Introduction
Liver regeneration occurs after events that could
lead to the loss of liver mass, including surgery, trauma,
infection and liver transplantation (1). During liver regeneration, and
following hepatectomy, liver cell proliferation and apoptosis
occur, and liver size is largely controlled by liver cell
proliferation (2,3). Subsequent hepatocyte stimulation is
triggered by multiple feedback signals, allowing hepatocytes to
enter a rapid growing phase. The clinical implications of liver
regeneration are extremely valuable, as it enables recovery from
the symptoms of surgical damage, such as reduced liver size and
dysfunction; conversely, poor liver regeneration is likely to
result in patient fatality (4).
Thus, the explicit functional mechanism of liver regeneration is
required in order to amplify its full clinical potential following
surgery. Previous studies have highlighted a variety of regulatory
factors for the hepatocyte cell cycle and the appropriate
progression of liver regeneration, including the release of
inflammatory cytokines (5),
hepatocyte growth factor (HGF) (6)
and metabolic regulatory factors, and cell cycle regulation. It has
also been demonstrated that certain types of non-coding RNAs such
as long non-coding RNAs (lncRNAs) and micro (mi)RNAs, play
important roles in liver regeneration (7,8),
though the precise molecular mechanisms of the associated pathways
remain unclear.
lncRNAs are non-coding transcripts (>200
nucleotides) which have been widely demonstrated as vital
regulators of numerous cellular responses, developmental processes
and disease (9). Emerging evidence
has shown that lncRNAs may be functionally significant in the
progression of liver regeneration (10,11).
For instance, lncRNA Metastasis Associated Lung Adenocarcinoma
Transcript 1 (MALAT1) was recently reported to promote hepatocyte
proliferation by stimulating the Wnt/β-catenin signaling pathway
(1).
Small nucleolar RNA host gene 12 (SNHG12) is a newly
identified lncRNA that is associated with various malignancies, and
is considered to be a useful tumor biomarker (12). It was recently demonstrated that
SNHG12 promoted cellular proliferation and metastasis, which
regulated the Wnt/β-catenin signaling pathway in papillary thyroid
carcinoma cells; the same interplay was detected in an in
vivo disease model (13). In
addition, other studies have revealed that SNHG12 plays a
regulatory role in glioma (14),
osteosarcoma (15), and gastric
carcinoma (16), by acting as a
sponge for associated micro (mi)RNAs. However, the precise
mechanism by which SNHG12 regulates hepatocyte proliferation in
liver regeneration is yet to be elucidated. The aim of the present
study was to investigate the underlying mechanism of
SNHG12-mediated hepatocyte proliferation during liver
regeneration.
Materials and methods
Animals and 2/3 partial hepatectomy
(PH) model
Prior to animal experimentation, the present study
was approved by the Institutional Animal Ethics Committee of the
Second Military Medical University. Male BALB/c mice (6–8 weeks of
age) were obtained from Beijing Vital River Laboratory Animal
Technology Co., Ltd. The mice were maintained in sterile conditions
with free access to food and water, under a 12-h light/dark cycle.
2/3 PH was performed according to previously described methods
(17), where the left and median
liver lobes were removed under isoflurane anesthesia (2% v/v).
Euthanasia was carried out by CO2 inhalation for 5 min.
Humane endpoints included weight loss of >20%, dehydration,
severe lameness, ruffled fur, and hunched appearance. None of mice
exhibited the humane endpoint criteria. The liver tissues were
collected and the liver/body weight ratio was calculated on days 0,
1, 2, 3, 4, 5, 6, 7, 8, 9 and 10 after 2/3 PH.
