Introduction
Colorectal adenocarcinoma is the second leading
cause of malignancy-related mortality worldwide (1,2), of
which the prevalence has been increasing in recent years.
Tumorigenesis of colorectal adenocarcinoma is a multi-step process
that involves multiple factors and genes regulating a number of
pathways. Therefore, it is important to investigate the roles of
these factors and genes in colorectal adenocarcinoma for cancer
prevention, early diagnosis and therapeutic development.
In a previous study using cDNA subtractive library
construction and microarray analysis, 86 differentially expressed
sequence tags (dbESTs) were identified in human colorectal
adenocarcinoma tissues (3,4). Among these dbESTs, ES274081 (GenBank
accession no. NM_001013649.3; gene name, hcrcn81) was
selected for further investigation. Using qRT-PCR, mRNA levels of
hcrcn81 in the colorectal cancer tissues were identified to
be lower compared with normal colorectal tissues from patients with
colorectal adenocarcinoma, indicating the involvement of
hcrcn81 in the development of human colorectal
adenocarcinoma (5).
The PI3K/Akt/mammalian target of rapamycin (mTOR)
pathway is crucial in the development and progression of colorectal
cancer by regulating cancer cell proliferation, resistance to
apoptosis, angiogenesis and metastasis (6). The mTOR protein is a key kinase
downstream of the growth factor receptor, PI3K and Akt signaling
pathway, which is involved in cell growth, survival, metabolism and
proliferation (7). In previous
years, the role of mTOR in cancer development and progression has
been elucidated. Activation of the mTOR signaling pathway often
results from genetic alterations of a number of negative regulators
of mTOR, including PTEN, tuberous sclerosis complex (TSC) 1 and
TSC2 (8). It has been demonstrated
that activation of the PI3K/Akt/mTOR pathway correlates with tumor
progression and poor survival in a variety of tumor types (9,10),
indicating that mTOR may be a promising molecular target for
colorectal cancer. The mTOR inhibitor, rapamycin, is a natural
macrolide antibiotic isolated from Streptomyces
hygroscopicus. Rapamycin binds FKBP-12 (FK506-binding protein)
and the resulting complex inhibits the protein kinase activity of
mTOR. Rapamycin was originally used as an antifungal and
immunosuppressive agent, however, the subsequent identification of
the inherent antiproliferative properties of rapamycin led to the
investigation of this compound as an anticancer agent (11). Therefore, to study the role of
hcrcn81 in the tumorigenesis of colorectal cancer, the
effect of rapamycin treatment on hcrcn81 expression was
analyzed.
Materials and methods
Cell lines and culture conditions
Human colorectal carcinoma cell lines, SW480 and
LoVo (both obtained from the American Type Culture Collection,
Manassas, VA, USA), were cultured in Dulbecco’s Modified Eagle’s
medium (DMEM; Hyclone Laboratories, Inc., Logan, UT, USA)
containing 10% fetal bovine serum, penicillin (100 IU/ml) and
streptomycin (100 μg/ml). Cells were grown at 37°C in a humidified
atmosphere with 5% CO2. Experiments were performed using
cells harvested from exponentially growing cultures.
Drug
Rapamycin stock solutions (5 mg/ml; Fermentek Ltd.,
Jerusalem, Israel) were prepared in DMSO. These solutions were
stored at −20°C prior to use and were diluted into six
concentrations in DMEM for subsequent experiments.
In vitro cellular assays
Rapamycin stock solutions were diluted in DMEM at
the concentrations of 0.05, 0.1, 0.2, 0.5, 1 and 10 μM. DMSO was
used as the solvent control, of which the final concentration was
0.1%. Cells were treated with rapamycin for 48 h.
RNA isolation
TRIzol reagent (Invitrogen Life Technologies,
Carlsbad, CA, USA) was used for total RNA isolation, according to
the manufacturer’s instructions. Total RNA yield was determined by
absorbance at 260 nm using a spectrophotometer. The quality of RNA
products was confirmed by sharp bands representing 28S and 18S rRNA
molecules and the intensity ratio of 2:1 of these 2 bands (28S:18S)
on 1% agarose gel.
