter Assay System on a luminometer (both from
Promega, Madison, WI, USA).
Chromatin immunoprecipitation
assay
Chromatin immunoprecipitation assay was performed as
previously described by using Magna ChIP™ kit
(Millipore, Billerica, MA, USA) (17). In brief, AGS cells were transfected
with SP1 plasmids for 48 h, then chromatin was immunoprecipitated
using anti-SP1 antibody (ab13370; Abcam, Cambridge, UK) at dilution
1:10. RT-PCR was performed to determine SP1 binding site. ChIP
primer sequences are provided: Forward, GGCAGGTGCAGCACGTGC and
reverse, TCAGAAAGGCAGGCGCTGC.
Immunofluorescence staining
AGS cells were seeded into 12-well plate, then
transfected with plasmids. After 48 h, the cells were fixed with
cold methanol and permeabilized with 1% Triton X-100 in PBS for 15
min. Anti-LC3b antibody (3868s, 1:200; Cell Signaling Technology,
Inc., Danvers, MA, USA) was placed in PBS at 4°C overnight,
following by incubation with Alexa Fluor 594 (CA11012s, 1:1,000;
Invitrogen Life Technologies, Carlsbad, CA, USA) for 1 h at room
temperature. The nucleic were stained using DAPI (C1005, 1:1,000;
Beyotime Institute of Biotechnology, Haimen, China) for 3 min at
room temperatuire. Finally, cell images were taken using a
microscope (Olympus BX61; Olympus, Tokyo, Japan), then the
LC3b-positive cells were calculated.
Real-time PCR
RNA was isolated using TRIzol reagent from cells,
500 ng of RNA was reverse-transcribed into cDNA using the RT system
by PrimeScript RT reagent kit (RR037A; Takara). Real-time PCR which
contain 4.6 µl CDNA, 5 µl SYBR Green (4887352001; Roche
Diagnostics, Basel, Switzerland) and 0.4 µl primers was performed
on ABI 7500 Fast Real Time PCR system. The following primers were
used in this study: GAPDH forward, CTGGGCTACACTGAGCACC and reverse,
AAGTGGTCGTTGAGGGCAATG; LC3b forward, GATGTCCGACTTATTCGAGAGC and
reverse, TTGAGCTGTAAGCGCCTTCTA; ATG4b forward, ATGGACGCAGCTACTCTGAC
and reverse, TTTTCTACCCAGTATCCAAACGG; ATG5 forward,
AAAGATGTGCTTCGAGATGTGT and reverse, CACTTTGTCAGTTACCAACGTCA; p62
forward, GCACCCCAATGTGATCTGC and reverse,
CGCTACACAAGTCGTAGTCTGG.
Western blotting
Cells were lysed in RIPA lysis buffer containing 10
mM Tris-HCl (pH 7.5), 1% SDS, 1 mM Na3VO4, 10
mM NaF and protease inhibitor cocktail (4693132001; Roche
Diagnostics). Protein samples (20 µg) were subjected to
immunoblotting, resolved on SDS-PAGE with 80 V, transferred onto
nitrocellulose membranes with 300 mA for 3 h and blocked by 5% skim
milk then probed with the various antibodies at 4°C overnight.
Second day, the membranes washed with TBST buffer and probed with
second antibodies which diluted with 5% skim milk at 1:1,500.
Detection was performed by measuring the chemiluminescent signal as
assayed by SuperSignal Ultra. SDS-PAGE (15%) for LC3b and 10%
SDS-PAGE for other protein. Antibodies anti-SP1 (9389s, 1:1,000;
Cell Signalling Technology, Inc.), anti-p62 (610833, 1:1,000; BD
Biosciences, Franklin Lakes, NJ, USA), anti-LC3b (3868s, 1:1,000;
Cell Signalling Technology, Inc.), GAPDH (1:5,000; Sigma-Aldrich;
Merck KGaA, Darmstadt, Germany) diluted with 5% BSA were used for
western blotting.
Immunohistochemistry analysis
A representative formalin-fixed paraffin-embedded
tissue block was chosen from the 10 gastric cancer tissues and 17
normal gastric tissues. Sections mounted on poly-L-lysine-coated
slide were incubated for 30 min at 60°C, deparaffinised by standard
methods, and placed in 0.05 m Tris-HCI buffer, pH 7.2. Antigen
retrieval was performed for 20 min in 10 mm sodium citrate buffer
(pH 6.0) heated at 95°C in a steamer, followed by cooling for 20
min. After blocking endogenous peroxidase activity with 0.3%
aqueous hydrogen peroxide for 5 min, the primary polyclonal
anti-sp1 and anti-p62 antibody was incubated with the sections at a
final dilution of 2 µg/ml for 30 min. The relative level was
quantified by Image-Pro Plus. The distribution of immunolabelling
was determined from a minimum of three representative high-power
(×400 magnification) fields and categorised into three groups: 0%,
negative; 1–25%, focal; and 26–100%, diffuse.
