Introduction
In 2018, breast cancer (BC) was expected to account
for 30% of all new cancer diagnoses in women, and remains the
principal cause of mortality from cancer in women worldwide
(1). Recently, emphasis has been
on the tumorigenesis of the immune system and immunotherapy for BC
(2).
Chemokines, known as chemotactic cytokines, are a
family of small molecular weight proteins (6–14 kDa) of the immune
system, which bind to G protein-coupled receptors and participate
in tissue maintenance and development, and in pathological
conditions. Chemokines can be divided into four types according to
their structural differences: CC, CXC, XC, and CX3C (3). Increasing evidence shows that CXCRs
and their ligands affect some cancer-associated processes,
including tumor cell activation, proliferation, invasion and
migration (4,5).
CXCR1-CXCR7 have been identified (6). CXCR1/2 share ~76% sequence homology
and bind to CXC ligand 8 (CXCL8) with similar affinities (7–9).
CXCR1/2 and CXCL8 play important roles in the initiation and
transfer of inflammatory mediators, as well as in the related tumor
growth and metastasis (10). The
CXCL9, −10, −11/CXCR3 axis contributes to tumor suppression by
regulating immune cell development, differentiation and activation
through paracrine signaling. However, the axis also promotes tumor
progression and metastasis through autocrine signaling (11). CXCR4, which is widely expressed on
the surface of epithelial and endothelial cells (12), is activated by CXCL12. This
generates signals for a number of processes leading to tissue
remodeling, including the homeostasis and development of normal
tissues, hemopoiesis and angiogenesis. These roles make CXCR4 an
important player in tumorigenesis (13). CXCR5 is specifically expressed in
Burkitt lymphoma and lymphoid tissues, and is important for B cell
migration through its binding to CXCL13 (14). It was reported that increased CXCR5
expression may result in increased survival and migration of MCF-7
BC cells that lack functional cellular tumor antigen p53 (TP53)
(15). CXCR6-positive cells are
cancer stem cells that can generate tumors through the process of
self-renewal and their ability to differentiate into multiple cell
types, which may also play a role in the mechanism of CXCR6 in
tumor progression (16). CXCR7 is
also expressed in different types of cancer and on tumor-associated
vasculature. Emerging evidence also reveals its participation in
metastasis and tumor progression (16).
Individual CXCRs have discrete roles in cancer
development and progression. Nevertheless, the contributions of
distinct CXCRs to BC tumorigenesis need to be explored. Thus, in
the present study, large databases were mined for transcription
expression information about CXCR family members in BC and normal
tissues. Finally, the expression levels of CXCR family members were
analyzed in diverse subtypes of BC and the prognostic value of CXCR
family members in BC was assessed.
Materials and methods
Oncomine database analysis
The transcript expression levels of different CXCRs
in a variety of cancers were analyzed using the publicly available
cancer microarray database Oncomine (http://www.oncomine.org), which contains numerous
datasets, including The Cancer Genome Atlas (TCGA) dataset. In the
present study, the thresholds were set as follows: Data type, mRNA;
gene rank, all; fold change, 2; P-value, 0.01. The differences in
expression between cancer specimens and normal control datasets for
CXCR family members were compared.
Gene expression-based outcome for
breast cancer (GOBO) database analysis
The transcript levels of specific CXCRs in BC
subtypes were analyzed using the GOBO database (http://co.bmc.lu.se/gobo). The GOBO database contains
gene expression data about 1,881 BC samples and 51 BC cell lines
from experiments conducted using Affymetrix microarrays (17).
Survival analysis using the
Kaplan-Meier (KM) plotter
The KM plotter tool (http://kmplot.com/analysis/) can be used to evaluate
the influence of the expression of 54,675 genes on survival in
10,461 cancer samples, including relapse-free survival (RFS) and
overall survival (OS), to determine the prognostic values of CXCRs
in 5,143 BC patients.
The effects of specific CXCRs on the prognosis of BC
were studied, according to the following criteria: All patients
with BC; the four molecular subtypes of BC, according to the 2013
St. Gallen criteria (18), were
basal-like [estrogen receptor (ER) and progesterone receptor (PR)
absent, human epidermal growth factor receptor-2 (HER2) negative],
luminal A [ER and PR positive, HER2 negative, Ki-67 ‘low’
(<14%)], luminal B [ER positive, HER2 negative and at least one
of the following: Ki-67 ‘high’ (≥20%), PR ‘negative or low’
(<20%) or ER positive, HER2 over-expressed or amplified, any
Ki-67, any PR], HER2 (HER2 over-expressed or amplified, ER and PR
absent); other characteristic molecular markers; related optimal
treatment for different BC molecular subtypes, for example,
basal-like BC with chemotherapy, luminal A BC with endocrine
therapy and luminal B BC with both chemotherapy and endocrine
therapy. The results are presented using KM curves, with the hazard
ratio (HR) with 95% confidence intervals (95% CI) and the P-value
for the log-rank test displayed.