Mouse primary hepatocytes
isolation
Mouse primary hepatocytes were isolated from the
livers of 2/3 PH model mice and control mice. Liver tissues were
fragmented using ophthalmic scissors, and the resulting tissue
pieces were washed with PBS containing with 100 U/ml of penicillin
and 100 mg/ml of streptomycin. The liver tissues were then digested
(0.25% trypsin; 0.04% EDTA), filtrated through a 4 µm cell strainer
and centrifuged for 5 min at 350 × g at 4°C. Then, mouse primary
hepatocytes (5–6×105/ml) were suspended and cultured in
high glucose DMEM (Gibco; Thermo Fisher Scientific, Inc.) with 10%
FBS (Gibco; Thermo Fisher Scientific, Inc.) and 1%
penicillin-streptomycin at 37°C in a humidified atmosphere of 5%
CO2, with medium replaced every 2 days. Cells were
subcultured for 3–4 days until confluence reached to 70–80%. Mouse
primary hepatocytes were treated with different concentrations of
HGF (5, 10, 20, 30, and 40 ng/µl).
Cell culture and transfection
The NCTC 1469 and BNL CL.2 mouse hepatocyte cell
lines were purchased from the American Type Culture Collection, and
cultured in DMEM containing 10% fetal bovine serum at 37°C with (5%
CO2). NCTC 1469 cells were treated with HGF (20 ng/µl)
or the Wnt inhibitor IWR-1 (60 µM) at 37°C for 24 h. Recombinant
lentivirus (Lv)-SNHG12 and the scrambled control (Lv-NC) were
constructed by Shanghai GeneChem Co., Ltd. The cells were
transfected using Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) by exposure to diluted viral supernatant
containing polybrene for 48 h, as previously described (18). SNHG12 small interfering (si)RNA was
purchased from ZHBY Biotech Co. Ltd. and scramble siRNA was used as
the control.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA from tissues and cells was extracted using
a TRIzol® kit (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's instructions. Reverse
transcription (RT) was carried out using a PrimeScript RT reagent
Kit (Takara Bio, Inc., Tokyo, Japan). The RT system of 10 µl was
carried out according to the manufacturer's instructions. The RT
conditions were 37°C for 15 min and 85°C for 5 sec. mRNA and lncRNA
expression levels were determined using the SYBR Green Supermix
(Invitrogen; Thermo Fisher Scientific, Inc.) on an Applied
Biosystems 7300 real-time PCR system. Thermocycling conditions were
95°C for 10 min, following by 35 cycles at 95°C for 10 sec, 58°C
for 15 sec and 72°C for 20 sec, and a final 72°C for 20 min.
β-actin was used as the internal control. All qPCR experiments were
performed at ≥3 times, and the primer sequences were as follows:
SNHG12 forward, 5′-CATCAAGACTGAGAAAAAGCACACC-3′ and reverse,
5′-TACCTTAAAGCACAGCTCCAGAAAC-3′; Axin2 forward,
5′-GATGTTGGAGAGTGAGCGGCAG-3′ and reverse,
5′-TGTTGGGTGGGGTAAGGGGAG-3′; β-actin forward,
5′-CAACTGGGACGACATGGAG-3′ and reverse,
5′-TAGCACAGCCTGGATAGCAAC-3′.
Western blot analysis
The total protein from liver tissues and hepatocytes
was extracted and homogenized in RIPA buffer (Beijing Solarbio
Science & Technology Co., Ltd.). The concentrations of the
extracted nuclear and cytoplasmic fractions were quantified using
the bicinchoninic acid assay method. A total of 50 µg protein per
sample was separated by SDS-PAGE (10%) and then transferred to a
PVDF membrane, prior to blocking with 5% non-fat milk in 1X TBST,
overnight at 4°C. The membrane was then incubated with
anti-β-catenin (1:1,000; product no. ab32572), anti-β-actin
(1:5,000; product no. ab8227), anti-histone 3 (1:2,000; product no.
ab62642; all from Abcam) primary antibodies for 2 h at room
temperature. After washing three times in 1X TBST, the membranes
were incubated with the corresponding HRP-conjugated secondary
antibody (1:5,000; cat. no. A6154-1 ml; Sigma-Aldrich; Merck KGaA)
for 1 h at room temperature. The membranes were washed once more,
exposed to ECL (Sangon Biotech Co., Ltd.), and then photographed by
ChemiDoc™ XRS+ Imaging system (Bio-Rad Laboratories, Inc.). The
protein expression levels were quantified using ImageJ software
version 1.46 (National Institutes of Health). Histone 3 and β-actin
were used as internal controls.
