Introduction
Osteosarcoma (OS) is a common primary malignant
tumor of the bone and primarily occurs in adolescents and young
adults (1). It accounts for ~2.4%
of all tumors in children and adolescents (2). The majority of cases of OS affect the
long bones, particularly the metaphysis, proximal tibia, proximal
humerus and distal femur (3).
Although significant advancements have been achieved in surgical
resection combined with radiotherapy and chemotherapy, the
prognosis of patients with OS has only been slightly improved, with
the median survival time reaching only 23 months (4,5). The
poor treatment outcome of patients with OS is due to local
recurrence and metastasis, especially pulmonary metastasis
(6). Although numerous molecular
targets have been identified to be implicated in OS formation and
progression, their detailed mechanisms remain unknown (7). Therefore, the mechanisms underlying
OS occurrence and development must be comprehensively understood to
identify effective therapeutic targets for treating OS.
MicroRNAs (miRs) are highly conserved, non-coding
and small RNAs that comprise 19–23 nucleotides (8). miRs are considered critical gene
regulators that directly bind to the 3′-untranslated regions
(3′-UTRs) by base pairing and therefore induce mRNA degradation
and/or translational suppression (9). miRs regulate ~60% of all human
protein coding genes, indicating that miRs serve important roles in
physiological and pathological behavior, including cell
proliferation, cycle, apoptosis, metastasis, embryogenesis and
differentiation (10). The
dysregulation of miRs has been recently reported in different types
of malignancies, including OS (11), prostate cancer (12), breast cancer (13) and colorectal cancer (14). Abnormally expressed miRs may serve
tumor suppressive roles or oncogenic roles in tumorigenesis and
tumor development, which depend on the biological functions of
their target genes (15).
Therefore, the expression and roles of miRs in OS must be further
investigated to identify potential therapeutic targets for the
therapy of patients with OS.
miR-376a-3p (miR-376a) is dysregulated in several
human cancer types and the dysregulation of miR-376a serves an
important role in various cell processes, including cell
proliferation, migration, invasion, metastasis and apoptosis
(16–19). However, to the best of our
knowledge, the expression pattern, clinical significance, specific
role and detailed mechanisms of miR-376a in OS remains unclear.
Therefore, the present study aimed to detect the miR-376a
expression in OS and its association with clinicopathological
characteristics and to determine the biological roles and
regulatory mechanism of miR-376a in OS cells.
Materials and methods
Tissue samples
A total of 49 primary OS tissues and corresponding
adjacent normal bone tissues were obtained from patients who
underwent surgical resection in the China-Japan Union Hospital of
Jilin University (Changchun, China), between October 2013 and March
2017. The patient characteristics are presented in Table I. No chemotherapy or radiotherapy
was administered prior to surgery. All tissues were snap-frozen in
liquid nitrogen (−196°C) at the time of surgery and were stored in
liquid nitrogen until further use. The present study was approved
by the Ethics Committee of the China-Japan Union Hospital of Jilin
University. In addition, written informed consent was provided by
all participants.
 | Table I.Association between miR-376a and
clinicopathological features of osteosarcoma patients. |
Table I.
Association between miR-376a and
clinicopathological features of osteosarcoma patients.
|
| miR-376a
expression |
|
|---|
|
|
|
|
|---|
| Clinicopathological
features | Low (n=25) | High (n=24) | P-value |
|---|
| Age (years) |
|
| 0.567 |
|
<20 | 16 | 13 |
|
|
≥20 | 9 | 11 |
|
| Gender |
|
| 0.377 |
|
Male | 14 | 17 |
|
|
Female | 11 | 7 |
|
| Tumor size
(cm) |
|
| 0.022 |
|
<8 | 7 | 15 |
|
| ≥8 | 18 | 9 |
|
| Location of the
primary tumor |
|
| 0.289 |
|
Tibia/femur | 22 | 18 |
|
|
Elsewhere | 3 | 6 |
|
| Lymph node
infiltration |
|
| 0.001 |
| No | 9 | 20 |
|
|
Yes | 16 | 4 |
|
Cell culture and transfection
A total of four human OS cell lines (HOS, SAOS-2,
MG-63 and U2OS), and a normal human osteoblast hFOB1.19 cell line
were acquired from the Shanghai Cell Bank of the Chinese Academy of
Sciences (Shanghai, China). All cell lines were cultured at 37°C in
a humidified atmosphere containing 5% CO2 and were
maintained in Dulbecco's modified Eagle's medium (DMEM)
supplemented with 10% fetal bovine serum (FBS; both from HyClone;
GE Healthcare Life Sciences, Logan, UT, USA), 100 U/ml penicillin
and 100 U/ml streptomycin (Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany).