Results
CXCR4 and CXCR3 are significantly
overexpressed in BC
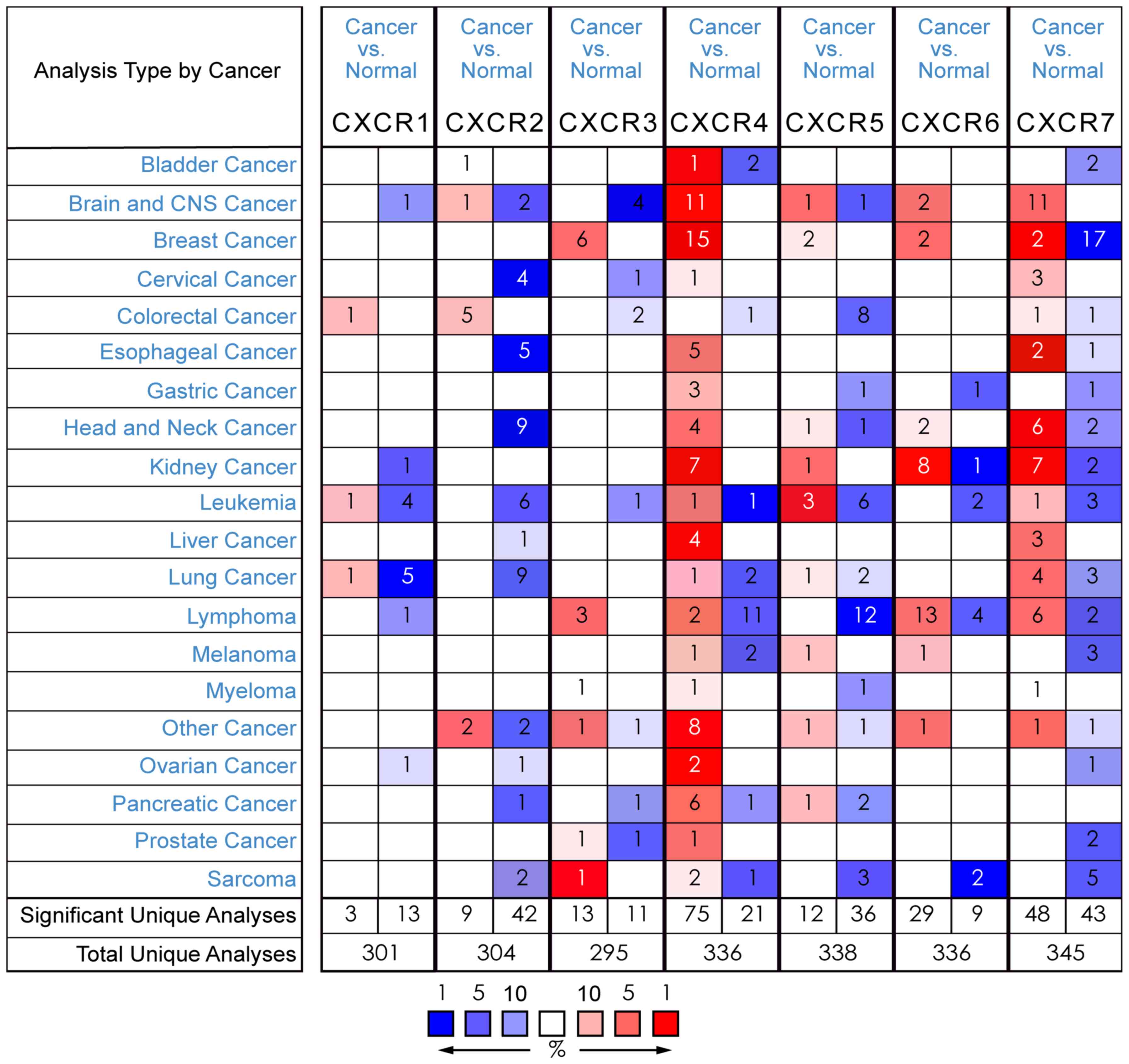
To date, it has been determined that there are seven
CXCR family members expressed in a variety of human cancer types
(Fig. 1). Utilizing the Oncomine
database for gene expression analysis in BC, 15 out of 50 analyses
met the threshold for CXCR4 in 9 out of 13 datasets, while 6 out of
43 analyses met the threshold for CXCR3 in 5 out of 10 datasets. No
fold change >2 was found between BC samples and normal tissues
for the transcript expression of CXCR1 (fold change=1.074; P=0.163;
Fig. 2A), CXCR2 (fold
change=−1.095; P=0.795; Fig. 2B),
CXCR5 (fold change=1.119; P=1.87×10−4; Fig. 2G), CXCR6 (fold change=1.713;
P=7.19×10−5; Fig. 2H)
and CXCR7 (fold change=−2.448; P=1.000; Fig. 2I).
The analysis showed that the CXCR3 transcript was
significantly elevated in BC samples compared with normal tissues.
The CXCR3 transcript level was 2.200-fold (P=1.52×10−9)
higher in BC samples in a large-sample dataset from TCGA database
(Fig. 2C). Similarly, in a
previous study by Curtis et al (19), CXCR3 was elevated by 2.857-fold
(P=4.20×10−11) in BC compared with normal samples
(Fig. 2D).
The transcript level of CXCR4 was 2.079-fold
(P=2.69×10−10) higher in BC compared with normal samples
in a 593-sample dataset from TCGA database (Fig. 2E). Furthermore, in a 59 sample
dataset from a previous study by Finak et al (20), CXCR4 mRNA expression was increased
by 7.230-fold (P=8.95×10−19) in BC tissues compared with
normal samples (Fig. 2F).
