Introduction
Lung cancer is one of the most common types of
cancer and the leading cause of cancer-associated mortality in
America (1). Of all lung cancer
cases, ~83% are of the non-small cell lung cancer (NSCLC)
histological type (2). Despite
advances in lung cancer treatment (3), NSCLC remains an aggressive type of
lung cancer due to its metastatic potential (3). Considering the important contribution
of NSCLC to the worldwide burden of cancer (4), it is important to further elucidate
the mechanisms and to explore novel treatments.
Nucleoside diphosphate kinase 4 (NME4), also known
as non-metastatic clone 23 human isoform 4 (NM23-H4), is one of the
members of the NM23 family (5).
The NME4 protein, which contains a mitochondrial target sequence
(6), has been shown to be
associated with mitophagy (7),
cell apoptosis (8), invasive
potential (9) and inflammatory
reactions (10). Kagan et
al (7) found that high
expression of wild-type NME4 in HeLa cells increases cardiolipin
externalization, thus regulating the elimination of mitochondria
via autophagy. In most cancer models, the loss of autophagy reduces
tumor growth, survival and proliferation (11). Genomic aberrations or altered gene
expression have been observed with respect to NME4 in several types
of cancer, such as gastric cancer (12,13),
colorectal carcinoma (14), renal
tumors (14), breast cancer
(15), testicular germ cell tumors
(16) and large cell anaplastic
lymphoma (17). However, rarely
has the relationship between NME4 and NSCLC been reported. A
previous study demonstrated that a decreased n-6/n-3 fatty acid
ratio reduces the invasive potential of human lung cancer cells by
downregulating the expression of cell adhesion/invasion-associated
molecules such as NME4 (9). In
addition, autophagy in NSCLC preserves mitochondrial quality and
regulates their abundance to promote tumorigenesis (18). The NME4 protein is involved in the
function of the outer and inner mitochondrial membranes (19) and is critical for mitochondrial
mitophagy (10). This indicates
that the NME4 gene may be implicated in the mechanisms of NSCLC
progression.
The present study was designed to further explore
the effect of NME4 on NSCLC in vitro. Firstly, The Cancer
Genome Atlas (TCGA) database and reverse transcription-quantitative
PCR (RT-qPCR) were used to assess the expression of NME4 in NSCLC
tissues and NSCLC cell lines. Then, short hairpin RNA (shRNA) was
used to silence the expression of NME4 in NSCLC cell lines; cell
proliferation, cell cycle, apoptosis, colony formation and MTT
assays were performed to clarify the possible role of NME4.
Materials and methods
Analysis of NME4 in NSCLC from
TCGA
TCGA (http://cancergenome.nih.gov/) is a collection of
microRNA-sequencing (seq), RNA-seq, single-nucleotide polymorphism
array, DNA methylation and exome sequencing data, among other data
types. TCGA can be used to analyze complex cancer genomics and
clinical parameters. In the present study, data of RNA-Seq in lung
adenocarcinoma for NME4 were extracted from TCGA, and the
expression levels of NME4 in each case was calculated according to
the distributions of the exon reads.
Cell lines and cell culture
The NCI-H1299 and A549 cell lines, which are
commonly used in NSCLC research, were purchased from the Cell Bank
of the Shanghai Institute of Cell Biology, Chinese Academy of
Sciences. Cells were cultured in RPMI-1640 medium (Invitrogen;
Thermo Fisher Scientific, Inc.) containing 10% FBS (Invitrogen;
Thermo Fisher Scientific, Inc.), 100 U/ml penicillin, 100 mg/ml
streptomycin and 2 mmol/l L-glutamine at 37°C in humidified air
containing 5% CO2. BEAS-2B cells were cultured in
DMEM/F-12 (Gibco; Thermo Fisher Scientific, Inc.), containing 5%
FBS (Invitrogen; Thermo Fisher Scientific, Inc.) and 100 U/ml
penicillin, 100 mg/ml streptomycin and 2 mmol/l L-glutamine at 37°C
in humidified air containing 5% CO2. The complete medium
in the present study was changed at least once every 2 days.
RNA extraction, reverse transcription
(RT) and quantitative PCR (qPCR)
TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) and M-MLV Reverse Transcriptase (Promega
Corporation) were used to extract the total RNA from cells and to
perform RT, respectively. RT was conducted at 73.5°C for 7 min, in
an ice water mixture for 5 min, at 43.5°C for 1 h and finally at
73.5°C for 3 min. Next, qPCR was performed on a MX3000P qPCR System
(Agilent Technologies, Inc.) using the Takara SYBR Master Mixture
(Takara Biotechnology Co., Ltd.). NME4 expression was quantified in
real time with SYBR Green and normalized to GAPDH expression, which
was used as an internal control. The denaturing, annealing and
extension conditions of each PCR cycle were 95°C for 30 sec, 95°C
for 5 sec and 60°C for 30 sec, respectively. The gene-specific
primer pairs were as follows: NME4 forward, 5′AGGGTACAATGTCGTCCGC3′
and reverse, 5′GACGCTGAAGTCACCCCTTAT3′; and GAPDH forward,
5′TGACTTCAACAGCGACACCCA3′ and reverse, 5′CACCCTGTTGCTGTAGCCAAA3′.
Each experiment was repeated twice in triplicate. The expression of
target genes was calculated using the 2−ΔΔCq method
(20).
Lentivirus-mediated knockdown vector
transduction
Lentiviral vectors for NME4-shRNA, which were
purchased from Shanghai GeneChem Co., Ltd., were used to examine
the function of NME4 (human NME4 cDNA; National Center for
Biotechnology Information accession no. NM_005009); the vector used
was hU6-MCS-CMV-EGFP. A total of two experimental groups for each
cell line were constructed. The shNME4 group was infected with
NME4-shRNA lentivirus (5′TGATTGGACACACCGACTC3′), while control
cells were infected with a lentivirus containing a scramble
sequence (5′TTCTCCGAACGTGTCACGT3′). NCI-H1299, A549 and BEAS-2B
cells in 6-well plates (2×106 cells/well) were infected
with lentiviral particles at a multiplicity of infection of 10
(5×106 TU/ml) for 16 h using polybrene (Sigma-Aldrich;
Merck KGaA), following which the medium was replaced with fresh
culture medium and cells were cultured for a further 56 h; the
knockdown efficiency was detected by RT-qPCR. Prior to
transduction, the medium of A549 cells was replaced with
Opti-Minimal Essential Medium (Gibco; Thermo Fisher Scientific,
Inc.) + polybrene, whereas the H1299 cell medium was replaced with
RPMI-1640 medium + polybrene; BEAS-2B cells were cultured with
DMEM/F12 + polybrene for transduction.