First-strand cDNA synthesis
Total RNA isolated from each sample was treated with
DNaseI to eliminate genomic DNA contamination prior to reverse
transcription (RT). The RT reaction was performed in a 20-μl volume
using the M-MuLV Reverse Transcriptase kit (Fermentas, Waltham, MA,
USA) for first-strand cDNA synthesis under the recommended
conditions. The synthesized cDNA product was immediately used for
quantitative real-time PCR or stored at −20°C.
Quantitative real-time PCR
Quantitative real-time PCR was performed for cDNA
amplification using SYBR Premix ExTaq (Takara Bio, Inc., Shiga,
Japan) and primers listed in Table
I, on a Bio-Rad C1000 real-time system (Bio-Rad, Hercules, CA,
USA), according to the manufacturer’s instructions and applied
international standards (12). For
each PCR, 2 μl cDNA obtained from 1 μg RNA template was used. The
thermal cycling conditions consisted of an initial denaturation
step at 95°C for 30 sec and 40 cycles of the following 3 steps:
denaturation at 95°C for 5 sec, annealing at 57°C for 30 sec and
elongation at 72°C for 30 sec. GAPDH was used as the internal
control. The amplified cDNA product was quantified using the
2−ΔΔCt method. Primers for hcrcn81 amplification
were designed to target its open reading frame, using Primer
Premier 5.0 software.
 | Table IPrimers for quantitative real-time
PCR. |
Table I
Primers for quantitative real-time
PCR.
| Gene | Primer sequence
(5′→3′) |
|---|
| hcrcn81 | F:
ACGCAACCCAGACTATGAAGAG |
| R:
CACCTTCTCACTCACCTTTCCT |
| GAPDH | F:
GGAAGGTGAAGGTCGGAGT |
| R:
TGAGGTCAATGAAGGGGTC |
Western blot analysis
Cells were treated with 10 μM rapamycin for 48 h.
DMSO was used as the solvent control, of which the final
concentration was 0.1%. Cell lysates were denatured in sample
buffer containing SDS. The same amount of the denatured protein (30
μg) was loaded on each lane and was separated on 12% SDS-PAGE and
then the protein product was transferred to PVDF (Bio-Rad)
membranes. Following blocking for 3 h in Tris-buffered saline
containing 0.1% Tween-20 and 3% bovine serum albumin, membranes
were incubated overnight at 4°C with primary antibody against
hcrcn81 (1:500). Membranes were then incubated with an
appropriate horseradish peroxidase-conjugated secondary antibody
and the corresponding protein product was visualized using ECL
reagent (Thermo Fisher Scientific, Waltham, MA, USA).
Statistical analysis
Statistical analysis was performed using the t-test
and Fisher’s exact test with SPSS version 19.0 software (SPSS,
Inc., Chicago, IL, USA). P<0.05 was considered to indicate a
statistically significant difference.