miR-376a mimic and miR mimic negative controls
(miR-NC) were synthesized and purchased from the Shanghai
GenePharma Co., Ltd. (Shanghai, China). The miR-376a mimic sequence
was 5′-AUCAUAGAGGAAAAUCCACGU-3′ and the miR-NC sequence was
5′-UUCUCCGAACGUGUCACGUTT-3′. SATB1 overexpression vector
pcDNA3.1-SATB1 and control empty vector pcDNA3.1 were ordered from
GeneCopoeia Inc., (Rockville, MD, USA). For transfection, MG-63 and
U2OS cells were plated into 6-well plates with a density of
5×105/well and grown in culture medium without
antibiotics. MG-63 and U2OS cells were transfected with miR-376a
mimic (100 pmol), miR-NC (100 pmol), pcDNA3.1-SATB1 (4 µg) or
pcDNA3.1 (4 µg) by using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA),
following the manufacturer's protocol. After transfection 48 h,
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR) was performed to detect miR-376a expression. MTT and
invasion assays were carried out at 24 h and 48 h posttransfection.
Western blot analysis was performed to detect SATB1 protein
expression at 72 h following transfection.
RT-qPCR analysis
The total RNA was isolated from the cells or tissues
using the TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). miR complementary DNA (cDNA) was produced using
a TaqMan MicroRNA Assay kit (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The temperature protocol for reverse
transcription was as follows: 16°C for 30 min, 42°C for 30 min and
85°C for 5 min. Quantification of miR-376a was performed using a
TaqMan MicroRNA assay (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The cycling conditions were as follows: 50°C for
2 min, 95°C for 10 min; 40 cycles of denaturation at 95°C for 15
sec; and annealing/extension at 60°C for 60 sec. To quantify the
SATB1 mRNA, the total RNA was reverse transcribed into cDNA using a
PrimeScripts RT reagent kit and qPCR was conducted using SYBR-Green
Premix Ex Taq II (both from Takara Biotechnology Co., Ltd., Dalian,
China). The temperature protocol for reverse transcription was as
follows: 37°C for 15 min and 85°C for 5 sec. The amplification was
performed with cycling conditions as follows: 5 min at 95°C,
followed by 40 cycles of 95°C for 30 sec and 65°C for 45 sec. The
primers were designed as follows: miR-376a, 5′-GTGCAGGGTCCGAGGT-3′
(forward) and 5′-ATCATAGAGGAAAATCCACG-3′ (reverse); U6,
5′-CTCGCTTCGGCAGCACA-3′ (forward) and 5′-AACGCTTCACGAATTTGCGT-3′
(reverse); SATB1, 5′-AGAGGAAGGCTTGGGAGTA-3′ (forward) and
5′-GGGCAGCAGAGCTATGTG-3′ (reverse); and GAPDH,
5′-CGGAGTCAACGGATTTGGTCGTAT-3′ (forward) and
5′-AGCCTTCTCCATGGTGGTGAAGAC-3′ (reverse). U6 snRNA and GAPDH were
used as the internal control for miR-376a and SATB1 mRNA,
respectively. Data were analyzed by 2−ΔΔCq method
(20).
MTT assay
Following transfection for 24 h, the cells were
re-suspended and inoculated into 96-well plates at a density of
3×103 cells per well. The cells were incubated at 37°C
under a humidified atmosphere containing 5% CO2 for 0–72
h. An MTT assay was performed every 24 h to detect cell
proliferation. A total of 10 µl MTT solution (Sigma-Aldrich; Merck
KGaA) was added to each well and the cells were incubated for 4 h
at 37°C. After the supernatant was discarded, 100 µl dimethyl
sulfoxide (Sigma-Aldrich; Merck KGaA), which was used to resolve
the crystalline formazan, was added to each well. The absorbance at
a wavelength of 490 nm was monitored with a microplate reader
(Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Invasion assay
The transfected cells were digested at 48 h
post-transfection, washed with PBS and suspended in FBS-free DMEM
medium. A total of 1×105 cells in FBS-free DMEM were
seeded into the upper chamber of Transwell plates (Corning Inc.,
Corning, NY, USA), which were pre-coated with Matrigel (BD
Biosciences, Franklin Lakes, NJ, USA). Subsequently, 500 µl DMEM
supplemented with 10% FBS was added to the lower chamber of the
Transwell plates. Following 24 h of culture at 37°C, the cells
remaining on the top of the chamber were removed using cotton
swabs. The invasive cells were fixed with 4% paraformaldehyde at
room temperature for 20 min and stained with 0.05% crystal violet
at room temperature for 20 min. A total of five fields per chamber
were randomly selected and the number of invasive cells was counted
under an inverted light microscope (×200; Olympus Corporation,
Tokyo, Japan).
