Introduction
Obsessive-compulsive disorder (OCD) is a mental
health disorder characterized by obsessive and compulsive thoughts
and behaviors. OCD typically arises in late adolescence or early
adulthood and can lead to chronic illness if it is left untreated
(1,2). Although there is no consensus
regarding its etiology, genetic studies have indicated that
heritability is associated with 26–61% of cases (3–5).
Eating disorders (EDs) are a group of mental health
syndromes characterized by significant disturbances in eating
behavior, and by distress or excessive concern with body shape or
weight. The most studied sub-types of EDs are anorexia nervosa (AN)
and bulimia nervosa (BN). The lifetime prevalence rates for EDs are
higher among women than men (6).
Although the cause of EDs remains unclear (7), it is hypothesized that genetic
factors have a significant role in the development of the disease
(8,9).
A number of clinical symptoms of EDs have also been
observed in OCD (10). In
addition, shared genetic liability and brain circuitries have been
identified among the obsessive psychiatric syndromes of AN, BN and
OCD (11). Additionally, genetic
analysis using genome-wide association studies and gene expression
data have revealed a number of genetic risks associated with ED and
OCD (12,13). However, thus far, no systematical
study has been performed to investigate the genetic risks shared by
the two diseases.
The present study integrated gene expression data
and a large-scale literature database to investigate the
association between OCD and ED at the genetic level, with the aim
of gaining an improved understanding of their common genetic basis
and identify novel potential genes associated with the two
diseases. In recent years, Pathway Studio (PS; pathwaystudio.com) has been widely used to study
modeled associations between proteins, genes, complexes, cells,
tissues and diseases (14).
Updated weekly, the PS relation database is the largest database
among known competitors in the field (15).
The present study intended to examine the hypothesis
that OCD and ED present significantly shared pathogenesis at the
genetic level. If the hypothesis was verified, then the aim was to
determine if risk genetic factors associated one disease were
worthy of study into its potential association with the other
disease.
Materials and methods
Data collection summary
Initially, large-scale ED-gene and OCD-gene relation
data were analyzed to identify shared genes and genetic pathways.
Then, expression data acquired from patients with OCD and ED, and
healthy controls, were used to identify potential novel common risk
genes for ED and OCD. Subsequently, gene-disease-drug-relation
network analysis was conducted to study the potential pathogenic
significance of the novel common genes to ED and OCD. The network
analysis was conducted using the ‘Shortest Path’ function module of
the PS database. The purpose of the network analysis was to
identify possible functional association and pathological pathways
between the novel common risk genes and OCE/ED.
OCD and ED-gene data acquisition
Disease-gene relation data for ED (all types) and
OCD were acquired from PS, as described previously (14). A genetic database, termed OCD_ED,
was developed through a complex analysis of the identified
association data. Besides the full lists of genes linked to the two
diseases (OCD_ED, OCD Related Genes; and OCD_ED, ED Related Genes),
the database also presented the supporting references for each
disease-gene relation (OCD_ED, Ref for OCD Related Genes and OCD_ED
Ref for ED Related Genes), including titles of the references and
the related parts of the study where a disease-gene association was
identified. The OCD_ED database is online available at
‘Bioinformatics Database’ (gousinfo.com/database/Data_Genetic/OCD_ED.xlsx).
The information can be used to locate the detailed description of
how a candidate gene is associated with OCD and/or ED.
Common risk genes
A gene expression dataset (GSE60190) of 133 subjects
(15 patients with ED, 16 patients with OCD and 102 non-psychiatric
controls) was used to test the genes associated with one of the two
diseases but not the other. The expression data is available online
at www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE60190.
For a gene associated with only OCD, one-way analysis of variance
was performed to compare the expression between healthy controls
and ED cases, in order to determine association with ED. Similarly,
the genes associated with only ED were tested for their potential
association with OCD. The Benjamini-Hochberg procedure was employed
to control the false discovery rate (FDR), and FDR-corrected
P-values were used to identify potential significant genes for
further analysis. All analyses were conducted using Matlab (R
2017a; The MathWorks, Inc., Natick, MA, USA).
Pathway analysis of potential risk
genes
For the target common risk genes identified through
expression analysis as described above, a shortest-path based
network analysis was performed to identify pathogenic pathways
between the target genes and the disease (ED/OCD). The analysis was
performed using PS.
Results
Shared genetic basis between OCD and
ED
Within the curated OCD_ED database, there were 81
genes associated with OCD, supported by 450 scientific references
(OCD_ED→OCD Related Genes and OCD_ED→Ref for OCD Related Genes).