Results
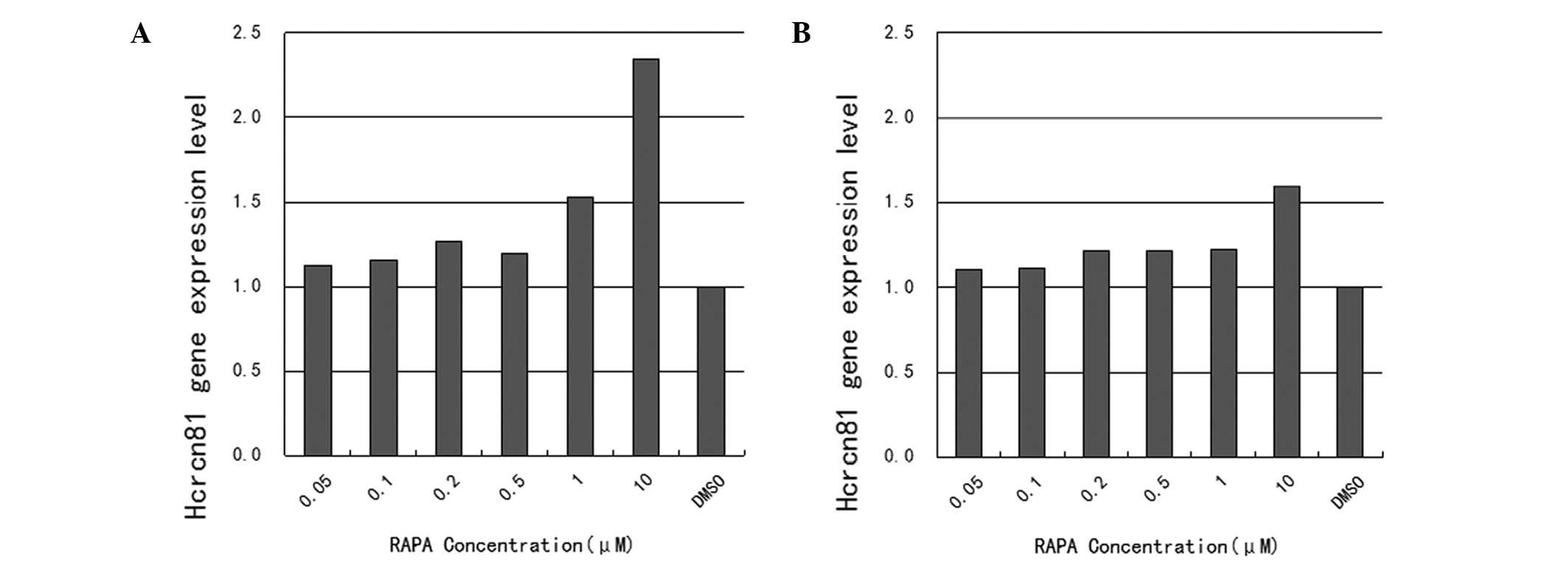
As demonstrated in Fig.
1, mRNA expression of hcrcn81 was upregulated in
rapamycin-treated cells, compared with cells treated with DMSO
alone. Specifically, upregulation rates were 112.1, 115.3, 126.5,
119.6, 152.5 and 234.6% for the tested rapamycin concentrations
ranging between 0.05 and 10 μM, respectively, in SW480 cells. In
this case, only upregulation in response to the two highest
concentrations, 1 and 10 μM, was found to be statistically
significant (P=0.013 and 0.036). The corresponding upregulation
rates in LoVo cells were 110.2, 111.3, 121.4, 121.6, 122.5 and
159.7% for the tested rapamycin concentrations ranging between 0.05
and 10 μM, respectively. The upregulation in response to the
highest concentration, 10 μM, was identified as statistically
significant (P=0.011).
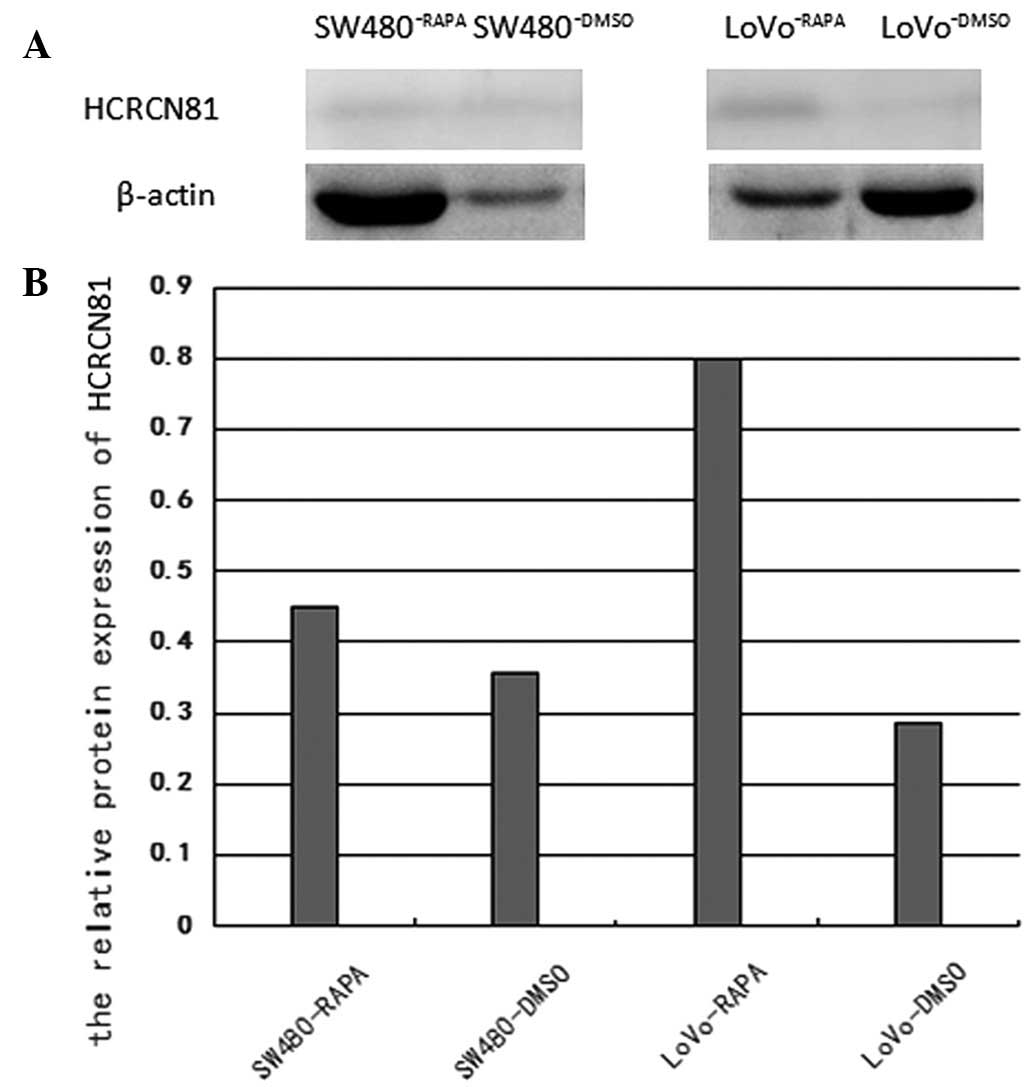
As revealed in Fig.
2, following treatment with 10 μM rapamycin for 48 h, the
protein expression of hcrcn81 was upregulated in SW480 and
LoVo cells lines tested with rapamycin, compared to that in the
SW480 and LoVo cell lines. The upregulation was 1.269-fold
(p=0.048) and 2.789-fold (p=0.024), respectively.
Discussion
In a previous study, we found that mRNA expression
of hcrcn81 was downregulated in human colorectal carcinoma
tissue samples by qRT-PCR. Specifically, among the 30 tested human
colorectal carcinoma tissue samples, 5 revealed upregulated
hcrcn81 mRNA expression, whereas 25 exhibited downregulated
hcrcn81 mRNA expression, accounting for 83% of the tested
samples. This observation indicated the potential involvement of
hcrcn81 in the pathogenesis of colorectal carcinoma. In
addition, the downregulation of hcrcn81 mRNA expression was
observed in 91% of the moderately differentiated samples (21/23),
but only 50% of poorly differentiated tissue samples (3/6). The
significantly higher prevalence of hcrcn81 downregulation
identified in moderately differentiated samples (P<0.05)
indicated a correlation of hcrcn81 expression with tumor
stage at the mRNA level (5). In
the present study, rapamwycin treatment was demonstrated to induce
hcrcn81 upregulation in human colorectal adenocarcinoma cell
lines at the mRNA and protein level.