In different breast cancer cell lines, the Neve
(21) expression and intensity of
CXCR3 and CXCR4 differ (Figs. 3E
and 4E), which may provide a
reference to the selection of cell lines for basic experiments.
 | Figure 3.Correlation analysis of CXCR4
expression in different molecular subtypes of BC. (A) In the
Oncomine analysis, the expression of CXCR4 in basal-like BC was
significantly higher than that in luminal-like BC. In the GOBO
analysis, the basal-like subtypes of BC expressed higher levels of
CXCR4 than the luminal A, luminal B and HER2 subtypes of BC based
on (B) HU and (C) PAM50 gene classifiers of BC. (D) The GOBO
analysis also revealed that the expression of CXCR4 in luminal-like
BC was significantly higher than that in the basal A or basal B
subtypes of BC. (E) The Neve expression (21) of CXCR4 in BC cell lines. CXCR, CXC
chemokine receptor; BC, breast cancer; lum, luminal; HER2, human
epidermal growth factor receptor 2; HR, hormone receptor; TN,
triple negative; GOBO, Gene Expression-Based Outcome for Breast
Cancer. |
 | Figure 4.Correlation analysis of CXCR3
expression in different molecular subtypes of BC. (A) In the
Oncomine analysis, CXCR3 expression showed no significant
difference between the basal-like and luminal-like subtypes of BC.
In the GOBO analysis, there was no significant difference between
the basal-like and luminal-like subtypes of BC based on the (B) HU
and (C) PAM50 gene classifiers of BC. (D) The GOBO analysis also
revealed that the expression of CXCR3 was not significantly
different in the TN, HER2 and HR subtypes of BC. (E) The Neve
expression (21) of CXCR4 in BC
cell lines. CXCR, CXC chemokine receptor; BC, breast cancer; lum,
luminal; HER2, human epidermal growth factor receptor 2; HR,
hormone receptor; TN, triple negative; GOBO, Gene Expression-Based
Outcome for Breast Cancer. |
Expression of CXCR4 and CXCR3 in
different molecular subtypes of BC
In the present study, CXCR4 and CXCR3 were
identified as being overexpressed in BC. To further explore the
expression of CXCR4 and CXCR3, their expression levels were
analyzed in different molecular subtypes of BC. The dataset from a
previous study by Farmer et al (22) revealed that CXCR4 was increased by
1.496-fold (P=5.75×10−4) in basal-like BC samples
compared to luminal-like BC samples (Fig. 3A), while CXCR3 showed no
significant difference between these two subtypes of BC (P=0.18;
Fig. 4A).
Basal-like subtype can be classified as basal A and
basal B. In GOBO analysis, CXCR4 expression in luminal-like BC was
statistically higher (P=0.01784) than in the basal A or basal B
subtypes of BC, and the basal-like subtype of BC expressed higher
levels of CXCR4 (P<0.00001) than in the luminal A or luminal B
subtype (Fig. 3B-D). The hormone
receptor sensitive subtype of BC was not significantly different
(P=0.12853) than the triple negative BC (TNBC) or HER2 subtypes in
terms of CXCR4 expression (Fig.
3D). But, CXCR3 showed no significant difference in expression
among the different molecular subtypes of BC (Fig. 4B-D).
CXCR4 expression predicts better
survival in patients with ER-negative/TP53-mutated basal-like BC,
in patients with basal-like BC treated with chemotherapy and in
patients with luminal A BC not treated with endocrine therapy
The prognostic value of CXCR4 in BC was evaluated
and indicated that high expression of CXCR4 was significantly
associated with a poorer RFS (HR=1.18; P=0.0028) in all patients
with BC (Fig. 5A). In Fig. 6A, a high CXCR4 transcript level was
not significantly correlated with better OS (HR=1.01; P=0.9) in all
patients with BC.
 | Figure 5.Prognostic value of CXCR4 in terms of
RFS in BC. (A) High mRNA expression of CXCR4 was significantly
associated with longer RFS in all patients with BC. High mRNA
expression of CXCR4 was also associated with longer RFS in patients
with (B) basal-like BC, but not in patients with (C) HER2, (D)
luminal A or (E) luminal B subtypes of BC. High mRNA expression of
CXCR4 was associated with longer RFS in patients with (F) TP53
mutation, but not in patients with (G) wild-type TP53. High mRNA
expression of CXCR4 was associated with longer RFS in patients with
(H) basal-like BC who had received chemotherapy, while it was not
associated with RFS in (I) patients who had not received
chemotherapy. High mRNA expression of CXCR4 was associated with
shorter RFS in patients with (J) luminal A BC who had received
endocrine therapy, and was not associated with RFS in (K) patients
who had not received endocrine therapy. CXCR, CXC chemokine
receptor; BC, breast cancer; RFS, relapse-free survival; HER2,
human epidermal growth factor receptor 2; TP53, cellular tumor
antigen p53; HR, hazard ratio. |
 | Figure 6.Prognostic value of CXCR4 in terms of
OS in BC. (A) No significant correlation was found between high
expression of CXCR4 and a longer OS in all patients with BC. High
mRNA expression of CXCR4 was associated with longer OS in patients
with (B) basal-like BC, but not in patients with the (C) HER2, (D)
luminal A or (E) luminal B subtypes of BC. High mRNA expression of
CXCR4 was associated with longer OS in patients with (F)
ER-negative BC, but not in those with (G) ER-positive BC. High mRNA
expression of CXCR4 was associated with longer OS in patients with
(H) basal-like BC who had received chemotherapy, while it was not
associated with OS in (I) patients who had not received
chemotherapy. CXCR, CXC chemokine receptor; BC, breast cancer;
HER2, human epidermal growth factor receptor 2; OS, overall
survival; ER, estrogen receptor; HR, hazard ratio. |
Subtype analysis showed that a high level of CXCR4
transcript expression was positively correlated with a longer RFS
in both TP53-mutated (HR=0.5; P=0.0048) and basal-like BC (HR=0.77;
P=0.043), while no statistical significance was found in TP53
wild-type or other molecular subtypes of BC (Fig. 5B-F and G). Similarly, the analysis
also revealed that high transcript levels of CXCR4 were
statistically correlated with better OS in patients with basal-like
BC (HR=0.52; P=0.01) and in ER-negative BC (HR=0.65; P=0.034;
Fig. 6B-F and G).