For ED, there were 71 associated genes supported by 204 references
(OCD_ED, ED Related Genes and OCD_ED, Ref for ED Related Genes). A
significant overlap (P=6.80×10−34; right-tail Fisher's
Exact test) of 21 genes were identified between the two groups of
genes, as shown in Fig. 1. More
information concerning these 21 genes is in OCD_ED, 21 cross
genes.
To test the functional profile of the 21 common
genes associated with OCD and ED, a Pathway Enrichment Analysis
(PEA) was conducted using PS. The 10 most significantly enriched
pathways (P<4.30×10−7; q=0.001 for FDR) are presented
in Table I. In total, 60
pathways/gene sets were enriched with P<1.00×10−3
including all 21 genes (OCD_ED, Common Pathways).
 | Table I.Genetic pathways enriched with 21
genes associated with obsessive-compulsive disorder and eating
disorder. |
Table I.
Genetic pathways enriched with 21
genes associated with obsessive-compulsive disorder and eating
disorder.
| Name | GO ID | Number of genes | Overlap | P-value (following
FDR) | P-value (prior to
FDR) |
|---|
| Response to drug | 0017035 | 509 | 12 |
2.57×10−10 |
8.78×10−15 |
| Grooming
behavior | 0007625 | 16 | 5 |
2.66×10−8 |
2.60×10−12 |
| Response to
cocaine | 0042220 | 44 | 6 |
2.66×10−8 |
3.15×10−12 |
| Feeding
behavior | 0007631 | 45 | 6 |
2.66×10−8 |
3.64×10−12 |
| Synaptic
transmission | 0007268 | 472 | 10 |
4.20×10−8 |
8.51×10−12 |
| Locomotory
behavior | 0007626 | 108 | 7 |
4.20×10−8 |
8.60×10−12 |
| TC 2.A.22.1 | None | 3 | 3 |
1.46×10−7 |
3.98×10−11 |
| Positive regulation
of ERK1 and ERK2 cascade | 0070374 | 134 | 7 |
1.46×10−7 |
3.98×10−11 |
| Positive regulation
of peptidyl-serine phosphorylation | 0033138 | 77 | 6 |
3.38×10−7 |
1.04×10−10 |
| Response to
ethanol | 0017036 | 161 | 7 |
4.27×10−7 |
1.46×10−10 |
The PEA approach revealed 7 pathways (13 overlapped
genes) associated with behavior, 5 (14 overlapped genes) with neuro
system, 4 (17 overlapped genes) with drug effects, 3 (5 overlapped
genes) with neuro transmitter, 2 (8 overlapped genes) with brain
function development, 1 (9 overlapped genes) with cell
proliferation, and 1 (6 overlapped genes) with protein
phosphorylation. For detailed information of these significantly
enriched pathways, please refer to OCD_ED, Common Pathways. The
results of the present study suggested that OCD and ED share
multiple genetic pathways, through which these 21 genes serve
various functions affecting the pathogenic development of the two
diseases.
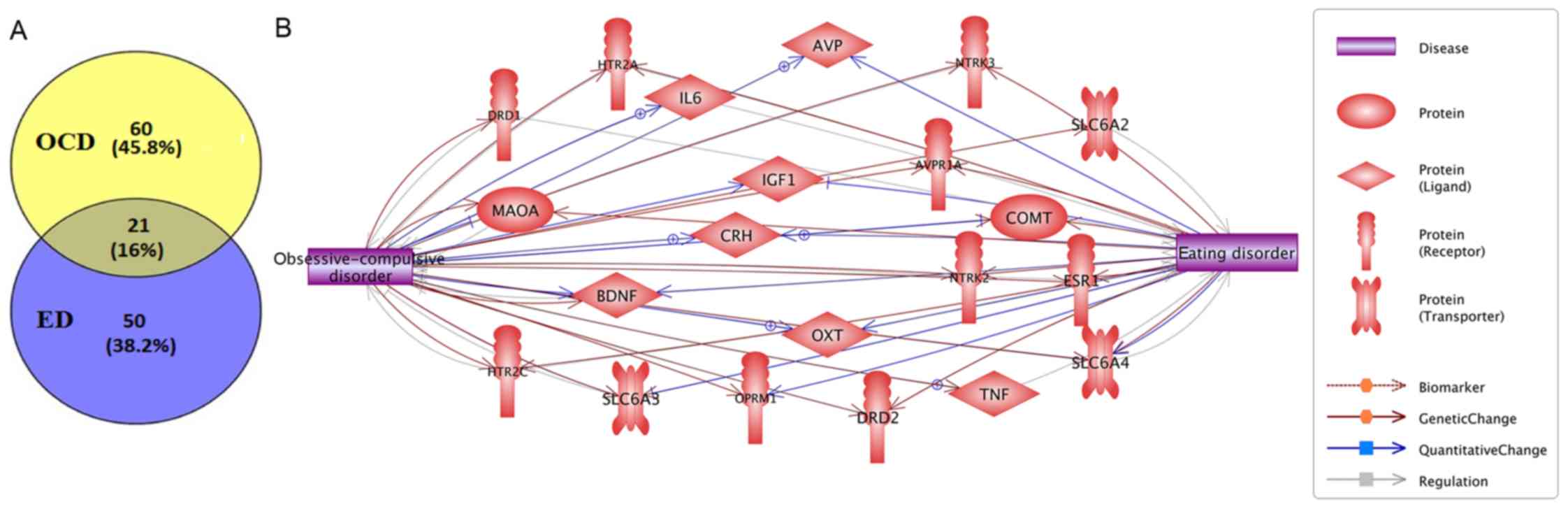
Potential co-regulations between OCD
and ED
Functional network analysis using PS demonstrated
that 17 out of the 21 common risk genes exhibit down- and
upregulation associated with OCD and ED (influenced by and
influencing OCD and ED), as presented in Fig. 2. Detailed information concerning
the network presented in Fig. 2 is
in OCD_ED, Co-Regulation Network; including the type of the
association, supporting references and related excerpts from the
references where an association has been identified. Fig. 2 demonstrates that OCD and ED may
influence the pathogenic development of each other through these
genetic pathways.