mTOR protein is a serine/threonine protein kinase
involved in the nutrient-sensitive signaling pathway, which is
crucial for the regulation of cell growth and proliferation. The
mTOR pathway is activated in various cell processes, including
tumorigenesis, insulin resistance, adipogenesis, angiogenesis and T
lymphocyte activation. In addition, the pathway is associated with
various human diseases, including cancer, obesity and type 2
diabetes (7). The activity of mTOR
is regulated by the concentration of amino acids, particularly
leucine and the levels of energy, growth factors and oxygen. In
addition to these key regulators, other cellular conditions and
signals, including inflammation, Wnt signaling, phosphatidic acid
and genotoxic stress, have also been found to be involved in the
regulation of the mTOR signaling pathway (7). Activation of the PI3K/Akt/mTOR
pathway inhibits apoptosis induced by a number of types of stimuli,
thereby promoting cell cycle progression, cell survival and
proliferation, which are important for tumor invasion and
metastasis. In addition, its role in neovascularization also
promotes tumorigenesis. Akt has been found to be overexpressed in
human colorectal carcinoma and Akt activation promotes cell
proliferation and regulates cell survival by inhibiting apoptosis
(13,14). In a previous study, Johnson et
al found that the expression levels of several key components
of the PI3K/Akt/mTOR pathway, including p85α, Akt1, Akt2,
phosphorylated-mTOR and phosphorylated-p70S6K, were significantly
elevated in colorectal carcinoma tissue samples, compared with
matched normal colorectal tissues from the same patient (14). Similarly, Vilar et al
reported that the PI3K/Akt/mTOR pathway is of special relevance in
mismatch repair-deficient colorectal cancer (15).
Rapamycin is known to induce apoptosis, indicating a
potential role of the mTOR pathway in the regulation of cell
survival (16). In addition,
rapamycin has been revealed to be effective in the clinical
treatment of several types of cancer. Boffa et al found that
rapamycin treatment inhibited cancer cell growth and metastatic
progression in non-small cell lung carcinoma (17). Medici and Olsen reported that
rapamycin treatment inhibited the proliferation of hemangioma
endothelial cells (18). Samkari
et al demonstrated that rapamycin treatment induced
expression of the anti-apoptotic protein, survivin, in
neuroblastoma (19). Sun and Jin
observed that rapamycin treatment repressed phosphorylation of
4E-BP-1 and p70-S6K induced by insulin in the human colorectal
carcinoma cell line, HT29 (20).
In the present study, rapamycin treatment was
demonstrated to induce hcrcn81 upregulation in the human
colorectal adenocarcinoma cell lines, SW480 and LoVo, at the mRNA
and protein levels. Specifically, upregulated mRNA expression of
hcrcn81 was observed following rapamycin treatment at all
concentrations ranging between 0.05 and 10 μM. However, in SW480
cells, upregulation in response to the two highest concentrations,
1 and 10 μM, was found to be statistically significant by Fisher’s
exact test (P=0.015 and 0.018). In LoVo cells, upregulation in
response to the highest concentration, 10 μM, was identified as
statistically significant by Fisher’s exact test (P=0.046).
In summary, the effective concentration of rapamycin
for significant hcrcn81 upregulation was 10 μM in the two
cell lines. The dose-dependent relationship of hcrcn81
upregulation by rapamycin treatment indicated the potential
involvement of hcrcn81 in the PI3K/Akt/mTOR pathway in
colorectal adenocarcinoma cells, which may be by regulation of mTOR
activity. It is possible that hcrcn81 is involved in the
induction of cell apoptosis, blockage of cell cycle progression and
inhibition of metastasis in cancer cells, similar to the effects of
rapamycin treatment. However, further studies must be performed to
comprehensively analyze the function of hcrcn81 in
carcinogenesis.
Acknowledgements
This study was supported by a grant from the Sichuan
University for Stomatological Key Laboratories
(SKLODSCU20090021).
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
2
|
Herrinton LJ, Liu L, Levin TR, Allison JE,
Lewis JD and Velayos F: Incidence and mortality of colorectal
adenocarcinoma in persons with inflammatory bowel disease from 1998
to 2010. Gastroenterology. 143:382–389. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen Y, Zhang Y, Zhou Z, Wang G and Yi Z:
Identification of differentially expressed genes in human
colorectal adenocarcinoma. World J Gastroenterol. 12:1025–1032.