Statistical analysis
All results are represented as mean ± SEM from three
independent experiments at least. Student's t-test was used to
analyze the differences in mean values between two groups by
GraphPad Prism 5.0 (GraphPad Software, Inc., La Jolla, CA, USA).
P<0.05 was considered to indicate a statistically significant
difference.
Results
Overexpression of SP1 inhibits
autophagy in gastric cancer
To explore a potential role of SP1 in gastric
cancer, we first overexpressed SP1 in two human gastric cancer cell
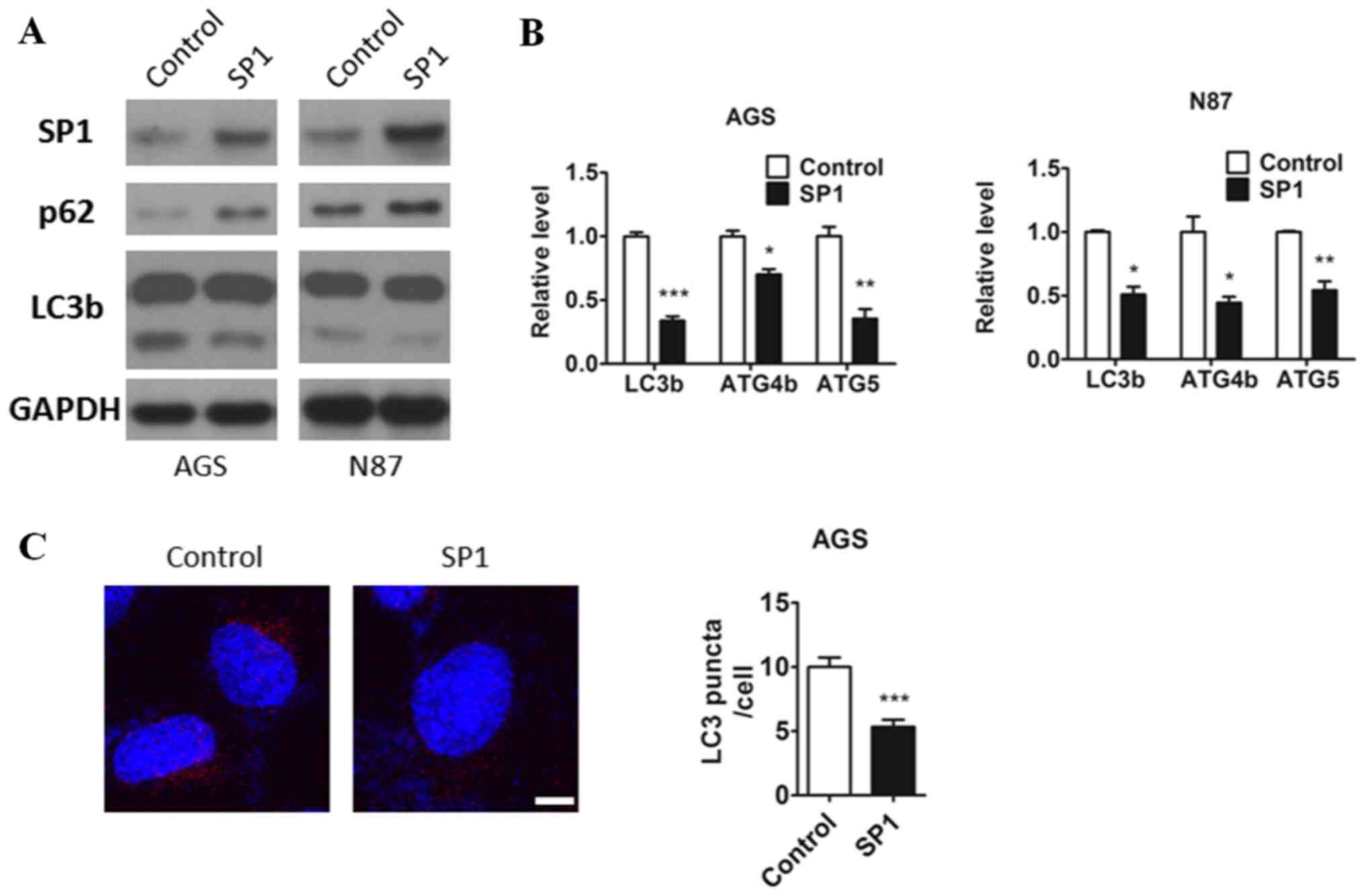
lines, AGS and N87. Interestingly, we observed decreased protein
level of autophagy marker LC3b and increased protein level of p62,
along with the increase of SP1expression (Fig. 1A). Many studies have proved that
those ATG genes expression promoted autophagy. LC3B localizes to
the autophagosome membrane which was the key regulator involved in
autophagosome formation (18).