Target genes of miR-376a
TargetScan Human 7.0 (http://www.targetscan.org/) and miRBase (http://www.mirbase.org/) were employed to predict the
putative targets of miR-376a.
Luciferase reporter assay
A wild-type (Wt) fragment of the 3′-UTR of SATB1
containing the predicted miR-376a binding sequences and its mutated
(Mut) sequence was chemically produced by Shanghai GenePharma Co.,
Ltd., and cloned into pMIR-GLO™ Luciferase vector (Promega
Corporation, Madison, WI, USA). The Wt and Mut luciferase plasmids
were defined as pMIR-SATB1-3′-UTR Wt and pMIR-SATB1-3′-UTR Mut,
respectively. Cells were seeded into 24-well plates with a density
of 1.5×105/well and cotransfected with miR-376a mimic or
miR-NC, and pMIR-SATB1-3′-UTR Wt or pMIR-SATB1-3′-UTR Mut, using
Lipofectamine 2000 according to the manufacturer's protocol.
Following transfection for 2 days, the dual-Luciferase Reporter
Assay System (Promega Corporation) was applied to measure the
luciferase activities, according to the manufacturer's protocol.
Firefly luciferase activity was normalized to Renilla
luciferase activity.
Western blot analysis
Tissue samples or cells were solubilized in cold
radioimmunoprecipitation assay lysis buffer (Beyotime Institute of
Biotechnology, Shanghai, China). The total protein concentration
was determined using a Bicinchoninic Acid Protein Assay kit (Pierce
Biotechnology, Inc., Rockford, IL, USA). Equal amounts of proteins
(20 µg) were separated using 10% SDS-PAGE, electrotransferred to
polyvinylidene fluoride membranes (EMD Millipore, Billerica, MA,
USA) and blocked at room temperature for 2 h in Tris-buffered
saline with 0.5% Tween (TBST) buffer containing 5% dried skimmed
milk. Following being washed three times with TBST, the membranes
were incubated with rabbit anti-human monoclonal SATB1 antibody
(cat. no. ab109122; 1:1,000; Abcam, Cambridge, MA, USA) or rabbit
anti-human monoclonal GAPDH antibody (cat. no. ab181603; 1:1,000;
Abcam) at 4°C overnight. Afterwards, the membranes were washed
three times with TBST and further incubated with goat anti-rabbit
horseradish peroxidase-conjugated secondary antibody (cat. no.
ab6721; 1:5,000; Abcam) at room temperature for 1 h. Finally, an
ECL Protein Detection kit (Pierce Biotechnology, Inc., Rockford,
IL, USA) was used to visualize the protein bands. GADPH was used as
the loading control. Protein expression was quantified using
Quantity One software version 4.62 (Bio-Rad Laboratories,
Inc.).
Statistical analysis
Data were presented as the mean ± standard deviation
of at least 3 independent experiments and analyzed with SPSS
software, version 21.0 (IBM SPSS, Armonk, NY, USA). The association
between miR-376a expression and clinicopathological characteristics
of OS was evaluated by a χ2 test. The two-tailed
Student's t-test and one-way analysis of variance combined with
Student-Newman-Keuls were used to compare differences between
groups. Spearman's correlation analysis was carried out to reveal
the correlation between expression levels of miR-376a and SATB1
mRNA in OS tissues. P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-376a expression is decreased in OS
tissues and cell lines
To understand the expression pattern of miR-376a in
OS, miR-376a expression was initially measured in 49 pairs of OS
tissues and adjacent normal bone tissues. RT-qPCR analysis revealed
that miR-376a was significantly downregulated in OS tissues
compared with the adjacent normal bone tissues (P<0.05; Fig. 1A). Subsequently, the clinical
significance of miR-376a expression in OS was investigated. The
median value of miR-376a expression was defined as the cutoff point
and this cutoff point was used to divide all OS patients into a low
miR-376a expression group (n=25) or a high miR-376a expression
group (n=24). A low miR-376a level was significantly correlated
with tumor size (P=0.022) and lymph node infiltration (P=0.001) in
OS but was not associated with age, gender, and location of the
primary tumor (all P>0.05; Table
I). In addition, the expression of miR-376a in four human OS
cell lines, namely, HOS, SAOS-2, MG-63 and U2OS, as well as a
normal human osteoblast hFOB1.19 cell line, was determined. As
indicated in Fig. 1B, the
expression level of miR-376a in all OS cell lines was significantly
decreased compared with in hFOB1.19 (P<0.05). These results
suggested that reduced expression of miR-376a may be associated
with OS progression.