Cell proliferation
NME4-shRNA-transfected cells and control cells were
collected with 0.25% trypsin-EDTA and resuspended in RPMI-1640
standard medium once they had achieved logarithmic growth. Cells
were then seeded at a density of 2,000 cells/well and further
incubated at 37°C in humidified air containing 5% CO2
for 5 days. GFP expression (from the hU6-MCS-CMV-EGFP vector) was
measured in each well using a Cellomics ArrayScan VT1 (Thermo
Fisher Scientific, Inc.) for a 5-day period. Data were mapped, and
cell proliferation curves were generated for each group.
Cell cycle analysis
A total of 2×106 cells were seeded in
6-well plates, incubated overnight and then starved in culture
medium without serum for 12 h to synchronize their cell cycle.
Next, cells were cultured in complete medium for 48 h, harvested,
washed and fixed in 70% ethanol overnight at 4°C. Cells were then
washed, and stained with propidium iodide (PI; 10 µg/ml) and RNase
A (100 µg/ml) at room temperature for 30 min, followed by flow
cytometry detection using a Guava easyCyte HT flow cytometer (EMD
Millipore) and Shortcut 3.1 software (Incyte Corporation). Cells
with sub-G1 DNA content were considered apoptotic cells. All
experiments were performed in triplicate.
Apoptosis analysis
The cells in each group were harvested with 0.25%
trypsin, washed once with ice-cold PBS and analyzed with the
Annexin V-Allophycocyanin Apoptosis Detection kit with PI
(eBioscience; Thermo Fisher Scientific, Inc.) to assess apoptosis.
Cells were centrifuged at 200 × g and room temperature for 10–15
min, resuspended and incubated at room temperature for 10–15 min
according to the manufacturers' instructions, followed by flow
cytometry detection of the apoptotic cells using Shortcut software.
Detection was performed at 72 h after NME4 knockdown.
Colony formation assay
Cell colony formation was examined by a colony
formation assay. Cells in each group were digested with 0.25%
trypsin and seeded into 6-well plates at a density of 800
cells/well. After 2 weeks of incubation, colonies that included
>50 cells were scored as surviving colonies. Colonies were
visualized under a fluorescence microscope (IX71; Olympus
Corporation). Cells were washed with PBS, fixed with 4%
paraformaldehyde (Sangon Biotech Co., Ltd.) for 30 min at room
temperature and stained with 500 µl Giemsa solution (ECM550;
Chemicon International; Thermo Fisher Scientific, Inc.) for 20 min
at room temperature. Following several washes with deionized
distilled water, the cells were allowed to air dry at room
temperature. Colonies were counted and images were captured with a
digital camera under light microscopy (magnification, ×100;
XDS-100; Shanghai CaiKang Optical Instrument Co., Ltd.). The assay
was repeated three times.
MTT assay
Cell proliferation was examined by MTT assay. Cells
(2,000 cells/ml) were seeded into 96-well plates and incubated at
37°C for 24, 48, 72, 96 or 120 h. At 4 h prior to each time point,
0.5% MTT solution (Thermo Fisher Scientific, Inc.) was added,
followed by incubation for 4 h at 37°C. The cell supernatants were
discarded, and the formazan crystals were dissolved in 100 µl
dimethyl sulfoxide. The optical density (OD) of each group was
measured using a microplate reader (M2009PR; Tecan Group, Ltd.) at
a wavelength of 490/570 nm.
Statistical analysis
Data were analyzed using SPSS 19.0 statistical
software (IBM Corp.). Data are expressed as the mean ± SD of three
experimental repeats. Comparisons between two groups were performed
by Student's t-test and between multiple groups were performed by
one-way ANOVA followed by Student-Newman-Keuls post hoc test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Information from TCGA database
To further elucidate the relationship between NME4
and NSCLC, a clinical study was performed using original data from
TCGA. It was found that NME4 was highly expressed in lung
adenocarcinoma compared to non-cancerous lung tissues (P<0.001;
Fig. 1A).
