Introduction
Lung cancer is the major cause of cancer-associated
mortality worldwide, with non-small cell lung cancer (NSCLC)
histology predominating over small cell lung cancer (SCLC). The
invasive and metastatic characteristics of lung tumor cells are
responsible for their high malignancy. Patients with lung cancer
frequently exhibit tumor cell invasion and metastasis prior to
diagnosis, which renders current treatments, including surgery,
radiotherapy and chemotherapy ineffective. Typically, the 5-year
survival rate following diagnosis is <20%. Therefore, it is
important to study the molecular basis of lung cancer cell invasion
and metastasis in order to design novel therapeutic agents that are
able to decrease the malignancy of lung cancer (1).
Protease-activated receptors (PARs) are
G-protein-coupled receptors (GPCRs) that signal in response to
extracellular protease. There are four human PARs (PAR1-4) which
have impotant roles in hemostasis and thrombosis as well as in
inflammatory and proliferative response (2). PAR1 was originally dubbed the
thrombin receptor since it was first found in a search for a
receptor that confers thrombin signaling on human platelets and
other cell types (3,4). Unlike typical ligand-receptor
interactions, thrombin cleaves the NH2 terminus of PAR1
at serine 42 (Ser42). Upon cleavage, the new
NH2-terminal peptide acts as a tethered ligand that
activates the receptor and initiates cellular signaling (5).
PAR1 is overexpressed in aggressive melanoma as well
as colon, prostate and invasive breast cancers (6–9).
Previous studies showed that the upregulation of PAR1 is strongly
associated with low survival rates in patients with gliomas
(10), breast cancer (11) and primary gallbladder carcinoma
(12).
The present study focused on the downregulation of
PAR1 expression by small interfering ribonucleic acids (siRNAs). By
using siRNA and Lipofectamine RNA interference (RNAi)MAX complex
formation in vitro, silencing was achieved at the protein
level as demonstrated by western blot analysis and at the mRNA
level as shown by polymerase chain reaction (PCR). Furthermore, the
growth and metastasis of A549 cells were decreased. PAR1 may be a
promising drug target in clinical cancer therapy.
Materials and methods
Cell lines and culture
The A549 cell line was obtained from Keygen Biotech
(Nanjing, China) and cultured in RPMI-1640 medium (Invitrogen,
Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS; TianHang
Biological Technology Co., Ltd., Hangzhou, China).
siRNAs
The sequences of three siRNA duplexes were purchased
from GenePharma, Shanghai, China. siRNA1 (5′-GAC ACU CUU UGU CCC
AUC UTT-3′), siRNA2 (5′-CUG UCA UGA UGU GCU CAA UTT-3′) and siRNA3
(5′-GGC AGU UGA UGG CAA GUA ATT-3′) were designed to target
different coding regions of the human PAR1 mRNA sequence (GeneBank
accession no. NM_2149). A BLAST (NCBI database; National Center for
Biotechnology Information, Bethesda, MD, USA) search was performed
to confirm the only targets of the three duplexes on PAR1. A
negative control (5′-UUC UCC GAA CGU GUC ACG UTT-3′) and a positive
control (GAPDH, 5′-GUA UGA CAA CAG CCU CAA GTT-3′) were also
obtained from GenePharma.
Efficiency of delivery in vitro
A549 cells were seeded in 6-well plates with
RPMI-1640 containing 10% FBS without antibiotics and allowed to
attach overnight. The following day, the cells were transfected
with the fluorescein amidite (FAM; GenePharma, Shanghai,
China)-labeled negative control siRNA, according to the
manufacturer’s instructions for Lipofectamine RNAiMAX transfection
reagent (Invitrogen Life Technologies, Carlsbad, CA, USA) -based
transfections when the confluence was 60–80%. Six hours following
transfection, the 6-well plates were observed under a fluorescence
microscope (Axiovert 40 CFL; Carl Zeiss, Jena, Germany) to observe
the fluorescence (green, negative control FAM). The final
concentration of the Lipofectamine RNAiMAX transfection reagent was
0.2% (5 μl). The final concentration of the negative control FAM
was 100 nM.
Transfection with siRNAs in vitro
Cells were transfected with siRNAs by the
aforementioned method. The final concentration of siRNAs was 100
nM. A control, a negative control and a GAPDH-positive control
group were also contained in the 6-well plates.