High CXCR4 mRNA expression was statistically
associated with a better RFS (HR=0.42; P=0.00095) and OS (HR=0.4;
P=0.035) in patients with basal-like BC treated with chemotherapy
(Figs. 5H and I, and 6H and I). However, high expression of
CXCR4 was significantly correlated with a poorer RFS (HR=1.44;
P=0.045) in patients with luminal A BC treated with endocrine
therapy (Fig. 5J and K).
High CXCR3 mRNA expression is
correlated with a longer survival in patients with
TP53-mutated/basal-like BC
High CXCR3 mRNA expression was statistically
correlated with a better RFS (HR=0.74; P=9.7×10−8) and
OS (HR=0.71; P=0.0015) in all patients with BC (Figs. 7A and 8A). Subtype analysis also showed that
high CXCR3 mRNA expression was significantly associated with a
longer RFS (HR=0.55; P=2.6×10−6) and OS (HR=0.51;
P=0.0073) in basal-like BC (Figs.
7B and 8B). Consistently,
patients with mutated TP53 and with high levels of CXCR3 transcript
expression were found to have longer RFS (HR=0.46; P=0.0014) and OS
(HR=0.43; P=0.032) (Figs. 7G and
I, and 8J and K). However,
high levels of CXCR3 transcript had a different effect on RFS and
OS for some characteristic markers. The analysis predicted a better
RFS in patients with luminal A BC (HR=0.8; P=0.01) and luminal B BC
(HR=0.7; P=0.00024; Fig. 7C-F and
I), as well as a longer OS in patients with ER-negative BC
(HR=0.59; P=0.0078) and lymph node positive BC (HR=0.66; P=0.04;
Fig. 8C-I).
 | Figure 7.Prognostic value of CXCR3 in terms of
RFS in BC. (A) High mRNA expression of CXCR3 was significantly
associated with longer RFS in all patients with BC. High mRNA
expression of CXCR3 was associated with longer RFS in patients with
the (B) basal-like, (C) luminal A and (D) luminal B subtypes of BC,
but not in patients with (E) HER2 BC. High mRNA expression of CXCR3
was associated with longer RFS both in (F) ER-positive and (G)
ER-negative patients. High mRNA expression of CXCR3 was associated
with longer RFS in patients with (H) TP53 mutated BC, but not in
patients with (I) TP53 wild type. CXCR, CXC chemokine receptor; BC,
breast cancer; RFS, relapse-free survival; TP53, cellular tumor
antigen p53; ER, estrogen receptor; HER2, human epidermal growth
factor receptor 2; HR, hazard ratio. |
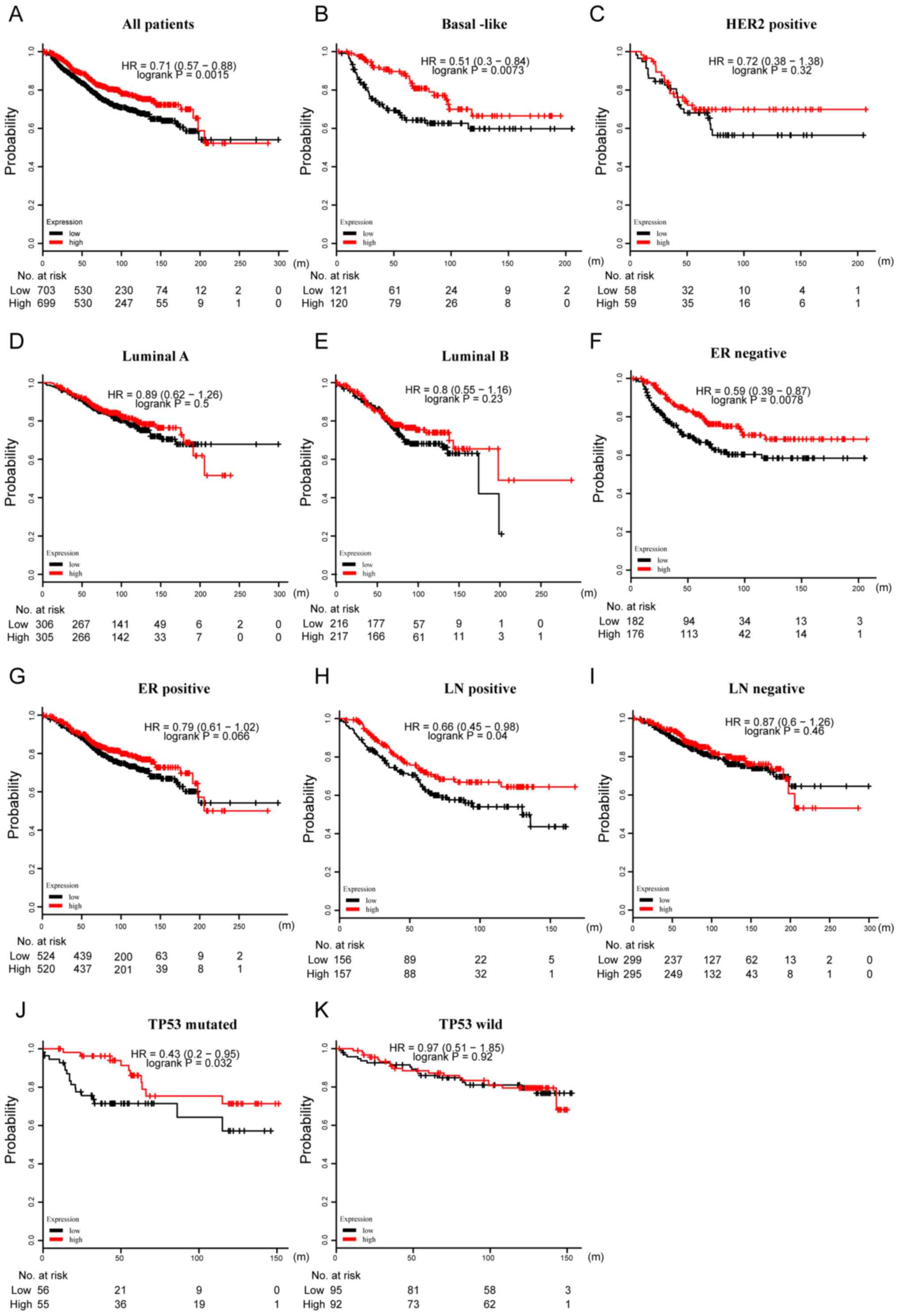 | Figure 8.Prognostic value of CXCR3 in terms of
OS in BC. (A) High mRNA expression of CXCR3 was significantly
associated with longer OS in all patients with BC. High mRNA
expression of CXCR3 was associated with longer OS in patients with
(B) basal-like BC, but not in patients with (C) HER2, (D) luminal A
or (E) luminal B subtypes of BC. High mRNA expression of CXCR3 was
associated with longer OS in patients with (F) ER-negative BC, but