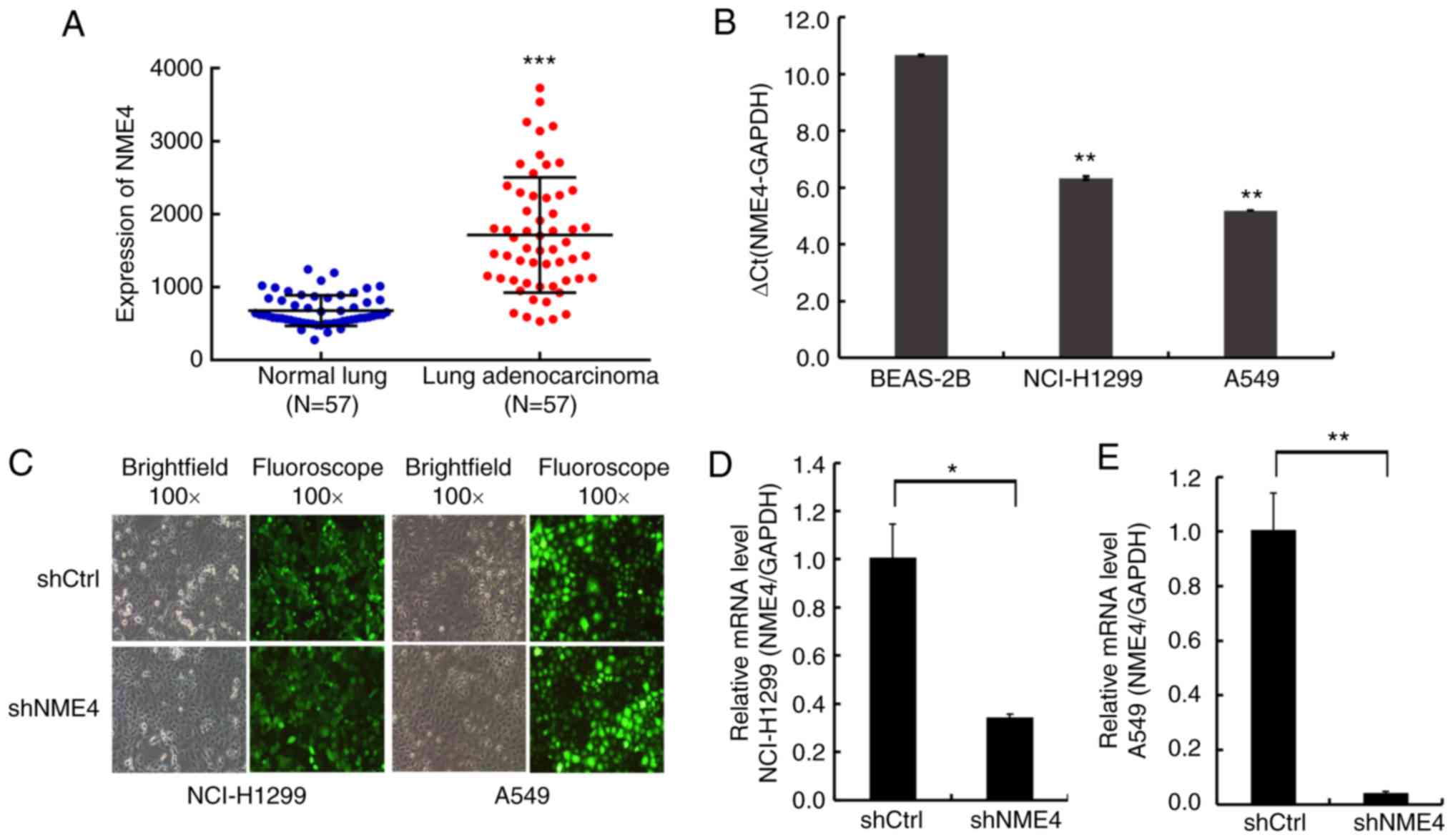 | Figure 1.Results for NME4 expression in TCGA
database and different cell lines. (A) NME4 was highly expressed in
lung adenocarcinoma compared with non-cancerous lung tissues, based
on TCGA database. ***P<0.001 vs. normal lung. (B) Expression
levels of NME4 mRNA in BEAS-2B, NCI-H1299 and A549 cell lines
(n=3). **P<0.01 vs. BEAS-2B. (C) After lentiviral transduction
for 72 h, the infection rate of cells reached >80% and the
status of the target cells was normal. (D) After lentiviral
transduction, relative NME4 mRNA expression was significantly
inhibited in the NCI-H1299 NME4-shRNA silenced cells (shNME4 group)
compared with the negative control cells, as assessed by RT-qPCR.
(E) After lentiviral transduction, relative NME4 mRNA expression
was significantly inhibited in the A549 NME4-shRNA silenced cells
compared with the negative control cells, as assessed by RT-qPCR.
GAPDH was used as an internal control. Comparisons between two
groups were performed by Student's t-test and between multiple
groups by ANOVA. Error bars indicate SD. *P<0.05, **P<0.01.
RT-qPCR, reverse transcription-quantitative PCR; NME4, nucleoside
diphosphate kinase 4; sh, short hairpin; TCGA, The Cancer Genome
Atlas. |
NME4 is overexpressed in NSCLC cell
lines and silenced upon lentivirus transduction
To investigate the NME4 mRNA expression levels in
NSCLC, RT-qPCR was performed in BEAS-2B, NCI-H1299, and A549 cells.
NME4 mRNA expression levels were high in NCI-H1299 and A549 cells
compared to BEAS-2B (Fig. 1B).
Then, NCI-H1299 and A549 cells were transfected with a NME4-shRNA
lentivirus or a scramble vector lentivirus. Upon lentiviral
infection for 72 h, the infection rate was >80% (Fig. 1C), making these cells suitable for
subsequent experiments. Upon NME4-shRNA lentiviral transduction,
RT-qPCR analysis revealed that NME4-shRNA decreased the mRNA
expression of endogenous NME4 significantly (P<0.05; Fig. 1D and E).
