Introduction
Resveratrol is a natural phenol produced by several
plants and is found in the skin of red grapes and thus red wine.
The anticancer activity of resveratrol has been studied since 1997,
when Jang et al reported that resveratrol application
prevented skin cancer development in a mouse model (1,2). A
number of researchers have suggested that resveratrol
administration prevents skin and colon cancer (3) in animal models with artificially
induced cancer. Recently, the effect of resveratrol against
hepatocellular carcinoma (HCC) has received much interest (4,5).
Experiments in cell cultures in vitro imply a
number of mechanisms in the pharmacological activity of
resveratrol. The responses of HepG2 cells to resveratrol were
reported to include the inhibition of CYP1A1 transcription
(6), upregulation of the
expression of Sirt1 and forkhead box O1 (FOXO1) (7), increase of JNK and ERK-1/2 MAP kinase
activity and perforin expression (8), inhibition of VEGF expression
(9) and downregulation of cyclin
D1 (10) and protein S (11). The HepG2 human HCC cell line is one
of the most suitable in vitro model systems to study
hepatocarcinogenesis and drug targeting. It has been demonstrated
that high doses of resveratrol treatment suppress HepG2
proliferation and subsequenctly induce cell death (12,13).
These molecular mechanisms may underlie the resveratrol-induced
apoptosis or self-protection response of the HepG2 cells.
However, the effects of resveratrol on the mRNA
expression levels of pten and bcl-xl are unknown. In the current
study, the role of resveratrol on the regulation of pten and bcl-xl
mRNA expression in HepG2 cells was investigated, using
semi-quantitative and quantitative PCR analysis. The current study
demonstrated that resveratrol treatment in HepG2 cells resulted in
pten mRNA reduction and bcl-xl mRNA increase.
Materials and methods
Materials
Dulbecco’s modified Eagle’s medium (DMEM), fetal
bovine serum (FBS), TRIzol reagent and all primers were purchased
from Invitrogen (Carlsbad, CA, USA). Resveratrol was purchased from
Sigma (St. Louis, MO, USA). M-MLV TransScript Reverse Transcriptase
and TransStart Green qPCR Supermix were obtained from TransGen
(Beijing, China).
Cell culture
The HepG2 human hepatocellular carcinoma cell line
was cultured in DMEM supplemented with 10% (v/v) FBS and maintained
at 37°C/5% CO2. The study was approved by the ethics
committee of Fujian Medical University, Fuzhou, China.
RNA extraction and reverse
transcription
HepG2 cells were treated with resveratrol (200
μmol/l) for 12 h and total RNA was extracted using TRIzol reagent
according to the manufacturer’s instructions. Total RNA was
reversed transcribed using M-MLV TransScript Reverse Transcriptase
according to the manufacturer’s instructions.
Quantitative real-time PCR
Amplification and quantification of pten, bcl-xl and
GAPDH genes were performed using TransStart Green qPCR Supermix
according to the manufacturer’s instructions. Real-time PCR primers
for each gene were designed at ‘www.genescript.com’. Two pairs of primers were
designed for pten and bcl-xl. The forward and reverse primer
sequences for each gene product are listed in Table I. Data were acquired with the
Bio-Rad (Hercules, CA, USA) cfx96 real-time PCR detection system.
For analysis, a standard curve was constructed for each gene in
four serial 2-fold dilutions of cDNA stocks and then the
amplification efficiency for each gene in each sample was
determined. The amounts of mRNA expression for pten and bcl-xl were
normalized to that of GAPDH, expressed as an arbitrary unit. The
arbitrary unit for either pten or bcl-xl in the control was set as
1. Thus, the percentage of relative mRNA expression for either pten
or bcl-xl in the sample was expressed as the arbitrary unit of the
sample divided by that of the control.
 | Table ISequences of proteins used in PCR. |
Table I
Sequences of proteins used in PCR.
| Gene (no.) | Primer sequences |
|---|
| GAPDH | F:
5′-TGATGACATCAAGAAGGTGGTGAAG-3′ |
| R:
5′-TCCTTGGAGGCCATGTGGGCCAT-3′ |
| pten (1) | F:
5′-ACCAGGACCAGAGGAAACCT-3′ |
| R:
5′-GCTAGCCTCTGGATTTGACG-3′ |
| pten (2) | F:
5′-AACTTGCAATCCTCAGTTTG-3′ |
| R:
5′-GCATCTTGTTCTGTTTGTGG-3′ |
| bcl-xl (1) | F:
5′-GAATGACCACCTAGAGCCTT-3′ |
| R:
5′-CATTTCCGACTGAAGAGTGA-3′ |
| bcl-xl (2) | F:
5′-ACAGCAGCAGTTTGGATG-3′ |
| R:
5′-GGATGTCAGGTCACTGAATG-3′ |
Statistical analysis
Data are expressed as means ± SEM. The two sample
t-test was used to compare differences between the treated groups
and their paired controls. P-values are indicated in the figures.