Cell proliferation assay
Cell Counting Kit (CCK)-8 reagent (Biotech Well) was
used to determine the level of cellular proliferation, per the
manufacturer's protocol. Briefly, NCTC 1469 or BNL CL.2 cells were
seeded into 96-well plates at a density of 3×105/ml, and
transfected with the aforementioned constructs for 48 h. CCK-8
reagent was then added to each well and the cells were incubated
for ~30 min at 37°C. The absorbance was measured by a microplate
reader (Bio-Rad Laboratories, Inc.) at a wavelength of 450 nm. The
number of cells was calculated in reference to a standard curve
obtained under the same conditions.
NCTC 1469 or BNL CL.2 cells were seeded at
3×103 cells/well in 96-well plates. After 2 days of
culture, proliferation was assessed using Cell Proliferation ELISA,
BrdU (colorimetric) kit (product no. 11647229001; Roche), per the
manufacturer's protocol. Briefly, BrdU-labeling solution (100 µM)
was added into each well and incubated for 4 h at 37°C. The
absorbance was measured at 450 nm by a microplate reader (Infinite
200; Tecan Group).
Immunohistochemistry (IHC)
The liver specimens were fixed using formalin (10%)
and embedded in paraffin. Serial sections of 5-µm thickness were
sliced from the paraffin embedded blocks, and used for subsequent
experiments. The sections were incubated overnight at 4°C with an
anti-proliferating cell nuclear antigen (PCNA) primary antibody
(1:500; product no. ab29; Abcam), then washed thrice prior to
subsequent incubation with an HRP-conjugated goat anti-rabbit
antibody (1:200; product no. 5571; Cell Signaling Technology,
Inc.). The sections were then stained with DAB (Sangon Biotech Co.,
Ltd.) for 5 min at 37°C to observe the expression of PCNA.
Luciferase reporter assay
NCTC 1469 or BNL CL.2 cells (5×104/well)
were seeded into 24-well plates. TOP-FLASH or FOP-FLASH constructs
and Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.)
were used to transfect the cells and luciferase activity was
measured using the Dual-Luciferase Reporter Assay System (Promega
Corporation) as previously described (19). Luciferase activity was normalized
to that in cells transfected with TOPFlash/FOPFlash.
Statistical analysis
Statistical analysis was conducted using SPSS
software version 16 (SPSS, Inc.). All data are presented as the
mean ± SD (standard deviation) from three or more separate
experiments. Statistical significances between two groups were
analyzed using one-way ANOVA followed by the Scheffé test (Figs. 2C-F, 3 and 5),
or two-tailed Student's t-test (Figs.
1, 2A and B, 4 and Fig.
S1). P<0.05 was considered to indicate a statistically
significant difference.
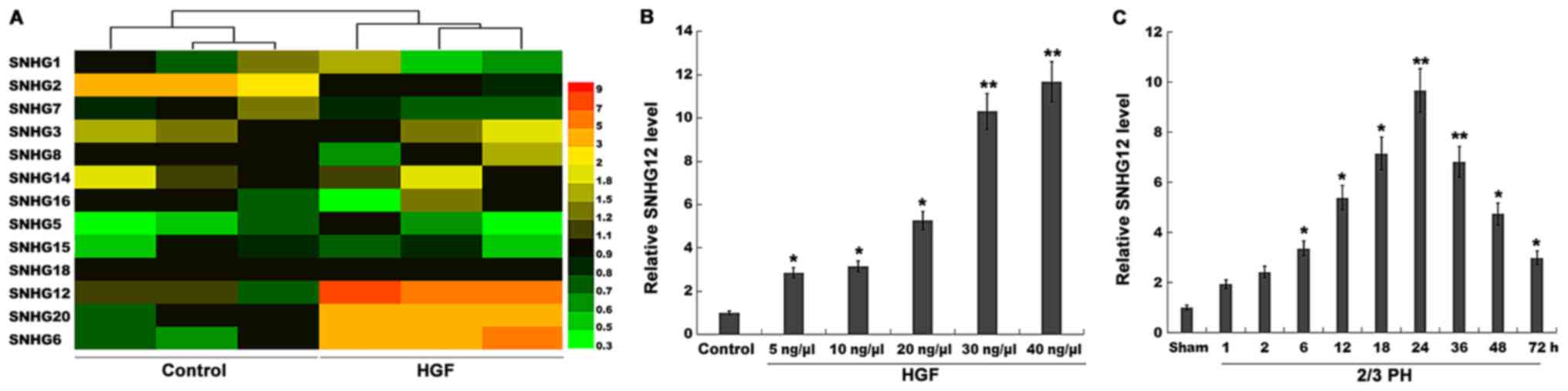 | Figure 1.SNHG12 expression is significantly
upregulated after 2/3 PH. (A) Heatmap of RT-qPCR data for the
expression of 13 liver cancer-related SNHGs (SNHG1, SNHG2, SNHG3,
SNHG5, SNHG6, SNHG7, SNHG8, SNHG12, SNHG14, SNHG15, SNHG16, SNHG18,
and SNHG20). NCTC 1469 cells were treated with HGF (20 ng/µl), and
then total RNA was extracted and used to perform RT-qPCR analysis.