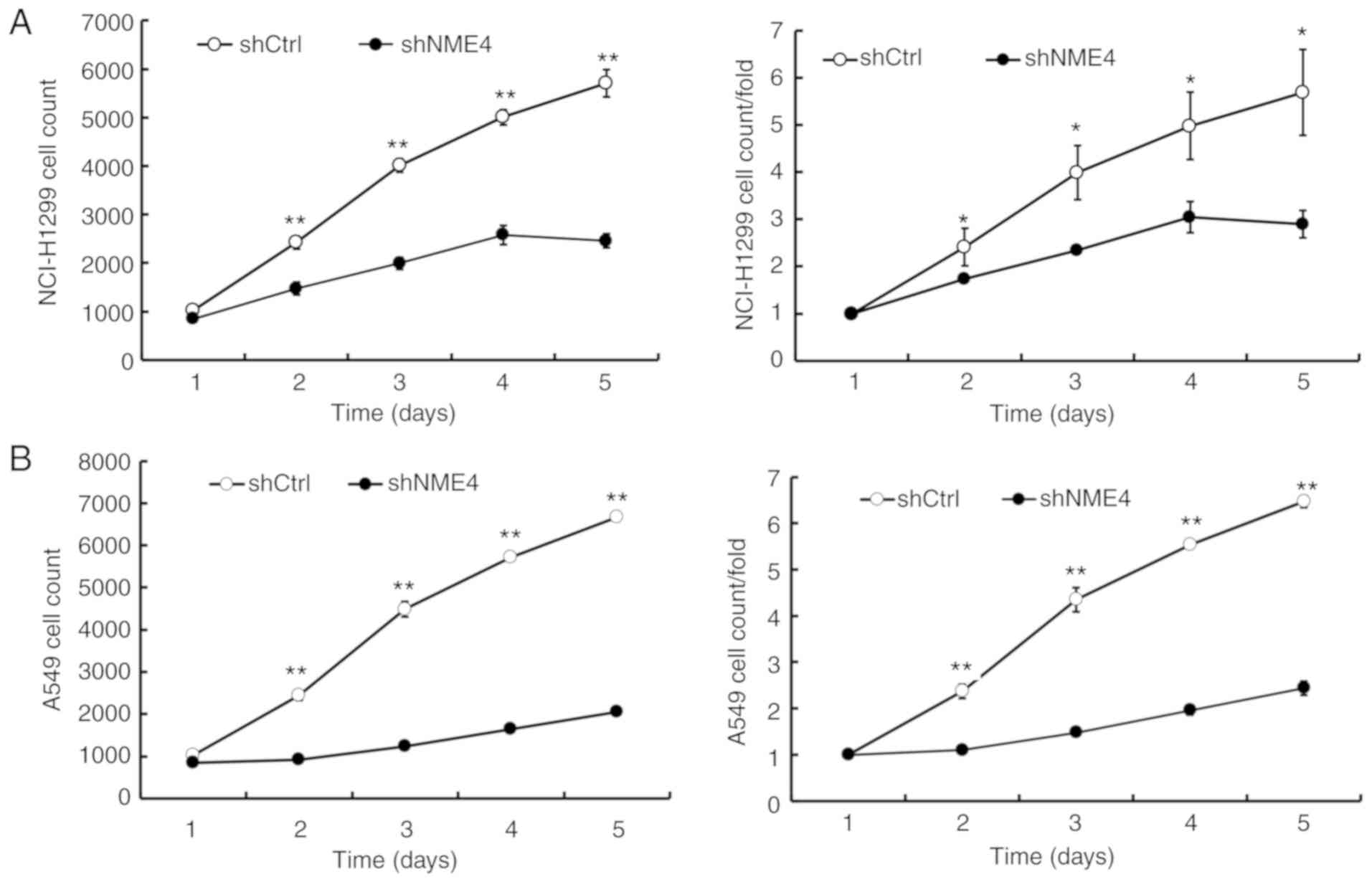
NME4 silencing inhibits NCI-H1299 and
A549 cell proliferation
Following lentiviral transduction, the proliferation
of NCI-H1299 and A549 cells was obviously inhibited in
NME4-shRNA-silenced cells compared with control cells, as shown by
GFP-based Cellomics ArrayScan VTI imaging (Fig. 2). The cell numbers in each group
were monitored for 5 days. Cell numbers were significantly reduced
in the NME4-shRNA-silenced cells (Fig.
2). These results suggested that NME4 was associated with
NCI-H1299 and A549 cell proliferation.
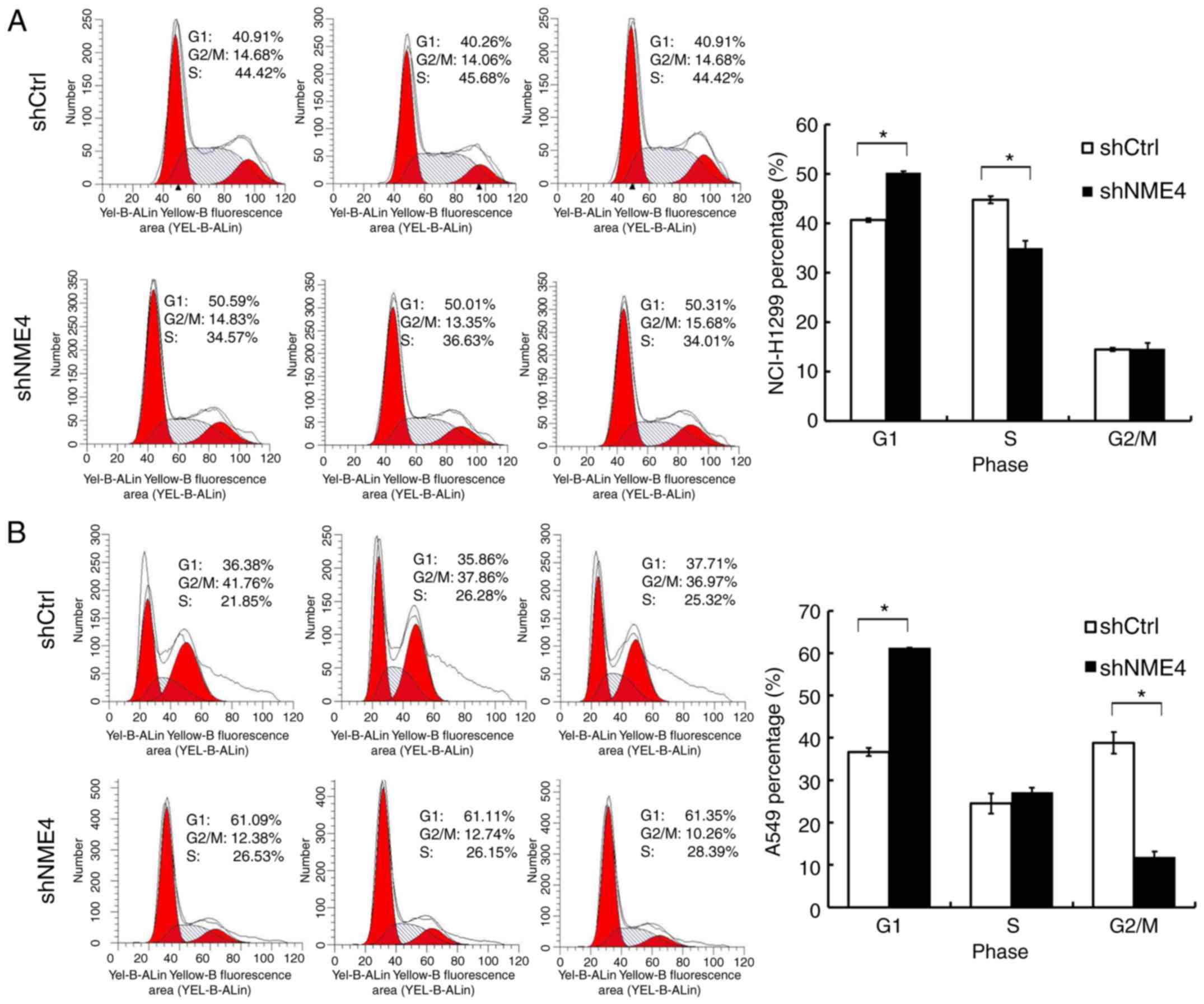
NME4 silencing induces cell cycle
progression
Cell cycle distribution was assessed to further
elucidate the growth-suppressing effect of NME4-shRNA on NCI-H1299
and A549 cells. Compared with the control group, NME4-shRNA
significantly increased the fraction of G1-phase cells in the
NME4-shRNA group (Fig. 3). The
results suggested that NME4 silencing may induce cell cycle arrest
at the G1 phase, and the effect of NME4 on the cell cycle was
time-dependent. G1 arrest was more pronounced in A549 cells
compared with NCI-H1299 cells (Fig.