not in patients with (G) ER-positive BC. High mRNA expression of
CXCR3 was associated with longer OS in patients with (H)
LN-positive BC, but not patients with (I) LN-negative BC. High mRNA
expression of CXCR3 was associated with longer RFS in patients with
(J) TP53-mutated BC, but not in patients with (K) TP53 wild-type
BC. CXCR, CXC chemokine receptor; BC, breast cancer; OS, overall
survival; TP53, cellular tumor antigen p53; ER, estrogen receptor;
HER2, human epidermal growth factor receptor 2; LN, lymph node; HR,
hazard ratio. |
Discussion
In the present study, the expression of CXCR family
members was examined in different tumors, and it was indicated that
CXCR4 and CXCR3 were highly expressed in BC. Subsequent analyses
were performed to determine whether a correlation was present
between the expression of CXCR4 and CXCR3 in the different
molecular subtypes of BC, and the survival rates associated with
their expression. The results indicated that the mRNA expression of
CXCR4 was statistically higher in patients with basal-like BC than
in other subtypes, which is consistent with a previous report
(23), and may explain why
basal-like BC has a poorer prognosis than other subtypes. Survival
analysis showed that high CXCR4 mRNA expression in BC promoted the
recurrence of BC, but did not have an impact on OS. There are many
confounding factors influencing OS, including non-cancer-related
mortality and participation in clinical trials. Two previous
meta-analyses (24,25) demonstrated that low CXCR4 mRNA
expression in patients with BC is associated with better survival,
including progression-free survival, disease-free survival and OS.
In the present study, it was found that the impact of low CXCR4
mRNA expression on RFS is consistent with these previous studies
(24,25), while the results showed no
difference in OS. The heterogeneity in OS between the combined
meta-analysis studies is high (I2=70% and P<0.00001;
I2=84.2% and P<0.001) (24,25).
There is a discrepancy in the OS rates among the present study and
previous studies (24,25). Therefore, larger sample-sizes and
high-quality trials are needed to show significance. In the present
study, CXCR4 was found to be involved in BC tumorigenesis and to
act as a prognostic biomarker for BC, based on its high expression
and correlation with survival.
A stratified analysis revealed that high CXCR4
levels in basal-like BC predicted a good clinical outcome, both in
terms of RFS and OS. Basal-like BC accounts for >70% of TNBC
cases. Previous studies using small-sample sizes found the opposite
result and support a negative role of CXCR4 in TNBC (26,27)
CXCR4 is thought to play an important role in promoting the
proliferation, recurrence and metastasis of BC, and may contribute
to an adverse prognosis (28).