Atg5 protein could conjugate with Atg12 and Atg8 (LC3) and involved
in the early stages of autophagosome formation (19). ATG4B (autophagin-1) shows the
catalytic efficiency for cleaving the C terminus of LC3B (20). And p62 protein is an
autophagy-specific substrate, the accumulation of p62 protein is
always employed as a readout for autophagy impairment (21). Therefore, those genes are widely
used as a markers of autophagic activity. To test whether SP1
alters the expression of the ATG genes (LC3b, ATG4b and ATG5) in
gastric cancer cells, we used qRT-PCR and demonstrated that the
mRNA expression of these genes were inhibited by the expression of
SP1 in AGS and N87 cell lines (Fig.
1B). Also, decreased formation of autophagosomes labeled by
LC3b antibody have been found in AGS cells expressing SP1 compared
to the control (Fig. 1C). So these
results indicated that SP1 may have a role in regulating autophagic
pathways in gastric cancer cells.
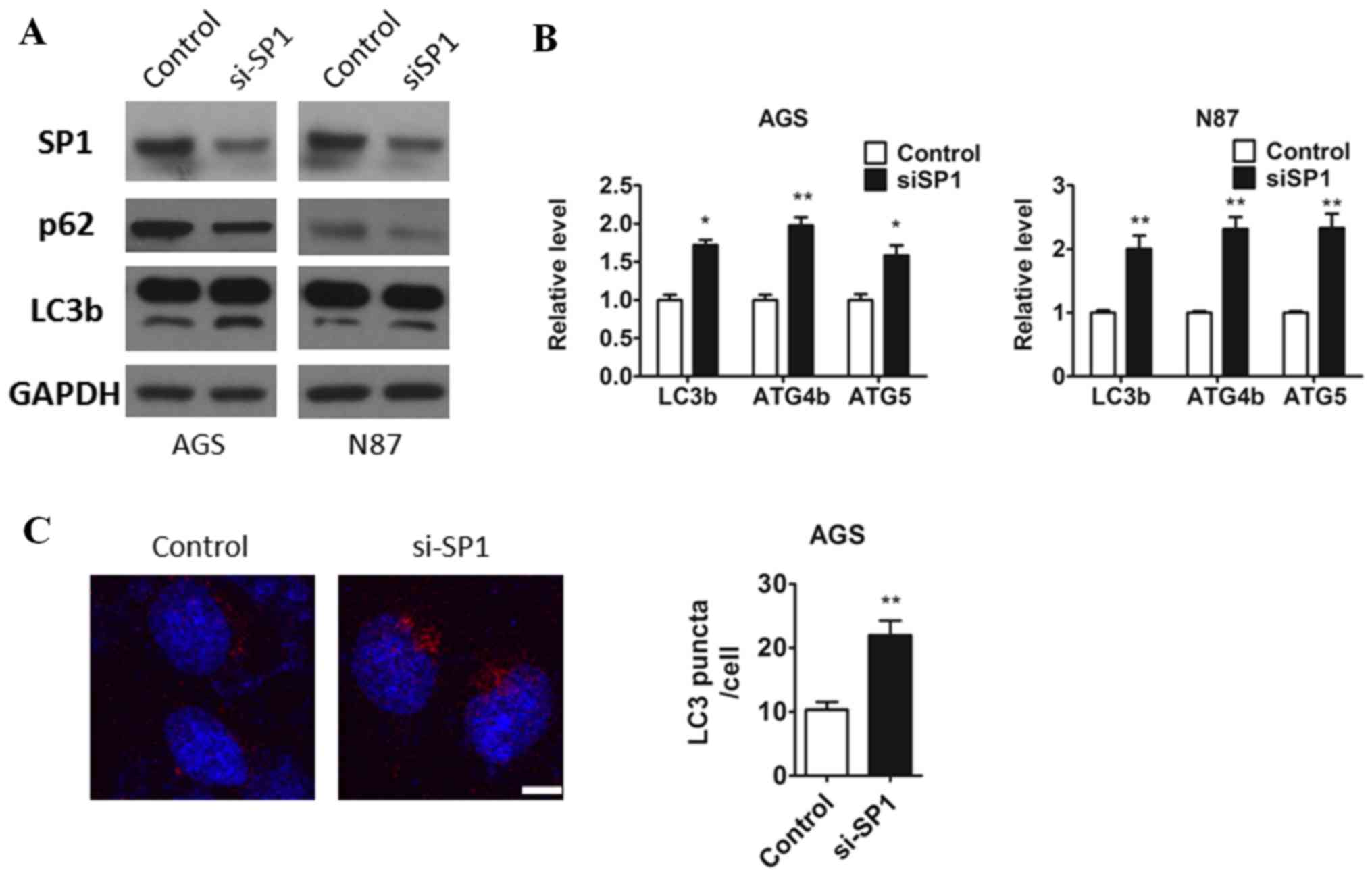
Inbihition of SP1 induces autophagy in
gastric cancer
As the data shown using SP1 overexpression, to
confirm our hypothesis that sp1 plays a role in autophagic pathways
and we respectively transfected control and si-sp1 oligos to both
AGS and N87 cells by Lipo 2000 and 48 h after transfection, cells
were lysed. As expected the protein level of sp1 was downregulated
in both AGS and N87 cell (Fig.