3), which may be due to the less efficient knockdown of NME4
(Fig. 1C and D). Whether the
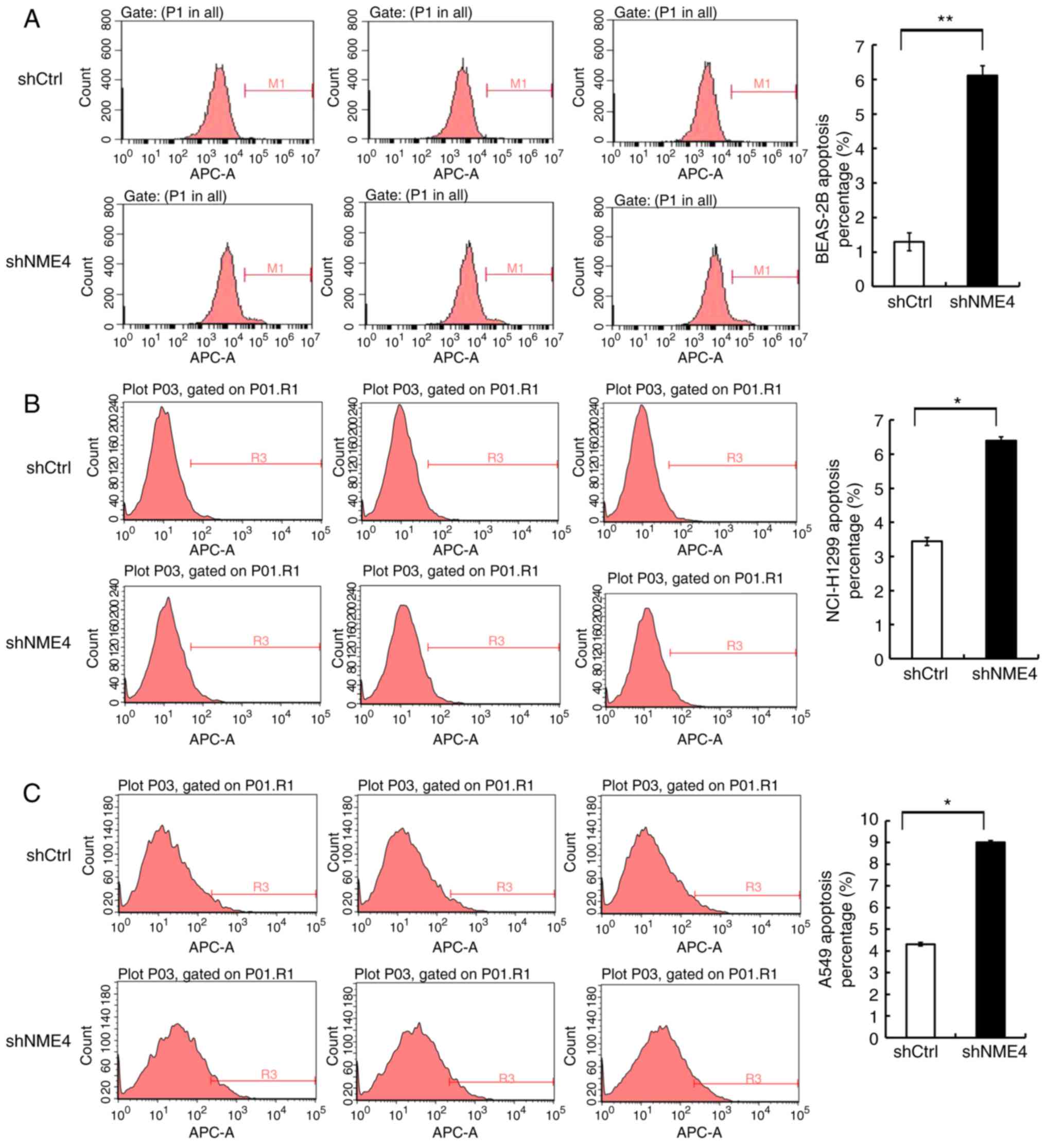
silencing of NME4 was associated with apoptosis in NCI-H1299 and
A549 cells was further investigated. The proportions of apoptotic
cells were increased by only 3–5% in NME4-silenced groups of
NCI-H1299 and A549 cells when compared with control cells,
comparable to the effects observed in shRNA-transduced BEAS-2B
cells (Fig. 4). These data do not
suggest that NME4 had an effect on the apoptosis of NCI-H1299 and
A549 cells. This level of increase in apoptosis may be due to the
state of the cells under the conditions of the assay. These results
may indicate that NME4 silencing interrupted cell cycle progression
and thus affected the progression of NSCLC, rather than increasing
cell apoptosis.