(B) RT-qPCR analysis of SNHG12 expression level in primary mouse
hepatocytes (2×105 cells/well) after treatment with
different concentrations of HGF (5, 10, 20, 30 and 40 ng/µl). (C)
RT-qPCR analysis of SNHG12 expression in liver tissues (n=5) at
different time-points after 2/3 PH. *P<0.05 and **P<0.01.
SNHG12, small nucleolar RNA host gene 12; PH, partial hepatectomy;
RT-qPCR, reverse transcription-quantitative PCR; HGF, hepatocyte
growth factor. |
Results
SNHG12 expression is significantly
upregulated after 2/3 PH
lncRNAs play crucial roles in the regulation of
omnigenous cellular activities, which may provide unique functions
and improve the regenerative ability of the liver. Recently, the
role of SNHGs in tumor progression was verified (20–22),
but their role in liver regeneration remains unexplored. In the
present study, 13 liver cancer-related SNHGs (SNHG1, SNHG2, SNHG3,
SNHG5, SNHG6, SNHG7, SNHG8, SNHG12, SNHG14, SNHG15, SNHG16, SNHG18
and SNHG20) were assayed using RT-qPCR. Since HGF is a key factor
in liver regeneration (23,24),
the expression of these SNHGs was assessed in NCTC 1469 cells
following treatment with HGF. Among the 13 SNHGs, the expression
level of SNHG2 was decreased, and that of SNHG6, 12 and 20 was
significantly increased after HGF treatment (Fig. 1A). SNHG12 was selected for further
analysis, since its level of upregulation was the most
significant.
Mouse primary hepatocytes were isolated and treated
with HGF. Fig. 1B revealed that in
primary hepatocytes, the expression level of SNHG12 was upregulated
in a dose-dependent manner following HGF treatment. Therefore, 2/3
PH was performed and the SNHG12 level was assessed once more. The
SNHG12 expression was also significantly increased at various
time-points after 2/3 PH (Fig.
1C); 2/3 PH in mice resulted in enhanced SNHG12 levels that
were detectable at 6 h, that peaked between 24 h, and that had
returned to almost normal levels by 72 h. Thus, the time-dependent
alterations in SNHG12 expression indicated that SNHG12 may
potentially be involved in liver regeneration.
SNHG12 promotes hepatocyte
proliferation in vitro and in vivo
To investigate the role of SNHG12 in hepatocyte
proliferation, a CCK-8 assay was performed using normal mouse liver
cell lines (NCTC 1469 cells and BNL CL.2 cells) following
SNHG12-overexpression or -knockdown. Fig. 2A and B revealed that SNHG12
expression was significantly increased in NCTC 1469 cells after
treatment with Lv-SNHG12, and decreased after treatment with
SNHG12-specific siRNA (siSNHG12). Furthermore, NCTC 1469 hepatocyte
proliferation was significantly increased by the overexpression of
SNHG12 (Fig. 2C), which also
promoted BNL CL.2 cell proliferation (Fig. 2D), whereas SNHG12-knockdown
decreased NCTC 1469 and BNL CL.2 cell proliferation (Fig. 2C and D).