1A). Consistently, we found that knockdown of SP1 in both AGS
and N87 cells promoted the transition of LC3-I to LC3-II, and the
protein level of p62 was diminished treated with si-SP1 oligos
compared to the cells treated with control oligols (Fig. 2A). Furthermore, we also observed
that SP1 deficiency led to an increasing of ATG genes (LC3b, ATG4b,
ATG5) mRNA expression in both AGS and N87 cells (Fig. 2B). To further confirm the changes
of autophagy accompany with sp1, AGS cells which were either
transfected with a control or sp1 plasmid were isolated and
immunofluorescence staining against LC3b antibody was applied and
it was found that the inhibition of SP1 resulted in a clear
increase in the autophagic flux (Fig.
2C). These results suggested that inhibition of the endogenous
SP1 affected the process of autophagy.
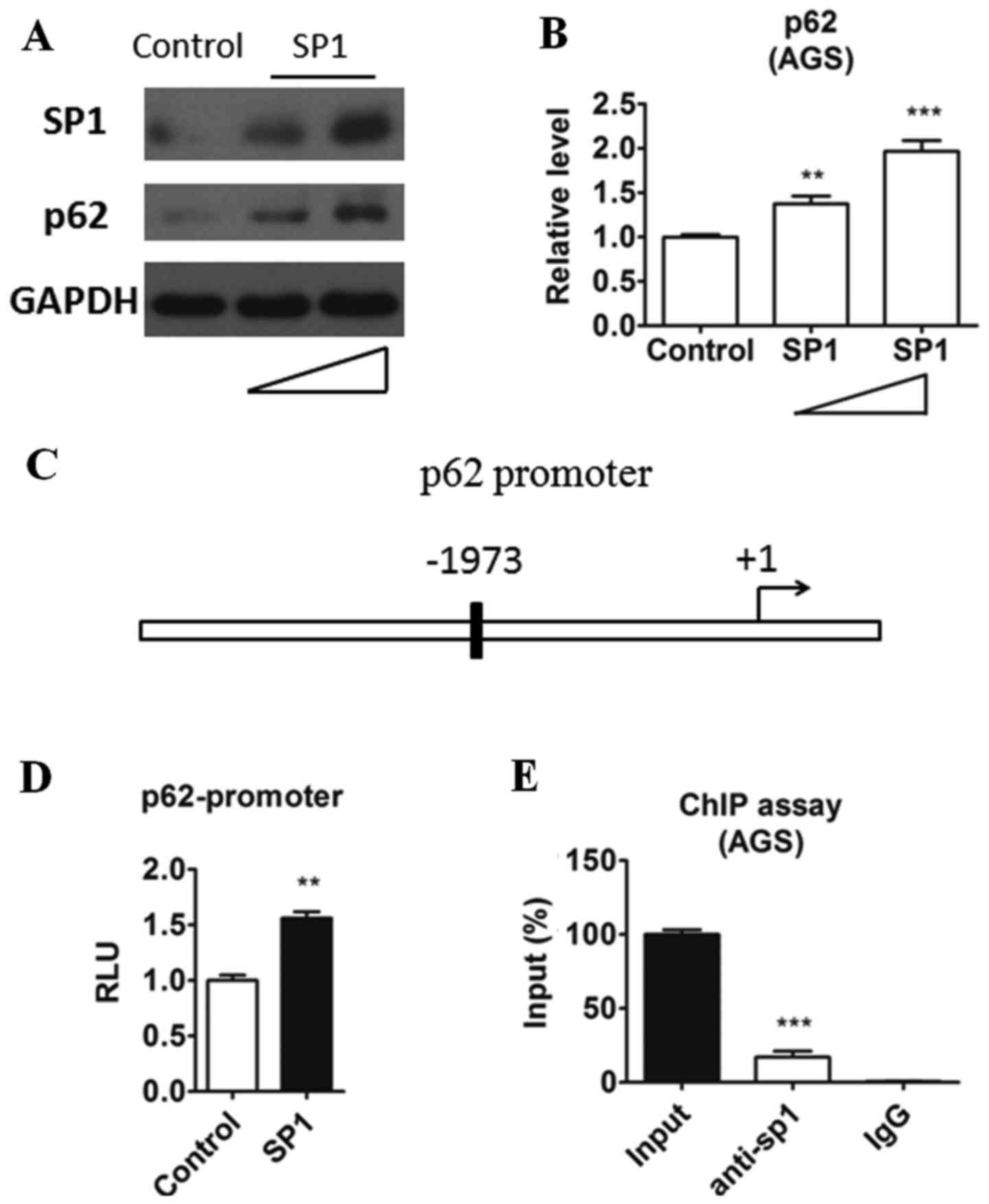
SP1 promotes p62 expression at
transcriptional level
According to the above results, we speculated that
the expression level of p62 was mediated by sp1. The
transcriptional activity of the p62 promoter was reported mediated
by some transcription factors, such as NRF2 (22). However, whether SP1 could affect
the expression of p62 and p62-mediated autophagy was unknown. To
confirm our hypothesis, we isolated AGS cells which were either
transfected with a control or sp1 plasmid and then quantified the
protein level of p62. As shown in Fig.
3A and B, a dose-dependent protein level change of p62 in
relation to SP1 was noticed. Overexpression of SP1 upregualted the
p62 expression in both protein and mRNA levels intriguingly. Sp1 is
a well-known transcriptional factor and regulate many genes
expression through binding to their promoter. Therefore we
speculated that sp1 may bind to the promoter of p62. By analyzing
the sequence before the p62 transcription start site, we found a