However, a recent study reported that CXCR4 inhibitors were not
efficient at inhibiting the growth of TNBC and even promoted the
metastatic spread in 25% of cases (29). Therefore, this previous study
indirectly supports the hypothesis that high CXCR4 expression
predicts a favorable prognosis in TNBC. High CXCR4 mRNA expression
does not always promote migration. For example, Ierano et al
(30) found that histone
deacetylase inhibitors induced CXCR4 mRNA expression but
antagonized CXCR4-mediated migration by inhibiting CXCR4 protein.
In addition, the present study revealed that, as in patients with
basal-like BC, high expression of CXCR4 predicted a better RFS and
OS in patients with ER-negative and TP53-mutated BC, respectively.
This may be because an ER-negative status and TP53 mutation are
features of TNBC (31). However,
not all basal-like BC is TNBC (32), and as such, there may be
discrepancies in the comparison between previous studies and the
present study. Thus, more studies are required to clarify the
relationship between CXCR4 expression and basal-like BC (or TNBC),
and to understand the underlying molecular mechanism.
Next, the relationship between CXCR4 expression in
different subtypes of BC was examined. Patients with basal-like BC
are predominantly treated with chemotherapy. The results of the
present study indicated that patients with basal-like BC and with
high CXCR4 expression after chemotherapy have a more favorable
prognosis, indicating that high expression of CXCR4 may increase
the sensitivity of chemotherapy in basal-like BC. At present, few
studies have reported on the relationship between the expression
level of CXCR4 and the efficacy of chemotherapy, and these studies
have used animal models or cell lines. For example, researchers
found that CXCR4 induces chemoresistance in acute myeloid leukemia
cells (OCI-AML3) and in colon cancer cells (HT-29 and SW480)
(33,34). Furthermore, Liang et al
(35) reported that the silencing
of CXCR4 sensitized TNBC cells to cisplatin. However, chemotherapy
for basal-like BC typically consists of paclitaxel and
anthracyclines. Another previous study showed that patients with BC
have decreased expression of CXCR4 and HER2 after neoadjuvant
chemotherapy, indicating that these two genes may be a part of the
mechanism of chemotherapy in BC (36). These previous studies may not
support the results of the present study; therefore, more clinical
trials are required to elucidate the role of CXCR4 in the efficacy
of different chemotherapy regimens in basal-like BC. Furthermore,
more experiments are required to understand the underlying
molecular mechanism. To the best of our knowledge, the present
study was the first to show that patients with luminal A BC and
with high levels of CXCR4 expression after endocrine therapy have a
shorter RFS than those with a low level of CXCR4 expression.
Therefore, the present study indicated that CXCR4 may be a cause of
resistance to endocrine therapy. Rhodes et al (37) found that the effects of CXCR4
overexpression were correlated with stromal cell-derived
factor-1-mediated activation of downstream signaling through ERK1/2
and p38 MAPK. CXCR4 overexpression was also associated with
increased ER-mediated gene expression, indicating that increased
CXCR4 signaling is sufficient to drive ER-positive breast cancer to
a metastatic and endocrine therapy-resistant phenotype through
enhanced MAPK signaling. As patients with luminal A BC are usually
only treated with adjuvant endocrine therapy (38), the efficacy of treatment can be
predicted using CXCR4 expression.
The number of studies about CXCR3 in BC is fewer
than that for CXCR4. CXCR3 has three isoforms, CXCR3-A, CXCR3-B and
CXCR3-alt. CXCR3-A and CXCR3-B are the predominant isoforms and
have different roles in BC; signaling through CXCR3-A promotes
tumor growth while CXCR3-B prevents cancer cell proliferation
(39). A previous study found that
CXCR3 inhibition is effective in both BC and host compartments
(40), while another previous
study revealed that CXCR3 deficiency induced cancer development by
promoting macrophage M2 polarization in a murine BC model (41). Thus, CXCR3 has multifaceted roles;
it mediates the recruitment of tumor-infiltrating lymphocytes into
the cancer microenvironment, resulting in a favorable clinical
outcome by inhibiting tumor development and metastasis, and high
expression of CXCR3 can promote tumor cell proliferation, migration
and invasion, contributing to poor survival rates for patients
(42). In the present study, it
was found that CXCR3 is a favorable factor in several subtypes of
BC, this is especially the case in basal-like BC.
In conclusion, CXCR4 and CXCR3 are significantly
highly expressed in BC in comparison with normal samples. CXCR4 was
found to be an adverse prognostic factor in BC; however, for
basal-like BC, CXCR4 predicted a better prognosis. CXCR3 was found
to be a favorable predictive factor in patients with BC.
Furthermore, CXCR4 promoted chemosensitivity in patients with
basal-like BC and induced resistance to endocrine therapy in
patients with luminal A BC.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Program for
the Cultivation of Youth talents in China Association of Chinese
Medicine (SR; grant no. QNRC2-C08; http://www.cacm.org.cn/), the Zhejiang Provincial
Program for the Cultivation of High-Level Innovative Health Talents
(SR; grant no. 2015-43; http://www.zjwjw.gov.cn/) and the Zhejiang Provincial
Program for the Cultivation of the Young and Middle-Aged Academic
Leaders in Colleges and Universities (SR; grant no. 2017-248;
http://www.zjedu.gov.cn/).
Availability of data and materials
The datasets generated and analyzed during the
current study are available in the Oncomine database (http://www.oncomine.org), the GOBO database
(http://co.bmc.lu.se/gobo) and the KM plotter tool
(http://kmplot.com/analysis/).