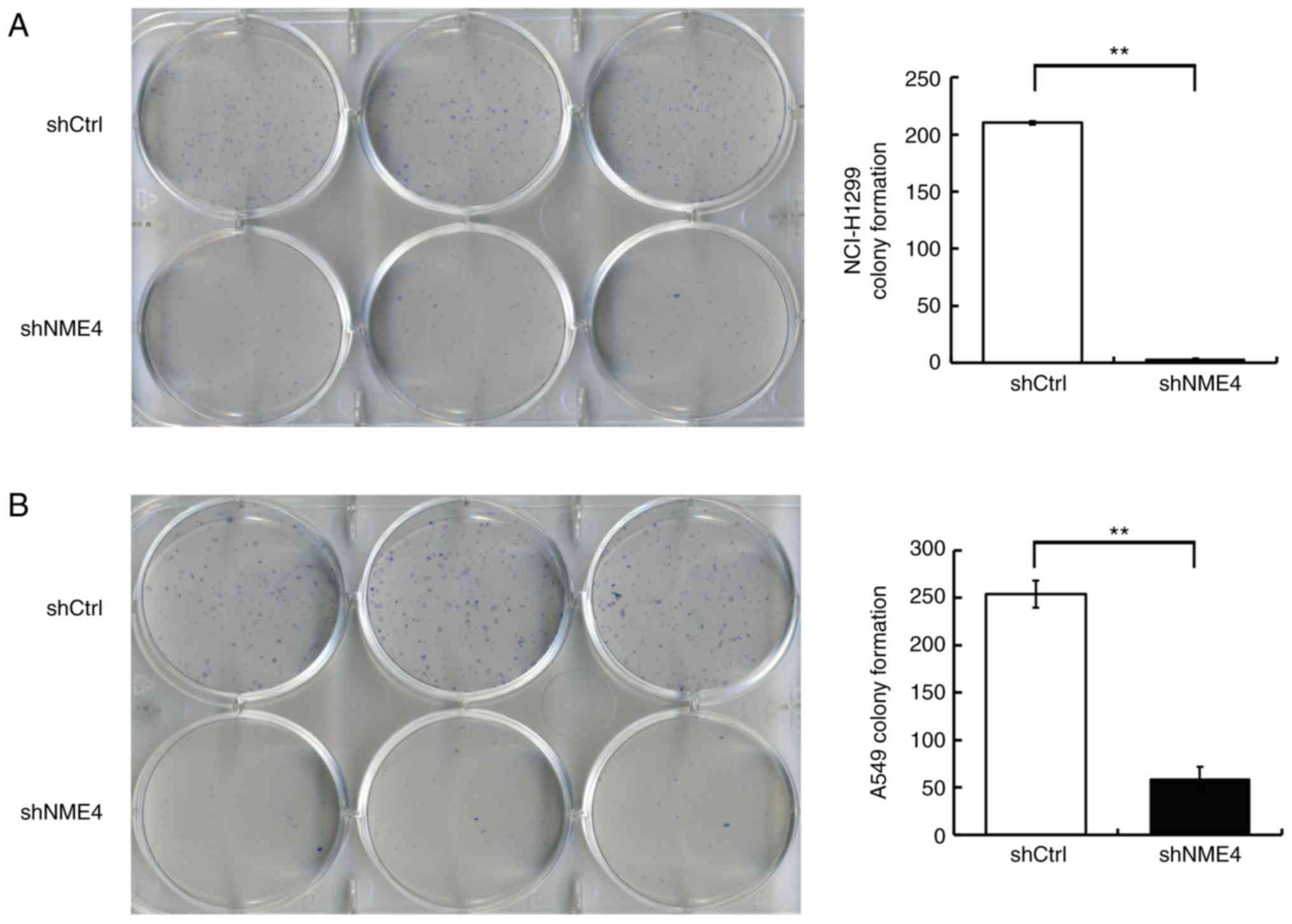
NME4 silencing reduces NCI-H1299 and
A549 cell colony formation
Silencing of NME4 suppressed the
anchorage-independent growth of NCI-H1299 and A549 cells in soft
agar (Fig. 5). Infection with
NME4-shRNA in NCI-H1299 and A549 cells significantly reduced the
number of cell clones (P<0.01), which confirmed that the
silencing of NME4 suppressed the proliferative potential of
NCI-H1299 and A549 cells, and revealed the critical role of NME4 in
the survival of NSCLC cells.
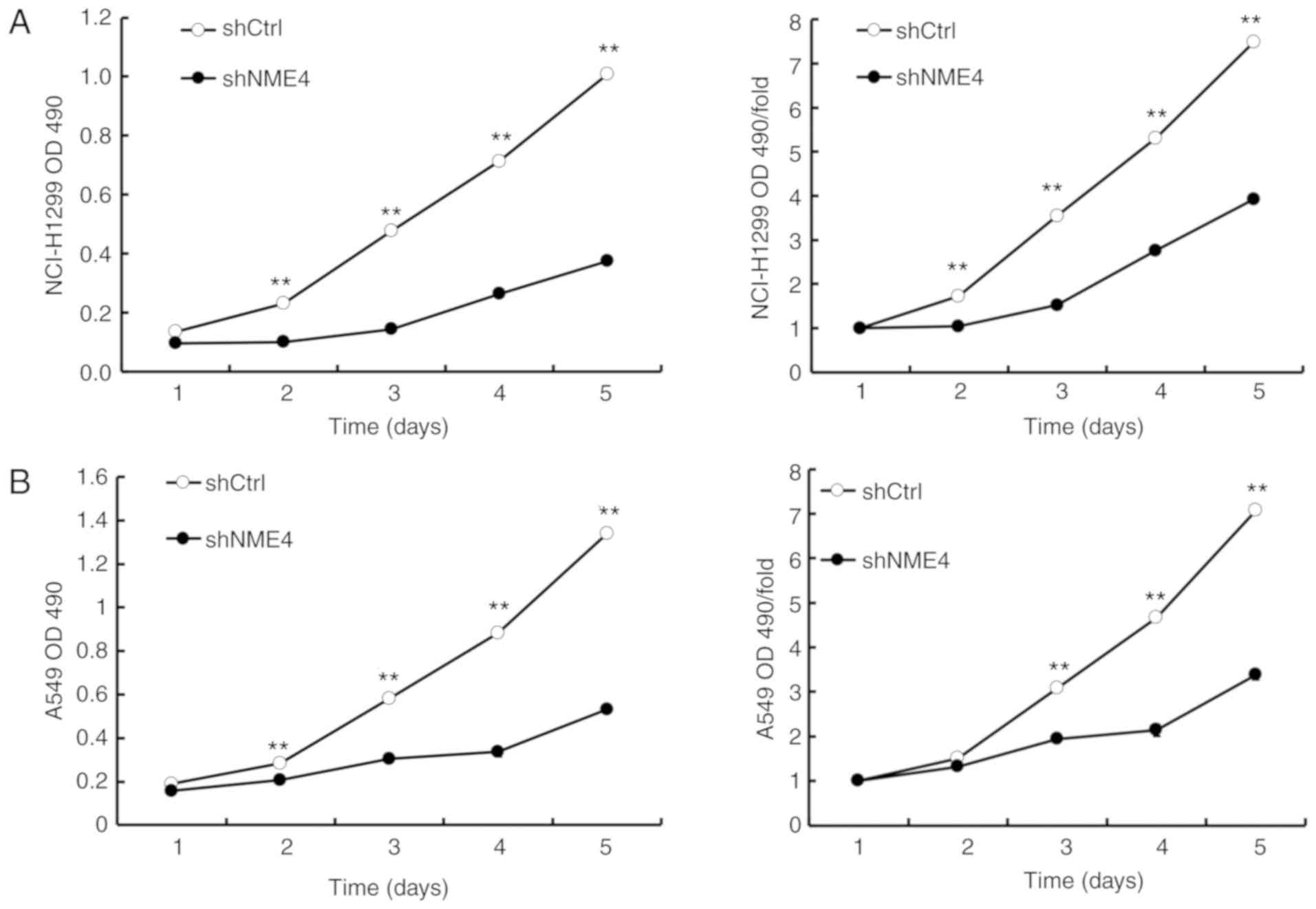
MTT assay
An MTT assay was performed to detect the
proliferation of NCI-H1299 and A549 cells upon transfection with
NME4-shRNA for 24, 48, 72, 96 or 120 h. The OD value at 490 nm
indicated the viability of the cells. The results of MTT assay
demonstrated that the silencing of NME4 significantly decreased the
proliferation of NCI-H1299 and A549 cells (Fig. 6), which indicates that NME4
contributes to NSCLC proliferation.
Discussion
As one of the most common cancer types, NSCLC also
has the highest mortality rate. Although the mainstays of lung
cancer treatment may slow down tumor growth, the genetic
versatility of tumor cells may induce resistance to the currently
available therapies. Thus, it is critical to develop novel,
effective and safe approaches for NSCLC treatment.
NME4 is one of the members of the NM23 family
(5), which have been postulated to
be involved in cell adhesion and migration (21), as well as possessing NDPK activity
and being involved in DNA repair mechanisms (12,22).
The NME4 protein contains a mitochondrial target sequence (19,23,24),
and is involved in the function of the outer and inner
mitochondrial membranes (19),
which is critical for mitochondrial mitophagy (7). Mitophagy can maintain the normal
metabolism of cells and prevent cellular stress responses and
genomic damage, thereby inhibiting tumor development. Furthermore,
tumor cells enhance their tolerance to hypoxia and low nutrition by
enhancing mitophagy so as to survive; mitophagy serves a
significant role in promoting tumor development (25,26).
Cancer cells require altered mitochondrial functions, including
organelle dynamics, to resist bioenergetic/biosynthetic
reprogramming for supporting proliferation, migration and invasion
(27). Since NME4 plays a role in
all of these, it is tempting to speculate that its altered
expression or function may affect the fate of a cancer cell. At the