potential SP1-bingding element (GGGCGG) located upstream of p62
(Fig. 3C). Then we cloned this
region into a luciferase reporter plasmid (WT) and evaluated the
transcriptional activity when SP1 overexpression in AGS cells. As
shown in Fig. 3D, SP1 could
increase p62 promoter transcriptional activity. To evaluated
whether SP1 could be recruited to the promoter of SP1, ChIP assay
was performed in AGS cells. In agreement with the results in
luciferase assay, we detected the recruitment of SP1 to the p62
promoter in ChIP assay (Fig. 3E).
Taken together, these results indicated that SP1 may positively
regulate p62 transcriptional activity by binding to its promoter in
gastric cancer cells.
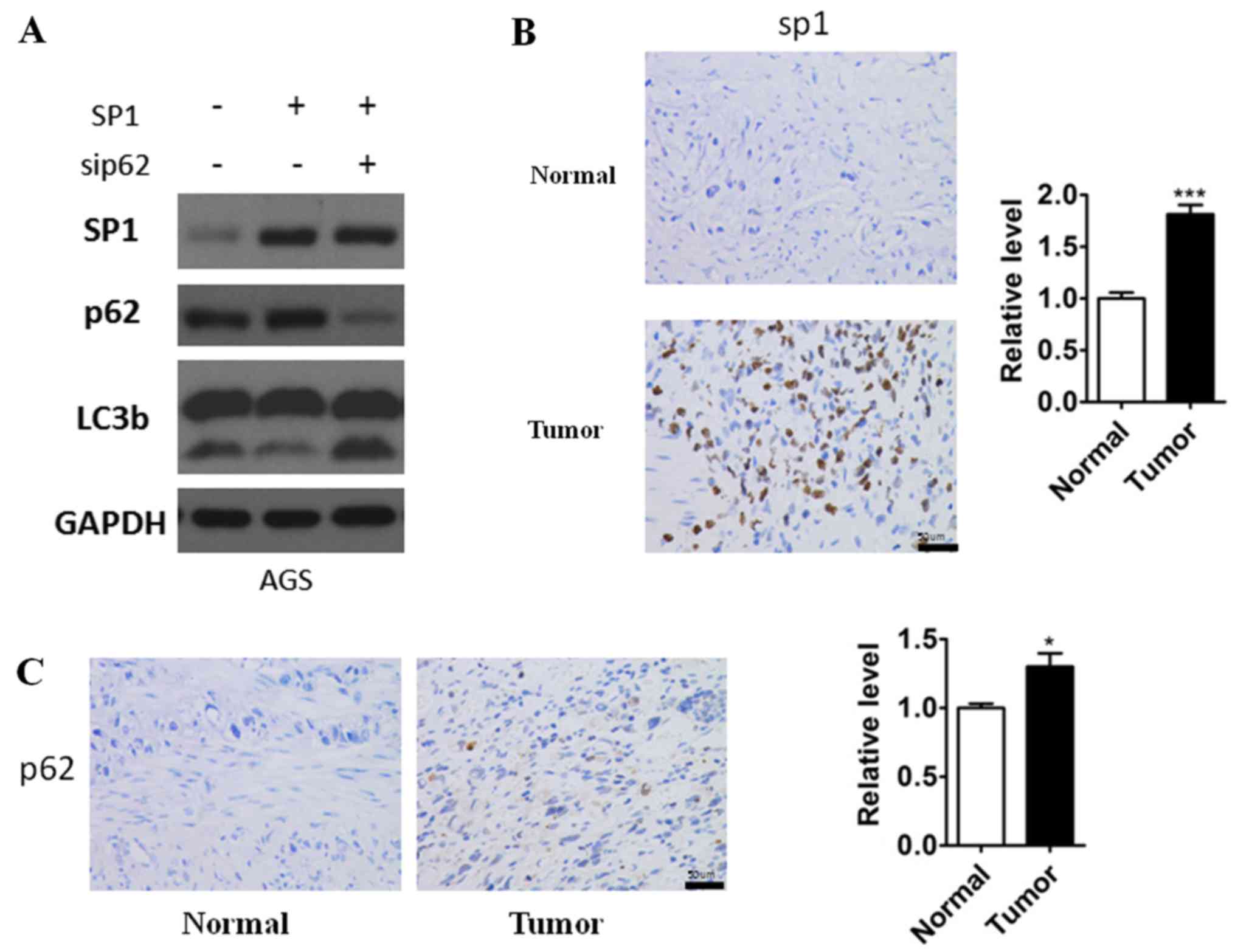
p62 is required for SP1-mediated
autophagy inhibition
According to the above results, we found that sp1
could affect both the expression of p62 and the autophagy in
gastric cancer cells. So we hypothesis that sp1 regulated autophagy
is p62-dependent in gastric cancer cells. To confirm our hypothesis
we extended our observation of SP1 regulatory network in AGS cells.
Overexpression of SP1 increased p62 expression and led to
downregulation of autophagic flux, while knockdown of p62 resulted
in upregulation of LC3b-II even though with the presence of high
SP1 expression (Fig. 4A). These
results, taken together, indicated that SP1 promoted gastric cancer
cells autophagy via activating p62, and in other words, p62 is
required for SP1-mediated autophagy inhibition. In addition, to
further confirm the results we had founded, the immunoreactivity of
SP1 and p62 was analyzed in the gastric cell nuclear staining As
shown in Fig. 4B and C we observed
increasingly high sp1and p62 expression in gastric cancer specimens
which was consistent to our study.
Discussion
Gastric cancer is one of the most popular and
aggressive cancers worldwide. Although factors of both genetics and