Gene expression analysis
Although there was a significant overlap between
ED-genes and OCD-genes (21 genes; P=6.80×10−34), certain
genes were linked to one disease only (60 for OCD and 50 for ED;
Fig. 1). These results were from
literature data analysis. The present study analyzed the
correlation between the 60 OCD-genes and ED, and the correlation
between 50 ED-genes and OCD, using a gene expression dataset
(GSE60190; 15 patients with ED, 16 patients with OCD and 102
non-psychiatric controls). Fig. 3
presents the ‘-log10’ transferred P-values (q=0.001 for FDR) for
each gene tested. The detailed results are presented in OCD_ED, 50
ED Genes for OCD and OCD_ED, 60 OCD Genes for ED, including the
P-values and FDR correction status. Note that 3 out of 50 ED-genes
and 2 out of 60 OCD-genes are not included in the expression
dataset utilized in the present study and thus, the corresponding
results were not available.
Of the 50 ED-genes, 3 [oxytocin receptor
(OXTR), glutamate decarboxylase 2 (GAD2) and
neuropeptide Y (NPY)] and out of the 60 OCD-genes one
[glutamate ionotropic receptor kainate type subunit 3
(GRIK3)] passed the FDR correction (q=0.001). According to
the PS database, OXTR, GAD2 and NPY has no direct
association with OCD, and GRIK3 has no direct association
with ED (no study has been identified that reports an association
between these genes and OCE or ED).
However, OXTR, GAD2 and NPY
demonstrated strong functional linkage to OCD, through multiple
genes and small molecules/drugs pathways (Fig. 4A). GRIK3 also demonstrated
an association with ED through the regulation of ED-related
disorders (Fig. 4B). The detailed
information of the associations in Fig. 4 is presented in OCD_EDà ‘3 Genes
for OCD’ and ‘1 Gene for ED’, including the type of association,
the underlying supporting reference, and the places in the study
where these associations have been identified and described.
Discussion
Previous studies have demonstrated that ED is
closely associated with OCD (16).
The present study integrated large-scale disease-gene relation data
and gene expression data to test the hypothesis that ED and OCD
exhibit significant shared genetic bases in terms of common risk
genes and pathways. Gene expression data analysis identified novel
potential common risk genes for ED and OCD. Results from functional
network analysis supported the association between these genes and
the two diseases.
The results of the present study demonstrated that
21 genes linked to OCD and ED present significant overlap
(P=6.76×10−34). These 21 genes are significantly
enriched within 60 pathways (P<1.00×10−3;
FDR-corrected, q=0.001; OCD_ED, Common Pathways). A number of these
pathways have been implicated in OCD and ED, including the pathway
associated with neuro system and neuro transmitter (17–21),
memory [Gene Ontology (GO) ID, 0007613] (22,23),
learning (GO ID, 0007612) (24,25)
and response to cocaine (GO ID, 0048148) (26,27).
These results suggested that OCD and ED share multiple genetic
pathways. Through these pathways, a large group of genes influence
the pathogenic development of the two diseases.