Values of P<0.05 were considered to indicate statistically
significant differences.
Results
Suppression of resveratrol on HepG2
proliferation
HepG2 cells were seeded equally in 6-well plates and
stimulated with 200 μmol/l resveratrol for various periods of time.
Consistent with Yan et al(5), high doses of resveratrol lead to
HepG2 apoptosis. The majority of the cells detached when treated
with 200 μmol/l resveratrol for more than 24 h. By contrast, the
morphology of cells treated for 12 h was almost the same as that of
the control, with the exception of cell density. As Fig. 1A shows, the density of stimulated
HepG2 cells is approximately half of that of the control cells,
supporting the hypothesis that resveratrol suppresses HepG2 cell
proliferation. Cells were then harvested and RNAs were extracted.
The RNAs were intact in control and treated cells (Fig. 1B).
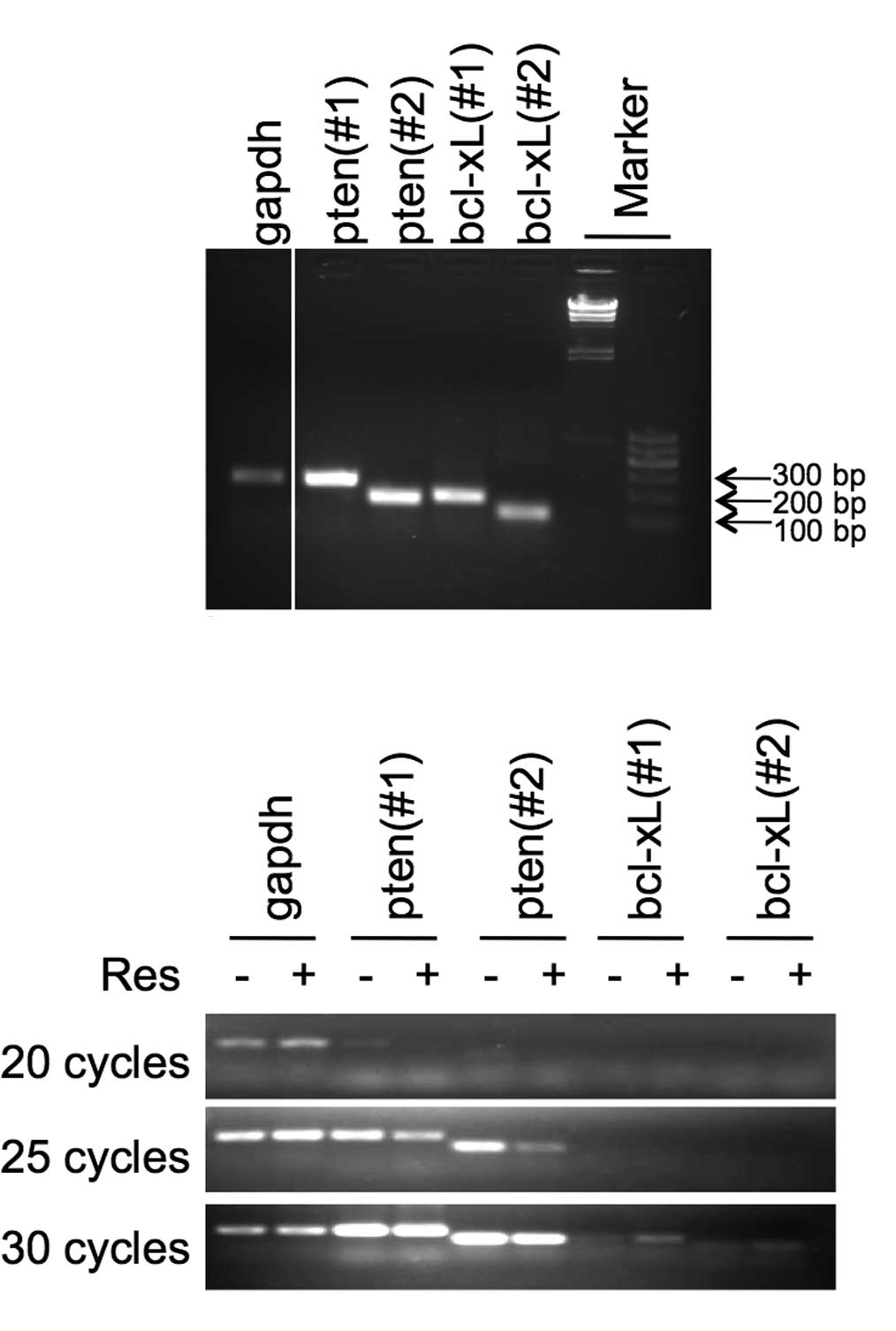
Semi-quantitative analysis shows reduced
pten and increased bcl-xl mRNA expression
Pten and bcl-xl are genes that regulate cell growth
and cell death, respectively. The effect of resveratrol on mRNA
expression of these genes was examined. RNAs were reversed
transcribed into cDNA, which was subjected to semi-quantitative
analysis. PCR products for each gene fragment from the cDNA are
shown in Fig. 2A. PCR products
were compared at 20, 25 and 30 cycles. As Fig. 2B shows, resveratrol exerted various
effects on mRNA expression of pten and bcl-xl. Compared with GAPDH
mRNA expression, pten mRNA expression was reduced, while bcl-xl
mRNA expression was increased. The data were consistent between the
two pairs of primers (numbers 1 and 2) for either pten or
bcl-xl.