epigenetic are usual causes of gastric cancer progression and
development (23), but the
underlying molecular mechanism is still unclear. Autophagy plays a
pivotal role in cancer progression. It is well accepted that
appropriate autophagic flux is critical for cancer cells (24). Here we identified a novel
regulatory mechanism that SP1 acted as a negative regulator of
autophagy by activating p62 transcription in gastric cancer cells.
The SP1-p62 axis may facilitate the tumorigenesis in gastric
cancer.
SP1 is one of the general zinc finger transcription
factor SP family, which plays an important role in cancer
progression (25). It had been
observed that the expression of SP1 was significantly elevated in
gastric cancer specimens, and proved to have a closed correlation
with patient survival (16). It is
suggesting that SP1 signaling pathway may have a positive effect on
gastric cancer progression and SP1 could be a prognostic marker for
diagnosis of gastric cancer. However, little was known about how
SP1 function in autophagy of gastric cancer. In the present study,
we showed that SP1 could inhibit autophagy level in gastric cancer
cells. SP1 deficiency led to a significant increase of autophagic
flux in gastric cancer cells. Mechanistically, we found SP1 could
increase the transcriptional activity of p62 through binding to the
GC box located in the upstream of SP1. We further confirmed that by
Luciferase assay and ChIP assay. Nonetheless, whether SP1 will
function to affect the other aspects of gastric cancer remain
further validation.
Alteration of autophagy-related genes was found in
gastric cancer (26,27). It had been found that autophagy
played a protective role at early stage of carcinogenesis (28,29).
Autophagy deficiency led to accumulation of p62 in cells (30). The p62 protein is a structural
component of the autophagesome (31,32).