miR-376a upregulation prohibits the
proliferation and invasion of OS cells
To determine whether miR-376a can affect the
malignant phenotypes of OS cells, MG-63 and U2OS cells, which
exhibited relatively lower miR-376a expression among the four OS
cell lines, were transfected with an miR-376a mimic or miR-NC.
Transfection with the miR-376a mimic significantly increased the
miR-376a levels in MG-63 and U2OS cells when compared with the
cells transfected with miR-NC (P<0.05; Fig. 2A). MTT and invasion assays were
used to examine the effects of miR-376a overexpression on OS cell
proliferation and invasion, respectively. The results revealed that
ectopic expression of miR-376a significantly inhibited the
proliferation (P<0.05; Fig. 2B)
and invasion (P<0.05; Fig. 2C)
of MG-63 and U2OS cells compared with the miR-NC groups. These
results suggested that miR-376a may serve a tumor suppressive role
in OS progression.
miR-376a directly targets SATB1 and
inhibits its expression in OS cells
To illustrate the molecular mechanism by which
miR-376a inhibits OS cell proliferation and invasion,
bioinformatics analysis was conducted to predict the potential
targets of miR-376a. SATB1 was predicted as a primary target of
miR-376a (Fig. 3A) and was chosen
for additional confirmation analysis due to its regulatory roles in
OS pathogenesis and development (21,22).
To confirm this prediction, luciferase reporter assays were
conducted in MG-63 and U2OS cells, which were transfected with
pMIR-SATB1-3′-UTR Wt or pMIR-SATB1-3′-UTR Mut, together with the
miR-376a mimic or miR-NC. Upregulation of miR-376a reduced the
luciferase activities of the plasmid carrying the Wt 3′-UTR of
SATB1 in MG-63 and U2OS cells (P<0.05) but had no effect on that
of the Mut 3′-UTR of SATB1 (Fig.
3B). Furthermore, the effects of miR-376a overexpression on
endogenous SATB1 expression were determined using RT-qPCR and
western blot analysis, respectively. As expected, SATB1 mRNA
(P<0.05; Fig. 3C) and protein
(P<0.05; Fig. 3D) levels were
significantly reduced in MG-63 and U2OS cells transfected with
miR-376a mimic. Therefore, SATB1 is a direct target gene of
miR-376a in OS.
SATB1 expression is upregulated in OS
and inversely correlated with miR-376a expression
SATB1 was validated as a direct target gene of
miR-376a in OS. Therefore, the SATB1 expression was detected in 49
pairs of OS tissues and adjacent normal bone tissues using RT-qPCR.
The expression level of SATB1 was significantly upregulated in OS
tissues compared with the adjacent normal bone tissues (P<0.05;
Fig. 4A). In addition, the SATB1
protein expression was also determined in several pairs of OS
tissues and adjacent normal bone tissues chosen at random using
western blot analysis. As demonstrated in Fig. 4B, the SATB1 protein expression in
OS tissues was significantly increased compared with the adjacent
normal bone tissues in the 4 cases tested (P<0.05; Fig. 4B and C). Furthermore, miR-376a
expression was inversely correlated with SATB1 mRNA levels in OS
tissues (P<0.0001; r=−0.5288; Fig.
4D). These results further suggested that SATB1 is a direct
target gene of miR-376a in OS.