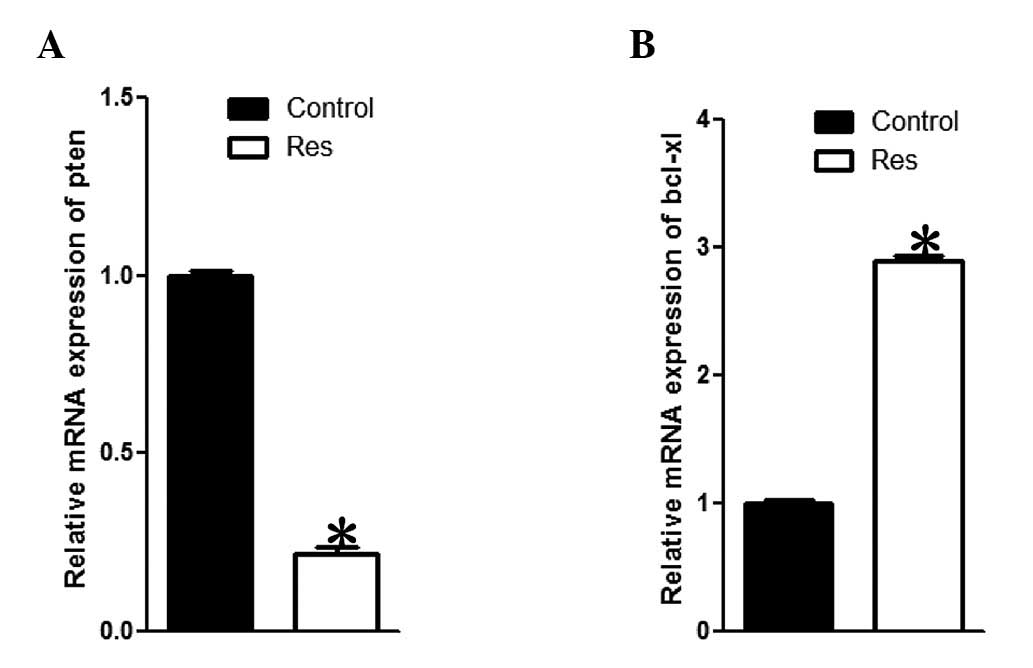
Quantitative analysis determines the fold
of pten mRNA reduction and bcl-xl mRNA increment
Quantitative PCR (qPCR) was performed to determine
the fold of mRNA expression alteration. The cDNA samples were
subjected to qPCR. As Fig. 3
shows, consistent with the semi-quantitative analysis, qPCR data
demonstrates a reduction of pten mRNA and an increase of bcl-xl
mRNA. The reduction fold for pten mRNA was determined to be ~4.59
(Fig. 3A), while the increment
fold for bcl-xl mRNA was determined to be almost 2.90 (Fig. 3B), which were statistically
significant.