The results of the BrdU ELISA assays also indicated
that hepatocyte proliferation was increased after
SNHG12-overexpression, and suppressed by SNHG12-knockdown (Fig. 2E and F). Subsequently, to assess
hepatocyte proliferation in vivo, the expression level of
PCNA was investigated by IHC in mouse-model tissues. At 36 h
post-2/3 PH, SNHG12-treated mice exhibited a higher number of
PCNA-positive nuclei in the hepatocytes (Fig. 3A and B). Furthermore, the role of
SNHG12 in regulating liver regeneration was assayed. As revealed in
Fig. 3C, after 2/3 PH the
liver/body weight ratio of mice overexpressing SNHG12 was
significantly greater than that of the control mice. Conversely,
SNHG12-inhibition suppressed hepatocyte proliferation in
vivo and reduced the liver/body weight ratio (Fig. 3A-C). These data indicated that
SNHG12 upregulation contributed to hepatocyte proliferation in
vitro and in vivo.
SNHG12 activates the Wnt/β-catenin
signaling pathway in hepatocytes
Previous studies have demonstrated the Wnt/β-catenin
pathway as the key factor in triggering liver regeneration after PH
(25). Furthermore, lncRNAs can
control liver regeneration by regulating Wnt/β-catenin signalling
(10). lncRNA SNHG12 was revealed
to promote cellular proliferation and metastasis by activating the
Wnt/β-catenin pathway in papillary thyroid cancer (13). Therefore, the present study sought
to investigate whether SNHG12 regulated Wnt/β-catenin in
hepatocytes following PH. A TOPflash/FOPflash assay was first
carried out to verify the role of SNHG12 in Wnt regulation.
Fig. 4A and B indicated that the
relative luciferase activity of BNL CL.2 and NCTC 1469 cells was
significantly increased following SNHG12 overexpression, whereas
SNHG12 knockdown suppressed the luciferase activity, compared with
the control. When Wnt signaling is activated, β-catenin is
dephosphorylated and translocates into the nucleus, where it
promotes the expression of downstream target genes (26,27).
In the present study, the effects of the SNHG12-induced nuclear
translocation of β-catenin were also investigated. SNHG12-induced
nuclear translocation was further investigated using
immunofluorescence (Fig. 4C). When
SNHG12 was overexpressed in NCTC 1469 cells, β-catenin nuclear
translocation was markedly increased (Fig. 4D), whereas SNHG12-knockdown
inhibited the nuclear translocation of β-catenin (Fig. 4E). Axin-2, a downstream target of
Wnt/β-catenin signaling, was also enhanced in BNL CL.2 cells after
SNHG12 overexpression, yet suppressed following SNHG12 knockdown,
compared with the control (Fig. 4F and
G). These results demonstrated that SNHG12 is a regulator of
Wnt/β-catenin signaling.
SNHG12 accelerates liver regeneration
by influencing Wnt signaling
To investigate the role of the SNHG12/Wnt pathway in
the regulation of liver regeneration following 2/3 PH, SNHG12 was
overexpressed and Wnt was concurrently inhibited using a specific
inhibitor (IWR-1); hepatocyte proliferation and liver regeneration
were then assessed in vitro and in vivo. As revealed
in Fig. 5A and B, the cell
proliferation of mouse hepatocytes was significantly increased
after SNHG12 overexpression, whereas IWR-1-induced Wnt inhibition
attenuated SNHG12-associated cellular proliferation. As
anticipated, SNHG12 overexpression was revealed in vivo, as
indicated by increased nuclear translocation of β-catenin (Fig. S1). The effect of SNHG12
overexpression on hepatocyte proliferation and liver regeneration
was thus investigated in vivo. After 2/3 PH, the
SNHG12-treated mice exhibited a larger number of PCNA-positive
hepatocyte nuclei than those in the control group (Fig. 5C). However, Wnt inhibition
partially suppressed SNHG12-associated hepatocyte proliferation
in vivo. Furthermore, the liver/body weight ratio of
SNHG12-treated mice was significantly increased. Conversely, Wnt
inhibition attenuated SNHG12-induced liver regeneration after 2/3
PH (Fig. 5D). These results
demonstrated that PH-induced upregulation of SNHG12 accelerated
hepatocyte proliferation and liver regeneration by activating
Wnt/β-catenin signaling.