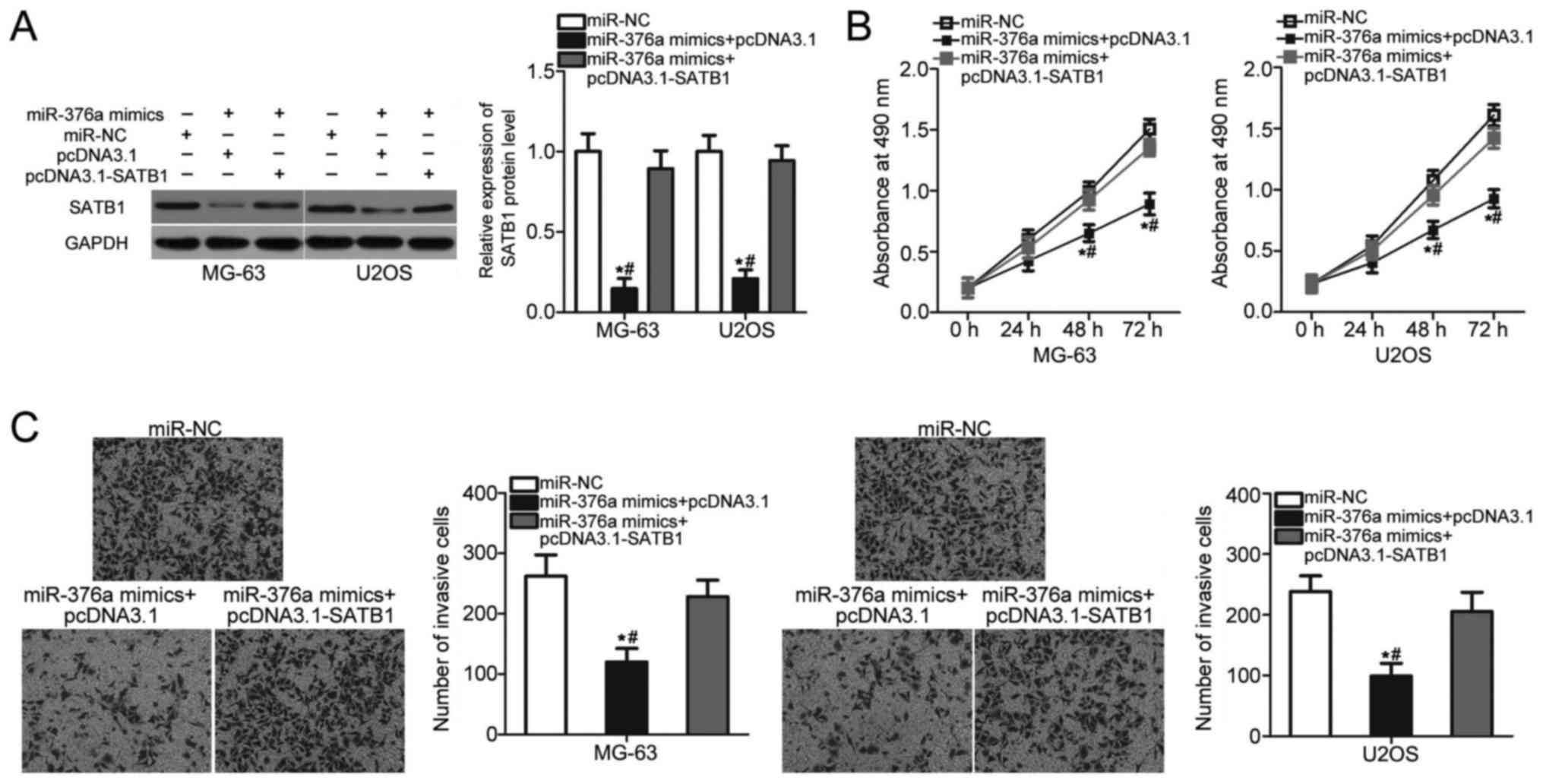
SATB1 overexpression attenuates the
suppressive effects of miR-376a on OS cell proliferation and
invasion
Rescue experiments were performed to assess whether
SATB1 mediated the inhibitory roles of miR-376a in OS.
miR-376a-overexpressing MG-63 and U2OS cells were co-transfected
with a SATB1 overexpression vector pcDNA3.1-SATB1 or control empty
vector pcDNA3.1. The decreased expression level of SATB1 induced by
miR-376a overexpression was recovered in MG-63 and U2OS cells
co-transfected with pcDNDA3.1-SATB1 (P<0.05; Fig. 5A). MTT and invasion assays revealed
that restored SATB1 expression eliminated the inhibitory effects of
miR-376a overexpression on the proliferation (P<0.05; Fig. 5B) and invasion (P<0.05; Fig. 5C) of MG-63 and U2OS cells. These
results suggested that miR-376a serves a tumor suppressive role in
OS partly by inhibiting SATB1 expression.
Discussion
Aberrantly expressed miRs are implicated in the
regulation of OS onset and development (23–25).