Discussion
The carcinogenesis and chemotherapy resistance of
cells in the majority of carcinomas are characterized by
uncontrollable cell growth and the inhibition of cell death. Pten
and bcl-xl are key protein factors in two pivotal pathways that
control cell growth and death, respectively. The pten gene is one
of the most commonly lost tumor suppressors in human cancer
(14). Pten preferentially
dephosphorylates phosphoinositide substrates such as PIP3, thereby
negatively regulating the Akt/PKB-mTORC1 pathway, which is a
pivotal signal contributing to cell growth and proliferation
(15). Mutations or deletions of
pten may both lead to increased cell proliferation and reduced cell
death (16,17). Frequent genetic inactivation of
pten occurs in prostate and endometrial cancer and reduced
expression is reported in numerous other tumor types (18). However, a number of tumors evade
death signals by expressing anti-apoptotic proteins, including the
pro-survival Bcl-2 family member, bcl-xl. Bcl-xl contains four
bcl-2 homology (BH) domains, which mediate its binding to Apaf-1
and subsequently the inhibition of apoptosis (19). Bcl-xl expression is enriched in
prostate carcinoma (20), bladder
cancer (21) and HCC (22). The downregulation of pten or
overexpression of bcl-xl has been reported to play a role in the
chemotherapy resistance in a variety of types of cancer.
In the current study, the effect of resveratrol on
pten and bcl-xl mRNA expression in HepG2 cells was examined. Data
revealed that resveratrol dynamically downregulated pten mRNA
expression and upregulated bcl-xl mRNA expression, theoretically
allowing the cells to survive. Therefore, the data suggest that
resveratrol does not exert its antitumor activity through the
inhibition of bcl-xl and induction of pten, and that resveratrol
activates a cellular protection system against cell death, which
underlies the chemoresistance to resveratrol in HepG2 cells.
Resveratrol is currently produced by chemical or
biotechnological synthesis (23,24)
and is sold as a nutritional supplement or cancer chemopreventive
agent. However, the in vivo effectiveness of resveratrol as
a chemotherapy reagent is largely limited by its poor systemic
bioavailability (25–27). Thus, high doses of resveratrol have
been recommended in several clinical trials investigating its
effects on colon cancer and melanoma (28,29).
Moreover, resveratrol has been reported to accumulate in the liver
(30). These data present new
concerns on the side-effects of resveratrol administration, which
may break the balance of pro- and anti-growth signals in
hepatocytes.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (Grant No. 31100995) and the Natural
Science Foundation of Fujian Province (Grant No. 2011J01187).
References
|
1
|
Jang M, Cai L, Udeani GO, et al: Cancer
chemopreventive activity of resveratrol, a natural product derived
from grapes. Science. 275:218–220. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Baur JA and Sinclair DA: Therapeutic
potential of resveratrol: the in vivo evidence. Nat Rev Drug
Discov. 5:493–506. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Santandreu FM, Valle A, Oliver J and Roca
P: Resveratrol potentiates the cytotoxic oxidative stress induced
by chemotherapy in human colon cancer cells. Cell Physiol Biochem.
28:219–228. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Weng CJ, Wu CF, Huang HW, Wu CH, Ho CT and
Yen GC: Evaluation of anti-invasion effect of resveratrol and
related methoxy analogues on human hepatocarcinoma cells. J Agric
Food Chem. 58:2886–2894. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yan F, Tian XM and Ma XD: Effects of
resveratrol on growth inhibition and gap-junctional intercellular
communication of HepG2 cells. Nan Fang Yi Ke Da Xue Xue Bao.
26:963–966. 2006.(In Chinese).
|
|
6
|
Ciolino HP, Daschner PJ and Yeh GC:
Resveratrol inhibits transcription of CYP1A1 in vitro by
preventing activation of the aryl hydrocarbon receptor. Cancer Res.
58:5707–5712. 1998.PubMed/NCBI
|
|
7
|
Wang GL, Fu YC, Xu WC, Feng YQ, Fang SR
and Zhou XH: Resveratrol inhibits the expression of SREBP1 in cell
model of steatosis via Sirt1-FOXO1 signaling pathway. Biochem
Biophys Res Commun. 380:644–649. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lu CC and Chen JK: Resveratrol enhances
perforin expression and NK cell cytotoxicity through
NKG2D-dependent pathways. J Cell Physiol. 223:343–351.
2010.PubMed/NCBI
|
|
9
|
Yu HB, Zhang HF, Zhang X, et al:
Resveratrol inhibits VEGF expression of human hepatocellular
carcinoma cells through a NF-κB-mediated mechanism.
Hepatogastroenterology. 57:1241–1246. 2010.PubMed/NCBI
|
|
10
|
Parekh P, Motiwale L, Naik N and Rao KV:
Downregulation of cyclin D1 is associated with decreased levels of
p38 MAP kinases, Akt/PKB and Pak1 during chemopreventive effects of
resveratrol in liver cancer cells. Exp Toxicol Pathol. 63:167–173.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hiroto Y, Tadokoro K, Tsuda T, et al:
Resveratrol, a phytoestrogen found in red wine, down-regulates
protein S expression in HepG2 cells. Thromb Res. 127:e1–7. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ma XD, Yan F, Ma AD and Wang HJ:
Resveratrol induces HepG2 cell apoptosis by depolarizing
mitochondrial membrane. Nan Fang Yi Ke Da Xue Xue Bao. 26:406–408.