Discussion
In the present study, the role of SNHG12-associated
hepatocyte proliferation and liver regeneration after 2/3 PH was
verified. The data demonstrated that: i) SNHG12 expression was
significantly increased following 2/3 PH; ii) SNHG12 enhanced
hepatocyte proliferation in vitro and in vivo, and
promoted liver regeneration in vivo; iii) SNHG12 contributed
to the activation of Wnt signaling; and iv) pharmacological
inhibition of Wnt partially attenuated SNHG12-induced liver
regeneration. These results demonstrated the function of the
SNHG12/Wnt axis in promoting hepatocyte proliferation and liver
regeneration after 2/3 PH, and indicate a novel therapeutic method
for liver failure and transplantation.
It is acknowledged that different cytokines, growth
factors and miRNAs regulate the genes responsible for cellular
proliferation during liver regeneration (28), whereas lncRNAs have rarely been
discussed in this context. In the present study, a comprehensive
analysis of the expression of 13 SNHG lncRNAs in HGF-treated cell
lines was performed, which indicated an upregulation in SNHG12
expression. The regulatory role of SNHG12 was then investigated in
association with hepatocyte proliferation. In vitro results
displayed a clear positive relationship between hepatocyte
proliferation and SNHG12. This relationship was also demonstrated
in vivo, as SNHG12 overexpression increased the liver mass
of 2/3 PH mice. A similar finding was previously reported, where
lncRNA MALAT1 was indicated as an activator of cellular
proliferation and cell cycle progression in hepatocytes (1). Collectively, the significance of
lncRNAs in liver regeneration is associated with their ability to
promote hepatocyte proliferation.
Previous studies have widely reported that the
Wnt/β-catenin pathway plays a primary role in organogenesis,
homeostasis and various human diseases (29). In terms of liver regeneration,
several studies have revealed the functional role of Wnt/β-catenin
signaling; the literature demonstrated that the Wnt/β-catenin
pathway is involved in Brahma-related gene 1-mediated liver
regeneration (30), and there is
direct evidence that Wnt agonists promote liver regeneration after
small-for-size liver transplantation in vivo (31). Given that the interplay between the
Wnt/β-catenin pathway and lncRNAs has been reported in numerous
diseases, including liver cancer, it is likely that SNHG12
accelerates liver regeneration by activating Wnt signaling. In the
present study, the activation of Wnt signaling was induced by
SNHG12 overexpression, whereas siRNA-SNHG12 suppressed the nuclear
translocation of β-catenin, decreasing Wnt/β-catenin activity.
Consistent with previous findings of lncRNA-LALR1-associated
activation of Wnt/β-catenin signaling in vivo (10), these results indicated a similar
regulatory role for SNHG12 in liver cells.
Finally, the use of the Wnt inhibitor IWR-1 revealed
that the promotion of hepatocyte proliferation was partially
attenuated, further confirming that SNHG12 may exert its
regenerative role by activating Wnt signaling. Subsequent in
vivo studies confirmed the aforementioned conclusions, where
the number of hepatocytes, as well as liver mass, were decreased
upon IWR-1 administration.
Collectively, the results of the present study
demonstrated the potential regulatory role of SNHG12 in liver
regeneration. In vitro and in vivo studies revealed
that SNHG12 promoted hepatocyte proliferation by activating the Wnt
signaling pathway. This indicates that SNHG12 may be used as a
biomarker of prognosis for liver diseases and the success of
resection.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the
National Natural Science Foundation of China (grant no. 81770613)
and the Science and Technology Commission of Shanghai Municipality
(grant no. 19411967000).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
YaZ and ZQ participated in the design of the main
research ideas and manuscript correction. YiZ edited the manuscript
and conducted the experiments. BL and XJ performed statistical
evaluation of the final data and agreed to publish the paper. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Institutional
Animal Ethics Committee of the Second Military Medical
University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing of
interests.