Accumulation of p62 can promote tumorigenesis. We here found that
the p62 expression level was increase in SP1-positve gastric
specimens further supporting the notion that SP1 could target p62
expression. Besides that, we also observed that SP1-mediated
autophagy inhibition was blocked when elimination of p62, but the
underlying mechanism is still not clear. The recent study of Mathew
et al added a number of interesting twists to the role of
p62 in cancer. These investigators showed that when cells with
impairment in both apoptosis and autophagy were metabolically
stressed, the accumulation of p62 led to enhanced tumorigenicity
through an as yet undefned mechanism involving increased aneuploidy
(30). These observations placed
p62 as the missing link between defcient autophagy and increased
tumorigenesis through the control of genome instability (33). An interesting feed-forward loop may
contribute to this process. The increased ROS production in these
cells may be responsible, at least in part, for the induction of
p62 expression. p62 overexpression then contributes to additional
ROS production as part of an amplifying loop, thereby promoting
genome instability and autophagy. Meanwhile it is plausible that
when cells are deprived of nutrients and oxygen, and p62
overexpression reaches a certain threshold level, NF-κB, which is a
critical regulator of survivalis, inhibited by the mopping up of
critical components of the NF-κB pathway into huge p62 aggregates.
The dysregulated expression of SP1 resulted in hyperactivation of
p62 may help the gastric cancer cells survival or growth under
adverse condition.
In conclusion, our results provide evidence that SP1
inhibits gastric cancer cell autophagy via activating of p62.
Further study will focus on the vivo effects of SP1 or p62 in
gastric tissues. Our finding would provide a novel strategy for the
treatment of gastric cancer.
Acknowledgements
This study was supported by the program of Zhejiang
Provincial Natural Science Foundation (grant no. LY17H160054).
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tahara E: Molecular aspects of invasion
and metastasis of stomach cancer. Verh Dtsch Ges Pathol. 84:43–49.
2000.PubMed/NCBI
|
|
3
|
Chan AO, Luk JM, Hui WM and Lam SK:
Molecular biology of gastric carcinoma: From laboratory to bedside.
J Gastroenterol Hepatol. 14:1150–1160. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Eisenberg-Lerner A, Bialik S, Simon HU and
Kimchi A: Life and death partners: Apoptosis, autophagy and the
cross-talk between them. Cell Death Differ. 16:966–975. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Maiuri MC, Zalckvar E, Kimchi A and
Kroemer G: Self-eating and self-killing: Crosstalk between
autophagy and apoptosis. Nat Rev Mol Cell Biol. 8:741–752. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shintani T and Klionsky DJ: Autophagy in
health and disease: A double-edged sword. Science. 306:990–995.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Choi AM, Ryter SW and Levine B: Autophagy
in human health and disease. N Engl J Med. 368:1845–1846. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cheng HY, Zhang YN, Wu QL, Sun XM, Sun JR
and Huang X: Expression of beclin 1, an autophagy-related protein,
in human cervical carcinoma and its clinical significance. Eur J
Gynaecol Oncol. 33:15–20. 2012.PubMed/NCBI
|
|
9
|
Wang K, Liu R, Li J, Mao J, Lei Y, Wu J,
Zeng J, Zhang T, Wu H, Chen L, et al: Quercetin induces protective
autophagy in gastric cancer cells: Involvement of Akt-mTOR- and
hypoxia-induced factor 1α-mediated signaling. Autophagy. 7:966–978.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tu SP, Quante M, Bhagat G, Takaishi S, Cui
G, Yang XD, Muthuplani S, Shibata W, Fox JG, Pritchard DM and Wang
TC: IFN-γ inhibits gastric carcinogenesis by inducing epithelial
cell autophagy and T-cell apoptosis. Cancer Res. 71:4247–4259.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Suske G: The Sp-family of transcription
factors. Gene. 238:291–300. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hagen G, Dennig J, Preiss A, Beato M and
Suske G: Functional analyses of the transcription factor Sp4 reveal
properties distinct from Sp1 and Sp3. J Biol Chem. 270:24989–24994.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shi Q, Le X, Abbruzzese JL, Peng Z, Qian
CN, Tang H, Xiong Q, Wang B, Li XC and Xie K: Constitutive Sp1
activity is essential for differential constitutive expression of
vascular endothelial growth factor in human pancreatic
adenocarcinoma. Cancer Res. 61:4143–4154. 2001.PubMed/NCBI
|
|
14
|
Kanai M, Wei D, Li Q, Jia Z, Ajani J, Le
X, Yao J and Xie K: Loss of Krüppel-like factor 4 expression
contributes to Sp1 overexpression and human gastric cancer
development and progression. Clin Cancer Res. 12:6395–6402. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jiang Y, Wang L, Gong W, Wei D, Le X, Yao
J, Ajani J, Abbruzzese JL, Huang S and Xie K: A high expression
level of insulin-like growth factor I receptor is associated with
increased expression of transcription factor Sp1 and regional lymph
node metastasis of human gastric cancer. Clin Exp Metastasis.