2006.
|
|
4
|
Zhang C and Chen Y: Electronic cloning and
validating of the suppression subtractive hybridization EST
ES274070 of human colorectal adenocarcinoma. US Chin J Lymphol
Oncol. 6:83–88. 2007.
|
|
5
|
Jiang Q, Zhang C and Chen Y:
NM_001013649.3 gene is down-regulated in human colorectal
adenocarcinoma. Mol Med Rep. 4:1279–1281. 2011.PubMed/NCBI
|
|
6
|
Zoncu R, Efeyan A and Sabatini DM: mTOR:
from growth signal integration to cancer, diabetes and ageing. Nat
Rev Mol Cell Biol. 12:21–35. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Laplante M and Sabatini DM: mTOR signaling
at a glance. J Cell Sci. 122:3589–3594. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Feng Z, Zhang H, Levine AJ and Jin S: The
coordinate regulation of the p53 and mTOR pathways in cells. Proc
Natl Acad Sci USA. 102:8204–8209. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gulhati P, Cai Q, Li J, et al: Targeted
inhibition of mammalian target of rapamycin signaling inhibits
tumorigenesis of colorectal cancer. Clin Cancer Res. 15:7207–7216.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chiang GG and Abraham RT: Targeting the
mTOR signaling network in cancer. Trends Mol Med. 13:433–442. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Miyake N, Chikumi H, Takata M, Nakamoto M,
Igishi T and Shimizu E: Rapamycin induces p53-independent apoptosis
through the mitochondrial pathway in non-small cell lung cancer
cells. Oncol Rep. 28:848–854. 2012.PubMed/NCBI
|
|
12
|
Bustin SA, Benes V, Garson JA and
Hellemans J: The MIQE guidelines: minimum information for
publication of quantitative real-time PCR experiments. Clin Chem.
55:611–622. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Roy HK, Olusola BF, Clemens DL, Karolski
WJ, Ratashak A, Lynch HT and Smyrk TC: AKT proto-oncogene
overexpression is an early event during sporadic colon
carcinogenesis. Carcinogenesis. 23:201–205. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Johnson SM, Gulhati P, Rampy BA, et al:
Novel expression patterns of PI3K/AKT/mTOR signaling pathway
components in colorectal cancer. J Am Coll Surg. 210:767–778. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Vilar E, Mukherjee B, Kuick R, et al: Gene
expression patterns in mismatch repair-deficient colorectal cancers
highlight the potential therapeutic role of inhibitors of the
phosphatidylinositol 3-kinase-AKT-mammalian target of rapamycin
pathway. Clin Cancer Res. 15:2829–2839. 2009.
|
|
16
|
Thimmaiah KN, Easton J, Huang S, Veverka
KA, Germain GS, Harwood FC and Houghton PJ: Insulin-like growth
factor I-mediated protection from rapamycin-induced apoptosis is
independent of Ras-Erk1-Erk2 and phosphatidylinositol 30-kinase-Akt
signaling pathways. Cancer Res. 63:364–374. 2003.PubMed/NCBI
|
|
17
|
Boffa DJ, Luan F, Thomas D, Yang H, Sharma
VK, Lagman M and Suthanthiran M: Rapamycin inhibits the growth and
metastatic progression of non-small cell lung cancer. Clin Cancer
Res. 10:293–300. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Medici D and Olsen BR: Rapamycin inhibits
proliferation of hemangioma endothelial cells by reducing
HIF-1-dependent expression of VEGF. PLoS One. 7:e429132012.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Samkari A, Cooper ZA, Holloway MP, Liu J
and Altura RA: Rapamycin induces the anti-apoptotic protein
survivin in neuroblastoma. Int J Biochem Mol Biol. 3:28–35.
2012.PubMed/NCBI
|
|
20
|
Sun J and Jin T: Both Wnt and mTOR
signaling pathways are involved in insulin-stimulated
proto-oncogene expression in intestinal cells. Cell Signal.
20:219–229. 2008. View Article : Google Scholar : PubMed/NCBI
|