References
|
1
|
Li C, Chang L, Chen Z, Liu Z, Wang Y and
Ye Q: The role of lncRNA MALAT1 in the regulation of hepatocyte
proliferation during liver regeneration. Int J Mol Med. 39:347–356.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tannuri AC, Tannuri U, Wakamatsu A, Mello
ES, Coelho MC and Dos Santos NA: Effect of the immunosuppressants
on hepatocyte proliferation and apoptosis in a young animal model
of liver regeneration: An immunohistochemical study using tissue
microarrays. Pediatr Transplant. 12:40–46. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mangnall D, Bird NC and Majeed AW: The
molecular physiology of liver regeneration following partial
hepatectomy. Liver Int. 23:124–138. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dutkowski P, Linecker M, Deoliveira ML,
Mullhaupt B and Clavien P: Challenges to liver transplantation and
strategies to improve outcomes. Gastroenterology. 148:307–323.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Selzner N, Selzner M, Tian Y, Kadry Z and
Clavien PA: Cold ischemia decreases liver regeneration after
partial liver transplantation in the rat: A
TNF-alpha/IL-6-dependent mechanism. Hepatology. 36:812–818. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pediaditakis P, Lopeztalavera JC, Petersen
BE, Monga SPS and Michalopoulos GK: The processing and utilization
of hepatocyte growth factor/scatter factor following partial
hepatectomy in the rat. Hepatology. 34:688–693. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shu J, Kren BT, Xia Z, Wong PY, Li L,
Hanse EA, Min MX, Li B, Albrecht JH, Zeng Y, et al: Genomewide
microRNA down-regulation as a negative feedback mechanism in the
early phases of liver regeneration. Hepatology. 54:609–619. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Castro RE, Ferreira DMS, Zhang X, Borralho
PM, Sarver AL, Zeng Y, Steer CJ, Kren BT and Rodrigues CM:
Identification of microRNAs during rat liver regeneration after
partial hepatectomy and modulation by ursodeoxycholic acid. Am J
Physiol Gastrointest Liver Physiol. 299:G887–G897. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Boon RA, Jae N, Holdt LM and Dimmeler S:
Long noncoding RNAs: From clinical genetics to therapeutic targets?
J Am Coll Cardiol. 67:1214–1226. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xu D, Yang F, Yuan JH, Zhang L, Bi HS,
Zhou CC, Liu F, Wang F and Sun SH: Long noncoding RNAs associated
with liver regeneration 1 accelerates hepatocyte proliferation
during liver regeneration by activating Wnt/β-catenin signaling.
Hepatology. 58:739–751. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yamamoto Y, Nishikawa Y, Tokairin T, Omori
Y and Enomoto K: Increased expression of H19 non-coding mRNA
follows hepatocyte proliferation in the rat and mouse. J Hepatol.
40:808–814. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhou B, Li L, Li Y, Sun H and Zeng C: Long
noncoding RNA SNHG12 mediates doxorubicin resistance of
osteosarcoma via miR-320a/MCL1 axis. Biomed Pharmacother.
106:850–857. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ding S, Qu W, Jiao Y, Zhang J, Zhang C and
Dang S: LncRNA SNHG12 promotes the proliferation and metastasis of
papillary thyroid carcinoma cells through regulating wnt/β-catenin
signaling pathway. Cancer Biomark. 22:217–226. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu X, Zheng J, Xue Y, Qu C, Chen J, Wang
Z, Li Z, Zhang L and Liu Y: Inhibition of TDP43-mediated
SNHG12-miR-195-SOX5 feedback loop impeded malignant biological
behaviors of glioma cells. Mol Ther Nucleic Acids. 10:142–158.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhou S, Yu L, Xiong M and Dai G: LncRNA
SNHG12 promotes tumorigenesis and metastasis in osteosarcoma by
upregulating Notch2 by sponging miR-195-5p. Biochem Biophys Res
Commun. 495:1822–1832. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang BF, Cai W and Chen B: LncRNA SNHG12
regulated the proliferation of gastric carcinoma cell BGC-823 by
targeting microRNA-199a/b-5p. Eur Rev Med Pharmacol Sci.