Therefore, key miRs in OS must be identified to develop promising
and effective therapeutic targets for patients with OS. In the
present study, the expression level and clinical significance of
miR-376a in OS was measured and the roles, and associated
regulatory mechanism of miR-376a were determined in OS. The results
demonstrated that miR-376a expression was downregulated in OS
tissues and cell lines. Decreased expression of miR-376a was
associated with tumor size and lymph node infiltration of OS
patients. In addition, miR-376a overexpression restricted cell
proliferation and invasion in OS. Furthermore, SATB1 was confirmed
as a direct target gene of miR-376a in OS. Additionally, SATB1
expression was significantly overexpressed in OS tissues and this
overexpression was inversely correlated with miR-376a levels in OS
tissues. Reintroduction of SATB1 expression rescued OS cells from
the tumor suppressive roles of miR-376a. These results revealed
that miR-376a may serve as a tumor suppressor in OS by directly
targeting SATB1.
Significant alterations in miR-376a expression have
been observed in multiple human cancer types. For instance,
miR-376a is under-expressed in glioma tissues, cell lines and serum
(16,26). Low miR-376a expression is
correlated with the World Health Organization grade and KPS score
of glioma patients. In addition, miR-376a is validated as an
independent factor that predicts the poor prognosis of glioma
patients (26). In hepatocellular
carcinoma, miR-376a is downregulated in tumor tissues and this
downregulation is associated with α-fetoprotein levels (17). In colorectal cancer, the expression
level of miR-376a in tumor tissues is decreased compared with the
adjacent normal mucosa. Low miR-376a expression is associated with
lymph node metastasis of colorectal cancer patients. Colorectal
cancer patients with low miR-376a expression present lower survival
rates compared with patients with high miR-376a levels (27). Low expression of miR-376a has also
been observed in prostate cancer (18) and non-small-cell lung cancer
(19). Conversely, miR-376a is
overexpressed in ovarian cancer. High miR-376a expression is
significantly associated with the clinical stage and the
International Federation of Gynaecological Oncologists stage of
ovarian cancer (28,29). These conflicting results indicate
that the expression patterns of miR-376a exhibit tissue specificity
and suggest that miR-376a may be developed as a meaningful
prognostic biomarker for predicting the prognosis of patients with
these human cancer types.
Differently expressed miRs have been associated with
the carcinogenesis and cancer progression of several human
malignancies. For instance, miR-376a suppresses the proliferation
and invasion of glioblastoma multiform cells (16). Zheng et al (17) reported that miR-376a overexpression
inhibits cell proliferation and promotes apoptosis in
hepatocellular carcinoma. Formosa et al (18) demonstrated that the resumption of
miR-376a expression attenuates the growth of prostate cancer cells
while inducing apoptosis. Wang et al (19) indicated that miR-376a re-expression
restricts cell proliferation and invasion while promoting apoptosis
in non-small-cell lung cancer. Conversely, miR-376a serves
oncogenic roles in ovarian cancer by promoting cell proliferation
and motility, inhibiting cell apoptosis in vitro and
increasing tumor growth in vivo (29). These studies suggested that
miR-376a may be a potential therapeutic target for patients with
these human malignancies.
Previous studies have identified several direct
targets of miR-376a, including SP1 (16) in glioblastoma multiform, p85α
(17) and H3K18 (30) in hepatocellular carcinoma,
frizzled-4 in prostate cancer (18), c-Myc (19) in non-small-cell lung cancer,
Krueppel-like factor 15 (29) and
Caspase-8 (29) in ovarian cancer.
SATB1, which is a cell type-specific nuclear matrix attachment
region binding protein, was a direct target gene of miR-376a in OS.
SATB1 is reportedly upregulated in several types of cancer,
including hepatocellular carcinoma (31), ovarian cancer (32), gastric cancer (33), prostate cancer (34) and colorectal cancer (35). The expression of SATB1 was also
upregulated in OS tissues and cell lines. The overexpression of
SATB1 contributes to OS progression by promoting cell
proliferation, migration and invasion, inhibiting apoptosis and
reducing the chemosensitivity of cells to arsenic trioxide
(21,22). These results suggest that knockdown
of SATB1 may be a valuable therapeutic strategy for the treatment
of patients with OS.
In conclusion, miR-376a expression was downregulated
in OS tissues and cell lines, and the low miR-376a expression in OS
was significantly associated with the tumor size and lymph node
infiltration. In addition, forced expression of miR-376a repressed
OS cell proliferation and invasion by directly targeting SATB1.