In addition, a 17-gene network was constructed,
through which OCD and ED may affect the pathogenic status of each
other. These 17 out of the 21 cross genes are downstream targets of
OCD/ED and also the upstream regulators of ED/OCD. The findings of
the present study supported the hypothesis that OCD and ED present
significant association at the genetic level.
Closer analysis of the 50 ED only genes using gene
expression data (GSE60190) demonstrated that 3 out of these 50 ED
genes, OXTR, GAD2 and NPY, were potential OCD markers
(FDR-corrected P<1.00×10−3). Functional network
analysis demonstrated that these 3 three genes presented strong
functional correlation with OCD, forming a genetic network
reinforced by 1,406 supporting references (Fig. 4A; see OCD_ED, →3 Genes for OCD).
These results supported multiple pathways between these three genes
and OCD. For example, NPY can significantly increase
arginine vasopressin (AVP) mRNA expression (28), while and AVP contributes to
OCD symptoms in humans (29). This
finding supports a NPY→-AVP→-OCD functional pathway.
More of these pathways may be identified from the literature
knowledge curated in the database OCD_ED→, 3 Genes for OCD.
On the other hand, gene expression data analysis and
shorted-path based network analysis suggested that GRIK3 may
be a risk gene for ED (FDR-corrected P<1.00×10−3;
OCD_ED, 60 OCD Genes for ED). GRIK3 presents a strong
expression pattern in the central nervous system, demonstrating
solid function in presynaptic neurotransmission (30). Haploinsufficiency of GRIK3
causes severe developmental delay (30), which is associated with ED
(31). GRIK3 also serves
important roles in cognitive defects (32), which are associated with ED
pathogenesis (33). Therefore, by
regulating the brain functions, GRIK3 may exert an influence
on the pathogenic development of ED.
Although the four genes (OXTR, GAD2, NPY and
GRIK3) identified in this study are proposed as novel common
risk genes for ED and OCD, and the findings were supported by
data-driven indirect evidence from gene expression data and pathway
analysis. Biological experiments are required to test these
potential associations.
In summary, results from the present study supported
the hypothesis that OCD and ED exhibit significant association at
the genetic level, which helps to explain their common pathological
symptoms. Additionally, four genes were suggested as novel
potential common risk genes for OCD and ED. To the best of our
knowledge, this is the first study integrating large-scale
disease-gene association data and gene expression data for a
systematical study of the associations between OCD and ED at the
genetic level. The findings of the present study may add novel
insights into the current field of OCD-ED correlation study, and
guarantee further studies using more data sets to test novel
potential risk genes for ED and OCD.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets analyzed during the present study are
available in the OCD_ED database online at ‘Bioinformatics
Database’ (gousinfo.com/database/Data_Genetic/OCD_ED.xlsx).
The expression dataset analysed during this study is available
online at ‘Gene Expression Omnibus’ (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE60190).
Authors' contributions
CX, HC and DL contributed to the design of the
present study, acquired and analyzed the data, and wrote and
revised the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Abramowitz JS, Taylor S and McKay D:
Obsessive-compulsive disorder. Lancet. 374:491–499. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Matsunaga H and Seedat S:
Obsessive-compulsive spectrum disorders: Cross-national and ethnic
issues. CNS Spectr. 12:392–400. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hanna GL, Fingerlin TE, Himle JA and
Boehnke M: Complex segregation analysis of obsessive-compulsive
disorder in families with pediatric probands. Hum Hered. 60:1–9.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fyer AJ, Lipsitz JD, Mannuzza S, Aronowitz
B and Chapman TF: A direct interview family study of
obsessive-compulsive disorder. Psychol Med. 35:1611–1621. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bloch MH and Pittenger C: The genetics of
obsessive-compulsive disorder. Curr Psychiatry Rev. 6:91–103. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hudson JI, Hiripi E, Pope HG Jr and
Kessler RC: The prevalence and correlates of eating disorders in
the national Comorbidity survey replication. Biol Psychiatry.
61:348–358. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rikani AA, Choudhry Z, Choudhry AM, Ikram
H, Asghar MW, Kajal D, Waheed A and Mobassarah NJ: A critique of
the literature on etiology of eating disorders. Ann Neurosci.
20:157–161. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Klump KL, Kaye WH and Strober M: The
evolving genetic foundations of eating disorders. Psychiatr Clin
North Am. 24:215–225. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bulik CM, Sullivan PF and Kendler KS:
Heritability of binge-eating and broadly defined bulimia nervosa.
Biol Psychiatry. 44:1210–1218. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
American Psychiatric Association.