22:1297–1306. 2018.PubMed/NCBI
|
|
17
|
Mitchell C and Willenbring H: A
reproducible and well-tolerated method for 2/3 partial hepatectomy
in mice. Nat Protoc. 3:1167–1170. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Huang Y, Jin C, Zheng Y, Li X, Zhang S,
Zhang Y, Jia L and Li W: Knockdown of lncRNA MIR31HG inhibits
adipocyte differentiation of human adipose-derived stem cells via
histone modification of FABP4. Sci Rep. 7:80802017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhou B, Guo H and Tang J: Long Non-Coding
RNA TFAP2A-AS1 inhibits cell proliferation and invasion in breast
cancer via miR-933/SMAD2. Med Sci Monit. 25:1242–1253. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lan T, Ma W, Hong Z, Wu L, Chen X and Yuan
Y: Long non-coding RNA small nucleolar RNA host gene 12 (SNHG12)
promotes tumorigenesis and metastasis by targeting miR-199a/b-5p in
hepatocellular carcinoma. J Exp Clin Cancer Res. 36:112017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Guo T, Wang H, Liu P, Xiao Y, Wu P, Wang
Y, Chen B, Zhao Q, Liu Z and Liu Q: SNHG6 Acts as a genome-wide
hypomethylation trigger via coupling of miR-1297-Mediated
S-adenosylmethionine-dependent positive feedback loops. Cancer Res.
78:3849–3864. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shan Y, Ma J, Pan Y, Hu J, Liu B and Jia
L: LncRNA SNHG7 sponges miR-216b to promote proliferation and liver
metastasis of colorectal cancer through upregulating GALNT1. Cell
Death Dis. 9:7222018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Michalopoulos GK and DeFrances MC: Liver
regeneration. Science. 276:60–66. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Oe H, Kaido T, Mori A, Onodera H and
Imamura M: Hepatocyte growth factor as well as vascular endothelial
growth factor gene induction effectively promotes liver
regeneration after hepatectomy in Solt-Farber rats.
Hepatogastroenterology. 52:1393–1397. 2005.PubMed/NCBI
|
|
25
|
Thompson MD and Monga SP: WNT/beta-catenin
signaling in liver health and disease. Hepatology. 45:1298–1305.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Qu Y, Olsen JR, Yuan X, Cheng PF, Levesque
MP, Brokstad KA, Hoffman PS, Oyan AM, Zhang W, Kalland KH and Ke X:
Small molecule promotes β-catenin citrullination and inhibits Wnt
signaling in cancer. Nat Chem Biol. 14:94–101. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zheng CH, Wang JB, Lin MQ, Zhang PY, Liu
LC, Lin JX, Lu J, Chen QY, Cao LL, Lin M, et al: CDK5RAP3
suppresses Wnt/β-catenin signaling by inhibiting AKT
phosphorylation in gastric cancer. J Exp Clin Cancer Res.
37:592018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ng R, Song G, Roll GR, Frandsen NM and
Willenbring H: A microRNA-21 surge facilitates rapid cyclin D1
translation and cell cycle progression in mouse liver regeneration.
J Clin Invest. 122:1097–1108. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Greka A and Mundel P: Cell biology and
pathology of podocytes. Annu Rev Physiol. 74:299–323. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li N, Kong M, Zeng S, Hao C, Li M, Li L,
Xu Z, Zhu M and Xu Y: Brahma related gene 1 (Brg1) contributes to
liver regeneration by epigenetically activating the Wnt/β-catenin
pathway in mice. FASEB J. 33:327–338. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ma Y, Lv X, He J, Liu T, Wen S and Wang L:
Wnt agonist stimulates liver regeneration after small-for-size
liver transplantation in rats. Hepatol Res. 46:E154–E64. 2016.
View Article : Google Scholar : PubMed/NCBI
|



