21:755–764. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang L, Wei D, Huang S, Peng Z, Le X, Wu
TT, Yao J, Ajani J and Xie K: Transcription factor Sp1 expression
is a significant predictor of survival in human gastric cancer.
Clin Cancer Res. 9:6371–6380. 2003.PubMed/NCBI
|
|
17
|
Martucci NM, Rea I, Ruggiero I,
Terracciano M, De Stefano L, Migliaccio N, Palmieri C, Scala G,
Arcari P, Rendina I and Lamberti A: A new strategy for label-free
detection of lymphoma cancer cells. Biomed Opt Express.
6:1353–1362. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kabeya Y, Mizushima N, Ueno T, Yamamoto A,
Kirisako T, Noda T, Kominami E, Ohsumi Y and Yoshimori T: LC3, a
mammalian homologue of yeast Apg8p, is localized in autophagosome
membranes after processing. EMBO J. 19:5720–5728. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mizushima N, Ohsumi Y and Yoshimori T:
Autophagosome formation in mammalian cells. Cell Struct Funct.
27:421–429. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yang Z, Wilkie-Grantham RP, Yanagi T, Shu
CW, Matsuzawa S and Reed JC: ATG4B (autophagin-1) phosphorylation
modulates autophagy. J Biol Chem. 290:26549–26561. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Klionsky DJ, Abdalla FC, Abeliovich H,
Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M,
Agostinis P, Aguirre-Ghiso JA, et al: Guidelines for the use and
interpretation of assays for monitoring autophagy. Autophagy.
8:445–544. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jain A, Lamark T, Sjøttem E, Larsen KB,
Awuh JA, Øvervatn A, McMahon M, Hayes JD and Johansen T: p62/SQSTM1
is a target gene for transcription factor NRF2 and creates a
positive feedback loop by inducing antioxidant response
element-driven gene transcription. J Biol Chem. 285:22576–22591.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Parkin DM, Pisani P and Ferlay J:
Estimates of the worldwide incidence of 25 major cancers in 1990.
Int J Cancer. 80:827–841. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Levine B: Cell biology: Autophagy and
cancer. Nature. 446:745–747. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hirose T and Horvitz HR: An Sp1
transcription factor coordinates caspase-dependent and -independent
apoptotic pathways. Nature. 500:354–358. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhao Z, Han F, Yang S, Wu J and Zhan W:
Oxamate-mediated inhibition of lactate dehydrogenase induces
protective autophagy in gastric cancer cells: Involvement of the
Akt-mTOR signaling pathway. Cancer Lett. 358:17–26. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu D, Gao M and Zhao S: Autophagy as a
novel strategy for treatment of gastric cancer: A hypothesis. Med
Sci Monit. 19:794–796. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lee HW, Jang KS, Choi HJ, Jo A, Cheong JH
and Chun KH: Celastrol inhibits gastric cancer growth by induction
of apoptosis and autophagy. BMB Rep. 47:697–702. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ge J and Chen Z, Huang J, Chen J, Yuan W,
Deng Z and Chen Z: Upregulation of autophagy-related gene-5 (ATG-5)
is associated with chemoresistance in human gastric cancer. PLoS
One. 9:e1102932014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mathew R, Karp CM, Beaudoin B, Vuong N,
Chen G, Chen HY, Bray K, Reddy A, Bhanot G, Gelinas C, et al:
Autophagy suppresses tumorigenesis through elimination of p62.
Cell. 137:1062–1075. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bjørkøy G, Lamark T, Brech A, Outzen H,
Perander M, Overvatn A, Stenmark H and Johansen T: p62/SQSTM1 forms
protein aggregates degraded by autophagy and has a protective
effect on huntingtin-induced cell death. J Cell Biol. 171:603–614.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ichimura Y, Kumanomidou T, Sou YS,
Mizushima T, Ezaki J, Ueno T, Kominami E, Yamane T, Tanaka K and
Komatsu M: Structural basis for sorting mechanism of p62 in
selective autophagy. J Biol Chem. 283:22847–22857. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mathew R, Kongara S, Beaudoin B, Karp CM,
Bray K, Degenhardt K, Chen G, Jin S and White E: Autophagy
suppresses tumor progression by limiting chromosomal instability.
Genes Dev. 21:1367–1381. 2007. View Article : Google Scholar : PubMed/NCBI
|