Diagnostic and statistical manual for mental disorders. American
Psychiatric Press. (Washington DC, USA). 2000.
|
|
11
|
Kaye WH, Bulik CM, Thornton L, Barbarich N
and Masters K: Comorbidity of anxiety disorders with anorexia and
bulimia nervosa. Am J Psychiatry. 161:2215–2221. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mathews CA, Badner JA, Andresen JM,
Sheppard B, Himle JA, Grant JE, Williams KA, Chavira DA, Azzam A,
Schwartz M, et al: Genome-wide linkage analysis of
obsessive-compulsive disorder implicates chromosome 1p36. Biol
Psychiatry. 72:629–636. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jaffe AE, Deep-Soboslay A, Tao R, Hauptman
DT, Kaye WH, Arango V, Weinberger DR, Hyde TM and Kleinman JE:
Genetic neuropathology of obsessive psychiatric syndromes. Transl
Psychiatry. 4:e4322014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nikitin A, Egorov S, Daraselia N and Mazo
I: Pathway studio-the analysis and navigation of molecular
networks. Bioinformatics. 19:2155–2157. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lorenzi PL, Claerhout S, Mills GB and
Weinstein JN: A curated census of autophagy-modulating proteins and
small molecules: Candidate targets for cancer therapy. Autophagy.
10:1316–1326. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fang A and Hofmann SG:
RelationshipAssociation between social anxiety disorder and body
dysmorphic disorder. Clin Psychol Rev. 30:1040–1048. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jimerson DC, Lesem MD, Kaye WH, Hegg AP
and Brewerton TD: Eating disorders and depression: Is there a
serotonin connection? Biol Psychiatry. 28:443–454. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jimerson DC, Lesem MD, Kaye WH and
Brewerton TD: Low serotonin and dopamine metabolite concentrations
in cerebrospinal fluid from bulimic patients with frequent binge
episodes. Arch Gen Psychiatry. 49:132–138. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xu P, He Y, Cao X, Valencia-Torres L, Yan
X, Saito K, Wang C, Yang Y, Hinton A Jr, Zhu L, et al: Activation
of serotonin 2C receptors in dopamine neurons inhibits binge-like
eating in mice. Biol Psychiatry. 81:737–747. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Karch S and Pogarell O: Neurobiology of
obsessive-compulsive disorder. Nervenarzt. 82:299–307. 2011.(In
German). View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Koo MS, Kim EJ, Roh D and Kim CH: Role of
dopamine in the pathophysiology and treatment of
obsessive-compulsive disorder. Expert Rev Neurother. 10:275–290.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Muller J and Roberts JE: Memory and
attention in obsessive-compulsive disorder: A review. J Anxiety
Disord. 19:1–28. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shafran R, Lee M, Cooper Z, Palmer RL and
Fairburn CG: Attentional bias in eating disorders. Int J Eat
Disord. 40:369–380. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Graybiel AM and Rauch SL: Toward a
neurobiology of obsessive-compulsive disorder. Neuron. 28:343–347.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wood AM, Dygdon JA and Conger AJ: Eating
disorders and sense of self: A learning theory conceptualization.
Eat Behav. 17:45–48. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Crum RM and Anthony JC: Cocaine use and
other suspected risk factors for obsessive-compulsive disorder: A
prospective study with data from the Epidemiologic Catchment Area
surveys. Drug Alcohol Depend. 31:281–295. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jonas JM and Gold MS: Cocaine abuse and
eating disorders. Lancet. 1:390–391. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nemoto T, Sugihara H, Mano A, Kano T and
Shibasaki T: The effects of ghrelin/GHSs on AVP mRNA expression and
release in cultured hypothalamic cells in rats. Peptides.
32:1281–1288. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
McDougle CJ, Barr LC, Goodman WK and Price
LH: Possible role of neuropeptides in obsessive compulsive
disorder. Psychoneuroendocrinology. 24:1–24. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Takenouchi T, Hashida N, Torii C, Kosaki
R, Takahashi T and Kosaki K: 1p34.3 deletion involving GRIK3:
Further clinical implication of GRIK family glutamate receptors in
the pathogenesis of developmental delay. Am J Med Genet A.
164A:456–460. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pritts SD and Susman J: Diagnosis of
eating disorders in primary care. Am Fam Physician. 67:297–304.
2003.PubMed/NCBI
|
|
32
|
Moldrich RX, Cheung NS, Pascoe CJ and
Beart PM: Excitotoxic injury profiles of low-affinity kainate
receptor agonists in cortical neuronal cultures. Eur J Pharmacol.
378:R1–R3. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Khouzam HR: A review of trazodone use in
psychiatric and medical conditions. Postgrad Med. 129:140–148.
2017. View Article : Google Scholar : PubMed/NCBI
|