Authors' contributions
SR, KG and MS contributed to the study design. KZ
and FS conducted the data collection. QY and LS performed the
statistical analysis. KG and GF interpreted the data. KG, SR and GF
prepared the manuscript. KG and GF performed the literature search.
SR and MS were responsible for funds collection.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nakasone ES, Hurvitz SA and McCann KE:
Harnessing the immune system in the battle against breast cancer.
Drugs Context. 7:2125202018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Balkwill FR: The chemokine system and
cancer. J Pathol. 226:148–157. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zlotnik A, Burkhardt AM and Homey B:
Homeostatic chemokine receptors and organ-specific metastasis. Nat
Rev Immunol. 11:597–606. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhu Q, Han X, Peng J, Qin H and Wang Y:
The role of CXC chemokines and their receptors in the progression
and treatment of tumors. J Mol Histol. 43:699–713. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Vandercappellen J, Van Damme J and Struyf
S: The role of CXC chemokines and their receptors in cancer. Cancer
Lett. 267:226–244. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Holmes WE, Lee J, Kuang WJ, Rice GC and
Wood WI: Structure and functional expression of a human
interleukin-8 receptor. Science. 253:1278–1280. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Murphy PM and Tiffany HL: Cloning of
complementary DNA encoding a functional human interleukin-8
receptor. Science. 253:1280–1283. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kunsch C and Rosen CA: NF-kappa B
subunit-specific regulation of the interleukin-8 promoter. Mol Cell
Biol. 13:6137–6146. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ha H, Debnath B and Neamati N: Role of the
CXCL8-CXCR1/2 axis in cancer and inflammatory diseases.
Theranostics. 7:1543–1588. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tokunaga R, Zhang W, Naseem M, Puccini A,
Berger MD, Soni S, McSkane M, Baba H and Lenz HJ: CXCL9, CXCL10,
CXCL11/CXCR3 axis for immune activation-A target for novel cancer
therapy. Cancer Treat Rev. 63:40–47. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gupta SK, Lysko PG, Pillarisetti K,
Ohlstein E and Stadel JM: Chemokine receptors in human endothelial
cells. Functional expression of CXCR4 and its transcriptional
regulation by inflammatory cytokines. J Biol Chem. 273:4282–4287.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Coke CJ, Scarlett KA, Chetram MA, Jones
KJ, Sandifer BJ, Davis AS, Marcus AI and Hinton CV: Simultaneous
activation of induced heterodimerization between CXCR4 chemokine
receptor and cannabinoid receptor 2 (CB2) reveals a mechanism for
regulation of tumor progression. J Biol Chem. 291:9991–10005. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Forster R, Mattis AE, Kremmer E, Wolf E,
Brem G and Lipp M: A putative chemokine receptor, BLR1, directs B
cell migration to defined lymphoid organs and specific anatomic
compartments of the spleen. Cell. 87:1037–1047. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mitkin NA, Hook CD, Schwartz AM, Biswas S,
Kochetkov DV, Muratova AM, Afanasyeva MA, Kravchenko JE,
Bhattacharyya A and Kuprash DV: p53-dependent expression of CXCR5
chemokine receptor in MCF-7 breast cancer cells. Sci Rep.
5:93302015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Szpakowska M, Meyrath M, Reynders N,
Counson M, Hanson J, Steyaert J and Chevigné A: Mutational analysis
of the extracellular disulphide bridges of the atypical chemokine
receptor ACKR3/CXCR7 uncovers multiple binding and activation modes
for its chemokine and endogenous non-chemokine agonists. Biochem
Pharmacol. 153:299–309. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ringner M, Fredlund E, Hakkinen J, Borg A
and Staaf J: GOBO: Gene expression-based outcome for breast cancer
online. PLoS One. 6:e179112011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Goldhirsch A, Winer EP, Coates AS, Gelber
RD, Piccart-Gebhart M, Thurlimann B and Senn HJ; Panel members, :
Personalizing the treatment of women with early breast cancer:
Highlights of the St Gallen international expert consensus on the
primary therapy of early breast cancer 2013. Ann Oncol.
24:2206–2223. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Curtis C, Shah SP, Chin SF, Turashvili G,
Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et
al: The genomic and transcriptomic architecture of 2,000 breast
tumours reveals novel subgroups. Nature. 486:346–352. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Finak G, Bertos N, Pepin F, Sadekova S,
Souleimanova M, Zhao H, Chen H, Omeroglu G, Meterissian S, Omeroglu
A, et al: Stromal gene expression predicts clinical outcome in
breast cancer. Nat Med. 14:518–527. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Neve RM, Chin K, Fridlyand J, Yeh J,
Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, et al: A
collection of breast cancer cell lines for the study of
functionally distinct cancer subtypes. Cancer Cell. 10:515–527.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Farmer P, Bonnefoi H, Becette V,
Tubiana-Hulin M, Fumoleau P, Larsimont D, Macgrogan G, Bergh J,
Cameron D, Goldstein D, et al: Identification of molecular apocrine
breast tumours by microarray analysis. Oncogene. 24:4660–4671.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang M, Liu HX, Teng XD, Wang HB, Cui J,
Jia SS, Gu XY and Li ZG: The differences in CXCR4 protein
expression are significant for the five molecular subtypes of
breast cancer. Ultrastruct Pathol. 36:381–386. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xu TP, Shen H, Liu LX and Shu YQ: The
impact of chemokine receptor CXCR4 on breast cancer prognosis: A
meta-analysis. Cancer Epidemiol. 37:725–731. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang F, Li S, Zhao Y, Yang K, Chen M, Niu
H, Yang J, Luo Y, Tang W and Sheng M: Predictive role of the
overexpression for CXCR4, C-Met, and VEGF-C among breast cancer
patients: A meta-analysis. Breast. 28:45–53. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen HW, Du CW, Wei XL, Khoo US and Zhang
GJ: Cytoplasmic CXCR4 high-expression exhibits distinct poor
clinicopathological characteristics and predicts poor prognosis in
triple-negative breast cancer. Curr Mol Med. 13:410–416. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chu QD, Panu L, Holm NT, Li BD, Johnson LW
and Zhang S: High chemokine receptor CXCR4 level in triple negative
breast cancer specimens predicts poor clinical outcome. J Surg Res.