4132006.(In Chinese).
|
|
13
|
Zhou R, Fukui M, Choi HJ and Zhu BT:
Induction of a reversible, non-cytotoxic S-phase delay by
resveratrol: implications for a mechanism of lifespan prolongation
and cancer protection. Br J Pharmacol. 158:462–474. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Steck PA, Pershouse MA, Jasser SA, et al:
Identification of a candidate tumour suppressor gene, MMAC1, at
chromosome 10q23.3 that is mutated in multiple advanced cancers.
Nat Genet. 15:356–362. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chappell WH, Steelman LS, Long JM, et al:
Ras/Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR inhibitors: rationale and
importance to inhibiting these pathways in human health.
Oncotarget. 2:135–164. 2011.PubMed/NCBI
|
|
16
|
Chu EC and Tarnawski AS: PTEN regulatory
functions in tumor suppression and cell biology. Med Sci Monit.
10:RA235–241. 2004.PubMed/NCBI
|
|
17
|
Li J, Yen C, Liaw D, et al: PTEN, a
putative protein tyrosine phosphatase gene mutated in human brain,
breast and prostate cancer. Science. 275:1943–1947. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Carracedo A, Alimonti A and Pandolfi PP:
PTEN level in tumor suppression: how much is too little? Cancer
Res. 71:629–633. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Adams JM and Cory S: The Bcl-2 protein
family: arbiters of cell survival. Science. 281:1322–1326. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Castilla C, Congregado B, Chinchon D,
Torrubia FJ, Japon MA and Saez C: Bcl-xL is overexpressed in
hormone-resistant prostate cancer and promotes survival of LNCaP
cells via interaction with proapoptotic Bak. Endocrinology.
147:4960–4967. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hameed DA, Abdel Raheem AM, Mosad E,
Hammouda HM, Kamel NA and Abdel Aziz MA: Bcl-XL and Bcl-2
expression in bilharzial squamous cell carcinoma of the urinary
bladder: which protein is prognostic? Urology. 72:374–378. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Watanabe J, Kushihata F, Honda K, et al:
Prognostic significance of Bcl-xL in human hepatocellular
carcinoma. Surgery. 135:604–612. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Farina A, Ferranti C and Marra C: An
improved synthesis of resveratrol. Nat Prod Res. 20:247–252. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Trantas E, Panopoulos N and Ververidis F:
Metabolic engineering of the complete pathway leading to
heterologous biosynthesis of various flavonoids and stilbenoids in
Saccharomyces cerevisiae. Metab Eng. 11:355–366. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Athar M, Back JH, Tang X, et al:
Resveratrol: a review of preclinical studies for human cancer
prevention. Toxicol Appl Pharmacol. 224:274–283. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Niles RM, Cook CP, Meadows GG, Fu YM,
McLaughlin JL and Rankin GO: Resveratrol is rapidly metabolized in
athymic (nu/nu) mice and does not inhibit human melanoma xenograft
tumor growth. J Nutr. 136:2542–2546. 2006.PubMed/NCBI
|
|
27
|
Wenzel E, Soldo T, Erbersdobler H and
Somoza V: Bioactivity and metabolism of trans-resveratrol orally
administered to Wistar rats. Mol Nutr Food Res. 49:482–494. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nguyen AV, Martinez M, Stamos MJ, et al:
Results of a phase I pilot clinical trial examining the effect of
plant-derived resveratrol and grape powder on Wnt pathway target
gene expression in colonic mucosa and colon cancer. Cancer Manag
Res. 1:25–37. 2009.
|
|
29
|
Boocock DJ, Faust GE, Patel KR, et al:
Phase I dose escalation pharmacokinetic study in healthy volunteers
of resveratrol, a potential cancer chemopreventive agent. Cancer
Epidemiol Biomarkers Prev. 16:1246–1252. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Notas G, Nifli AP, Kampa M, Vercauteren J,
Kouroumalis E and Castanas E: Resveratrol exerts its
antiproliferative effect on HepG2 hepatocellular carcinoma cells,
by inducing cell cycle arrest and NOS activation. Biochim Biophys
Acta. 1760:1657–1666. 2006. View Article : Google Scholar : PubMed/NCBI
|