Therefore, the results of the present study suggest that miR-376a
may be a promising therapeutic target for treating patients with
OS.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
HJ designed the present study. GZ and LM performed
the functional experiments. LM analyzed the data of this study. All
authors have read and approved the final draft.
Ethics approval and consent to
participate
The present study was approved by the Research
Ethics Committee of China-Japan Union Hospital of Jilin University
(Jilin, China) and was performed in accordance with the Declaration
of Helsinki and the guidelines of the Ethics Committee of
China-Japan Union Hospital of Jilin University. Written informed
consent was obtained from all patients for the use of their
clinical tissues.
Patient consent for publication
Written informed consent was obtained from all
patients for the use of their clinical tissues.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Maximov VV and Aqeilan RI: Genetic factors
conferring metastasis in osteosarcoma. Future Oncol. 12:1623–1644.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mirabello L, Troisi RJ and Savage SA:
Osteosarcoma incidence and survival rates from 1973 to 2004: Data
from the surveillance, epidemiology, and end results program.
Cancer. 115:1531–1543. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ritter J and Bielack SS: Osteosarcoma. Ann
Oncol. 21 Suppl 7:vii320–vii325. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liebner DA: The indications and efficacy
of conventional chemotherapy in primary and recurrent sarcoma. J
Surg Oncol. 111:622–631. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fagioli F, Aglietta M, Tienghi A, Ferrari
S, del Prever Brach A, Vassallo E, Palmero A, Biasin E, Bacci G,
Picci P and Madon E: High-dose chemotherapy in the treatment of
relapsed osteosarcoma: An Italian sarcoma group study. J Clin
Oncol. 20:2150–2156. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bacci G, Balladelli A, Palmerini E,
Alberghini M, Pollastri P, Galletti S, Mercuri M and Picci P:
Neoadjuvant chemotherapy for osteosarcoma of the extremities in
preadolescent patients: The Rizzoli Institute experience. J Pediatr
Hematol Oncol. 30:908–912. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dong J, Liu Y, Liao W, Liu R, Shi P and
Wang L: miRNA-223 is a potential diagnostic and prognostic marker
for osteosarcoma. J Bone Oncol. 5:74–79. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Berindan-Neagoe I and Calin GA: Molecular
pathways: microRNAs, cancer cells, and microenvironment. Clin
Cancer Res. 20:6247–6253. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sassen S, Miska EA and Caldas C: MicroRNA:
Implications for cancer. Virchows Arch. 452:1–10. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cai W, Jiang H, Yu Y, Xu Y, Zuo W, Wang S
and Su Z: miR-367 regulation of DOC-2/DAB2 interactive protein
promotes proliferation, migration and invasion of osteosarcoma
cells. Biomed Pharmacother. 95:120–128. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fu F, Wan X, Wang D, Kong Z, Zhang Y,
Huang W, Wang C, Wu H and Li Y: MicroRNA-19a acts as a prognostic
marker and promotes prostate cancer progression via inhibiting
VPS37A expression. Oncotarget. 9:1931–1943. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang Y, Jiang Z, Ma N, Wang B, Liu J,
Zhang L and Gu L: MicroRNA-223 Targeting STIM1 inhibits the
biological behavior of breast cancer. Cell Physiol Biochem.
45:856–866. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Du B, Wu D, Yang X, Wang T, Shi X, Lv Y,
Zhou Z, Liu Q and Zhang W: The expression and significance of
microRNA in different stages of colorectal cancer. Medicine
(Baltimore). 97:e96352018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Macfarlane LA and Murphy PR: MicroRNA:
Biogenesis, function and role in cancer. Curr Genomics. 11:537–561.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li Y, Wu Y, Sun Z, Wang R and Ma D:
MicroRNA376a inhibits cell proliferation and invasion in
glioblastoma multiforme by directly targeting specificity protein
1. Mol Med Rep. 17:1583–1590. 2018.PubMed/NCBI
|
|
17
|
Zheng Y, Yin L, Chen H, Yang S, Pan C, Lu
S, Miao M and Jiao B: miR-376a suppresses proliferation and induces
apoptosis in hepatocellular carcinoma. FEBS Lett. 586:2396–2403.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Formosa A, Markert EK, Lena AM, Italiano
D, Finazzi-Agro' E, Levine AJ, Bernardini S, Garabadgiu AV, Melino
G and Candi E: MicroRNAs, miR-154, miR-299-5p, miR-376a, miR-376c,
miR-377, miR-381, miR-487b, miR-485-3p, miR-495 and miR-654-3p,
mapped to the 14q32.31 locus, regulate proliferation, apoptosis,
migration and invasion in metastatic prostate cancer cells.