159:689–695. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Dayer R, Babashah S, Jamshidi S and
Sadeghizadeh M: Upregulation of CXC chemokine receptor 4-CXC
chemokine ligand 12 axis ininvasive breast carcinoma: A potent
biomarker predicting lymph node metastasis. J Cancer Res Ther.
14:345–350. 2018.PubMed/NCBI
|
|
29
|
Lefort S, Thuleau A, Kieffer Y, Sirven P,
Bieche I, Marangoni E, Vincent-Salomon A and Mechta-Grigoriou F:
CXCR4 inhibitors could benefit to HER2 but not to triple-negative
breast cancer patients. Oncogene. 36:1211–1222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ierano C, Basseville A, To KK, Zhan Z,
Robey RW, Wilkerson J, Bates SE and Scala S: Histone deacetylase
inhibitors induce CXCR4 mRNA but antagonize CXCR4 migration. Cancer
Biol Ther. 14:175–183. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Duffy MJ, Synnott NC and Crown J: Mutant
p53 in breast cancer: Potential as a therapeutic target and
biomarker. Breast Cancer Res Treat. 170:213–219. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Rakha EA, Elsheikh SE, Aleskandarany MA,
Habashi HO, Green AR, Powe DG, El-Sayed ME, Benhasouna A, Brunet
JS, Akslen LA, et al: Triple-negative breast cancer: Distinguishing
between basal and nonbasal subtypes. Clin Cancer Res. 15:2302–2310.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chen Y, Jacamo R, Konopleva M, Garzon R,
Croce C and Andreeff M: CXCR4 downregulation of let-7a drives
chemoresistance in acute myeloid leukemia. J Clin Invest.
123:2395–2407. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Heckmann D, Maier P, Laufs S, Wenz F,
Zeller WJ, Fruehauf S and Allgayer H: CXCR4 expression and
treatment with SDF-1α or plerixafor modulate proliferation and
chemosensitivity of colon cancer cells. Transl Oncol. 6:124–132.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liang S, Peng X, Li X, Yang P, Xie L, Li
Y, Du C and Zhang G: Silencing of CXCR4 sensitizes triple-negative
breast cancer cells to cisplatin. Oncotarget. 6:1020–1030. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yang SX, Loo WT, Chow LW, Yang XH, Zhan Y,
Fan LJ, Zhang F, Chen L, Wang QL, Xiao HL, et al: Decreased
expression of C-erbB-2 and CXCR4 in breast cancer after primary
chemotherapy. J Transl Med. 10 (Suppl 1):S32012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Rhodes LV, Short SP, Neel NF, Salvo VA,
Zhu Y, Elliott S, Wei Y, Yu D, Sun M, Muir SE, et al: Cytokine
receptor CXCR4 mediates estrogen-independent tumorigenesis,
metastasis, and resistance to endocrine therapy in human breast
cancer. Cancer Res. 71:603–613. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Park S, Lee SK, Paik HJ, Ryu JM, Kim I,
Bae SY, Yu J, Kim SW, Lee JE and Nam SJ: Adjuvant endocrine therapy
alone in patients with node-positive, luminal A type breast cancer.
Medicine. 96:e67772017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ma B, Khazali A and Wells A: CXCR3 in
carcinoma progression. Histol Histopathol. 30:781–792.
2015.PubMed/NCBI
|
|
40
|
Zhu G, Yan HH, Pang Y, Jian J, Achyut BR,
Liang X, Weiss JM, Wiltrout RH, Hollander MC and Yang L: CXCR3 as a
molecular target in breast cancer metastasis: Inhibition of tumor
cell migration and promotion of host anti-tumor immunity.
Oncotarget. 6:43408–43419. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Oghumu S, Varikuti S, Terrazas C, Kotov D,
Nasser MW, Powell CA, Ganju RK and Satoskar AR: CXCR3 deficiency
enhances tumor progression by promoting macrophage M2 polarization
in a murine breast cancer model. Immunology. 143:109–119. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Bronger H, Karge A, Dreyer T, Zech D,
Kraeft S, Avril S, Kiechle M and Schmitt M: Induction of cathepsin
B by the CXCR3 chemokines CXCL9 and CXCL10 in human breast cancer
cells. Oncol Lett. 13:4224–4230. 2017. View Article : Google Scholar : PubMed/NCBI
|