same time, it has been reported that NME4 may constitute an
important link between energy metabolism and cellular regulation.
Abnormal expression of the NME4 gene may induce an imbalance of
nucleotide pools in the mitochondria, resulting in the failure of
checkpoint controls and the accumulation of subsequent genetic
alterations, thus contributing to tumorigenesis (12).
Therefore, it was hypothesized that NME4 may
participate in an important mechanism in NSCLC. NME4 was
significantly overexpressed in NSCLC tissues, based on TCGA data.
Furthermore, the present study demonstrated that NME4 mRNA
expression levels were high in human NSCLC cell lines via qPCR. The
present study also established low expression of NME4 in A549 and
NCI-H1299 cells using lentivirus-mediated technology. When NME4 was
expressed at reduced levels, the cell proliferation rate and colony
formation of A549 and NCI-H1299 cells markedly decreased. Low
expression of NME4 also resulted in cell cycle arrest at the G1
phase in these NSCLC cell lines. Based on these observations, it
was concluded that NME4 may serve as a novel tumor promoter capable
of enhancing NSCLC progression. To the best of our knowledge, this
was the first study to identify that NME4 expression may exhibit
tumor-promoting potential in NSCLC. However, some limitations exist
in the present analysis. First, only two types of NSCLC cell lines
were used, which may result in a loss of comprehensiveness. Second,
the less efficient knockdown of NME4 in the NCI-H1299 cell line may
weaken the interpretation of the role of this gene from the results
of the present study.
In conclusion, the present study determined that
NME4 serves critical roles in NSCLC development. Although the
detailed mechanisms remain to be elucidated, the critical role of
NME4 in NSCLC development may provide evidence for the development
of novel therapeutics against NME4 for the treatment of NSCLC.
Acknowledgements
Not applicable.
Funding
This work was supported by the National Natural
Science Foundation of China (grant nos. 81403476, 81704154 and
81573758), Development Project of Shanghai Peak
Disciplines-Integrated Chinese and Western Medicine, Shanghai
Health and Family Planning Commission Program for Traditional
Chinese Medicine (grant no. 2016JP001), Young Elite Scientists
Sponsorship Program by China Association for Science and Technology
(grant no. 2018QNRC001) and the Scientific Research Project of
Shanghai Science and Technology commission (grant no.