Oncogene. 33:5173–5182. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang Y, Cong W, Wu G, Ju X, Li Z, Duan X,
Wang X and Gao H: MiR-376a suppresses the proliferation and
invasion of non-small-cell lung cancer by targeting c-Myc. Cell
Biol Int. 42:25–33. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang H, Qu S, Li S, Wang Y, Li Y, Wang Y,
Wang Z and Li R: Silencing SATB1 inhibits proliferation of human
osteosarcoma U2OS cells. Mol Cell Biochem. 378:39–45. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang H, Su X, Guo L, Zhong L, Li W, Yue
Z, Wang X, Mu Y, Li X, Li R and Wang Z: Silencing SATB1 inhibits
the malignant phenotype and increases sensitivity of human
osteosarcoma U2OS cells to arsenic trioxide. Int J Med Sci.
11:1262–1269. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li X, Sun X, Wu J and Li Z: MicroRNA-613
suppresses proliferation, migration and invasion of osteosarcoma by
targeting c-MET. Am J Cancer Res. 6:2869–2879. 2016.PubMed/NCBI
|
|
24
|
Qu Q, Chu X and Wang P: MicroRNA-195-5p
suppresses osteosarcoma cell proliferation and invasion by
suppressing naked cuticle homolog 1. Cell Biol Int. 41:287–295.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ren X, Shen Y, Zheng S, Liu J and Jiang X:
miR-21 predicts poor prognosis in patients with osteosarcoma. Br J
Biomed Sci. 73:158–162. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Huang Q, Wang C, Hou Z, Wang G, Lv J, Wang
H, Yang J, Zhang Z and Zhang H: Serum microRNA-376 family as
diagnostic and prognostic markers in human gliomas. Cancer Biomark.
19:137–144. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mo ZH, Wu XD, Li S, Fei BY and Zhang B:
Expression and clinical significance of microRNA-376a in colorectal
cancer. Asian Pac J Cancer Prev. 15:9523–9527. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Meng X, Joosse SA, Muller V, Trillsch F,
Milde-Langosch K, Mahner S, Geffken M, Pantel K and Schwarzenbach
H: Diagnostic and prognostic potential of serum miR-7, miR-16,
miR-25, miR-93, miR-182, miR-376a and miR-429 in ovarian cancer
patients. Br J Cancer. 113:1358–1366. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yang L, Wei QM, Zhang XW, Sheng Q and Yan
XT: MiR-376a promotion of proliferation and metastases in ovarian
cancer: Potential role as a biomarker. Life Sci. 173:62–67. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zheng Y, Chen H, Yin M, Ye X, Chen G, Zhou
X, Yin L, Zhang C and Ding B: MiR-376a and histone deacetylation 9
form a regulatory circuitry in hepatocellular carcinoma. Cell
Physiol Biochem. 35:729–739. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tu W, Luo M, Wang Z, Yan W, Xia Y, Deng H,
He J, Han P and Tian D: Upregulation of SATB1 promotes tumor growth
and metastasis in liver cancer. Liver Int. 32:1064–1078. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xiang J, Zhou L, Li S, Xi X, Zhang J, Wang
Y, Yang Y, Liu X and Wan X: AT-rich sequence binding protein 1:
Contribution to tumor progression and metastasis of human ovarian
carcinoma. Oncol Lett. 3:865–870. 2012.PubMed/NCBI
|
|
33
|
Lu X, Cheng C, Zhu S, Yang Y, Zheng L,
Wang G, Shu X, Wu K, Liu K and Tong Q: SATB1 is an independent
prognostic marker for gastric cancer in a Chinese population. Oncol
Rep. 24:981–987. 2010.PubMed/NCBI
|
|
34
|
Mao L, Yang C, Wang J, Li W, Wen R, Chen J
and Zheng J: SATB1 is overexpressed in metastatic prostate cancer
and promotes prostate cancer cell growth and invasion. J Transl
Med. 11:1112013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Baba H, Ishikawa T, Mogushi K, Ishiguro M,
Uetake H, Tanaka H and Sugihara K: Identification of SATB1 as a
specific biomarker for lymph node metastasis in colorectal cancer.
Anticancer Res. 36:4069–4076. 2016.PubMed/NCBI
|