17401930300).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YW and JD conceived and designed the study. WW and
MD performed the majority of the experiments, performed preliminary
analysis and drafted the manuscript. YW reviewed the data analysis
and revised the manuscript. JC, FX, CY, CM, LY and WT participated
in the completion of the experiments. All the authors read and
approved the final manuscript and agree to be accountable for all
aspects of the work.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Miller KD, Siegel RL, Lin CC, Mariotto AB,
Kramer JL, Rowland JH, Stein KD, Alteri R and Jemal A: Cancer
treatment and survivorship statistics, 2016. CA Cancer J Clin.
66:271–289. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Novello S, Barlesi F, Califano R, Cufer T,
Ekman S, Levra MG, Kerr K, Popat S, Reck M, Senan S, et al:
Metastatic non-small-cell lung cancer: ESMO clinical practice
guidelines for diagnosis, treatment and follow-up. Ann Oncol. 27
(Suppl 5):v1–v27. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Milon L, Rousseau-Merck MF, Munier A,
Erent M, Lascu I, Capeau J and Lacombe ML: nm23-H4, a new member of
the family of human nm23/nucleoside diphosphate kinase genes
localised on chromosome 16p13. Hum Genet. 99:550–557. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Schlattner U, Tokarska-Schlattner M, Epand
RM, Boissan M, Lacombe ML, Klein-Seetharaman J and Kagan VE:
Mitochondrial NM23-H4/NDPK-D: A bifunctional nanoswitch for
bioenergetics and lipid signaling. Naunyn Schmiedebergs Arch
Pharmacol. 388:271–278. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kagan VE, Jiang J, Huang Z, Tyurina YY,
Desbourdes C, Cottet-Rousselle C, Dar HH, Verma M, Tyurin VA,
Kapralov AA, et al: NDPK-D (NM23-H4)-mediated externalization of
cardiolipin enables elimination of depolarized mitochondria by
mitophagy. Cell Death Differ. 23:1140–1151. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fujita Y, Fujiwara K, Zenitani S and
Yamashita T: Acetylation of NDPK-D regulates its subcellular
localization and cell survival. PLoS One. 10:e01396162015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xia SH, Wang J and Kang JX: Decreased
n-6/n-3 fatty acid ratio reduces the invasive potential of human
lung cancer cells by downregulation of cell
adhesion/invasion-related genes. Carcinogenesis. 26:779–784. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Schlattner U, Tokarska-Schlattner M, Epand
RM, Boissan M, Lacombe ML and Kagan VE: NME4/nucleoside diphosphate
kinase D in cardiolipin signaling and mitophagy. Lab Invest.
98:228–232. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Amaravadi R, Kimmelman AC and White E:
Recent insights into the function of autophagy in cancer. Genes
Dev. 30:1913–1930. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Seifert M, Welter C, Mehraein Y and Seitz
G: Expression of the nm23 homologues nm23-H4, nm23-H6, and nm23-H7
in human gastric and colon cancer. J Pathol. 205:623–632. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xu ZY, Chen JS and Shu YQ: Gene expression
profile towards the prediction of patient survival of gastric
cancer. Biomed Pharmacother. 64:133–139. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hayer J, Engel M, Seifert M, Seitz G and
Welter C: Overexpression of nm23-H4 RNA in colorectal and renal
tumours. Anticancer Res. 21:2821–2825. 2001.PubMed/NCBI
|
|
15
|
Patocs A, Zhang L, Xu Y, Weber F, Caldes
T, Mutter GL, Platzer P and Eng C: Breast-cancer stromal cells with
TP53 mutations and nodal metastases. N Engl J Med. 357:2543–2551.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Skotheim RI, Autio R, Lind GE, Kraggerud
SM, Andrews PW, Monni O, Kallioniemi O and Lothe RA: Novel genomic
aberrations in testicular germ cell tumors by array-CGH, and
associated gene expression changes. Cell Oncol. 28:315–326.
2006.PubMed/NCBI
|
|
17
|
Gaiser T, Thorns C, Merz H, Noack F,
Feller AC and Lange K: Gene profiling in anaplastic large-cell
lymphoma-derived cell lines with cDNA expression arrays. J
Hematother Stem Cell Res. 11:423–428. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Guo JY and White E: Autophagy, metabolism,
and cancer. Cold Spring Harb Symp Quant Biol. 81:73–78. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Milon L, Meyer P, Chiadmi M, Munier A,
Johansson M, Karlsson A, Lascu I, Capeau J, Janin J and Lacombe ML:
The human nm23-H4 gene product is a mitochondrial nucleoside
diphosphate kinase. J Biol Chem. 275:14264–14272. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Postel EH: Multiple biochemical activities
of NM23/NDP kinase in gene regulation. J Bioenerg Biomembr.
35:31–40. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kowluru A, Tannous M and Chen HQ:
Localization and characterization of the mitochondrial isoform of
the nucleoside diphosphate kinase in the pancreatic beta cell:
Evidence for its complexation with mitochondrial succinyl-CoA
synthetase. Arch Biochem Biophys. 398:160–169. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Boissan M, Dabernat S, Peuchant E,
Schlattner U, Lascu I and Lacombe ML: The mammalian Nm23/NDPK
family: From metastasis control to cilia movement. Mol Cell
Biochem. 329:51–62. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Janin J, Dumas C, Moréra S, Xu Y, Meyer P,
Chiadmi M and Cherfils J: Three-dimensional structure of nucleoside
diphosphate kinase. J Bioenerg Biomembr. 32:215–225. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Redmann M, Dodson M, Boyer-Guittaut M,
Darley-Usmar V and Zhang J: Mitophagy mechanisms and role in human
diseases. Int J Biochem Cell Biol. 53:127–133. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gebert N, Joshi AS, Kutik S, Becker T,
McKenzie M, Guan XL, Mooga VP, Stroud DA, Kulkarni G, Wenk MR, et
al: Mitochondrial cardiolipin involved in outer-membrane protein
biogenesis: Implications for Barth syndrome. Curr Biol.
19:2133–2139. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Porporato PE, Payen VL, Pérez-Escuredo J,
De Saedeleer CJ, Danhier P, Copetti T, Dhup S, Tardy M, Vazeille T,
Bouzin C, et al: A mitochondrial switch promotes tumor metastasis.
Cell Rep. 8:754–766. 2014. View Article : Google Scholar : PubMed/NCBI
|