PCR
Total RNA was extracted from the cells with TRIzol
reagent (Invitrogen) according to the manufacturer’s instructions
and quantified by ultraviolet absorbance spectroscopy. Reverse
transcription was performed using 500 ng of total RNA. The reaction
mixture contained 5X PrimeScript® buffer (Takara,
Dalian, China), total RNA and RNase-free water, and the reaction
was performed according to the manufacturer’s instructions of the
PrimeScript® RT Master Mix Perfect Real Time (Takara).
Relative quantitative analysis of the cDNA was performed using the
ABI PRISM®7500 (Applied Biosystems, Grand Island, NY,
USA) and the SYBR®-Green Premix Ex Taq™ kit (Takara)
according to the manufacturer’s instructions. PCRs were performed
in a total volume of 20 μl, including 2 μl cDNA and 0.2 μM primers.
The primers used were: PAR1, sense, 5′-GTG ATT GGC AGT TTG GGT
CT-3′ and antisense, 5′-GCC AGA CAA GTG AAG GAA GC-3′; GAPDH,
sense, 5′-CAG TCC ATG CCA TCA CTG CCA-3′ and antisense, 5′-CAG TGT
AGC CCA GGA TGC CCT T-3′. Amplification was conducted at 95°C for
30 sec, then 40 cycles at 95°C for 5 sec and 60°C for 30 sec. At
the end of each PCR, melting curve analysis was performed to
confirm that the amplified product was specific. All the reactions
were performed in triplicate. Sample values were normalized to the
expression of the housekeeping gene GAPDH, and the relative
expression was calculated using the AB 7500 system SDS software
(Applied Biosystems).
Western blot analysis
Cell extracts were prepared with 200 μl mixture of
radioimmunoprecipitation assay (RIPA) lysis buffer (Beyotime,
Jiangsu, China) and 1 mM phenylmethylsulfonyl fluoride. The total
protein was extracted. Samples containing equivalent amounts of
protein (20 μg) were applied to a 10% SDS-PAGE gel by
electrophoresis. The separated proteins were transferred onto
polyvinylidene fluoride membranes (Millipore, Bedford, MA, USA) and
incubated overnight at 4°C. Blotting membranes were blocked for 1 h
at room tempreture, washed three times, and then incubated with
mouse anti-PAR1 (1:100; Santa Cruz Biotechnology, Inc., Santa Cruz,
CA, USA) in Tris-buffered saline and Tween 20 (TBST) overnight at
4°C. GAPDH antibodies (1:2,000; KangChen Biotech, Shanghai, China)
were used as an internal control. Following several washes with
TBST buffer, the membranes were incubated for 2 h with horseradish
peroxidase-linked secondary antibody (1:1,000; Jackson
ImmunoResearch Laboratories, Inc., West Grove, MA, USA). The
membranes were then processed with enhanced chemiluminescence
western blotting detection reagents (Pierce, Rockford, IL, USA).
Chemifluorescence was detected using the ChemiDoc™ XRS+ imaging
system (Bio-Rad, Hercules, CA, USA).
Cell viability assays
Cell viability was measured by the WST-8 assay
following optimized manufacturer’s instructions (Dojindo, Kumamoto,
Japan). Briefly, one day prior to transfection, the A549 cells were
seeded at a density of 5,000 cells/100 μl/well in 96-well culture
plates and incubated in a humidified incubator at 37°C. The cells
were then treated with PAR1 siRNA at five different concentrations
(0, 10, 25, 50 and 100 nM). A negative control group was also
included. Following 48 h of incubation, 10 μl
2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium
(WST-8) was added to each well. The cells were then incubated for 2
h prior to measuring the optical density (OD) at 540 nm. Each group
contained five duplicates. The percentage of viable cells was
determined using the formula: Ratio (%) = [OD (treatment) - OD
(blank)/OD (control) - OD (blank)] × 100.
Wound healing
A549 cells were seeded at 3×106
cells/well in 6-well plates. A linear wound was generated in the
monolayer with a sterile 10 μl plastic pipette tip. The experiment
was performed on PAR1 siRNA-transfected, negative siRNA-transfected
and control groups. After 0, 12, 24, 48 and 72 h of incubation,
images of the cells were captured by the TE2000 Nikon microscope
(Nikon Corporation, Tokyo, Japan), using NIS-Elements F software,
version 3.0 (Nikon Corporation). The mobility was calculated using
the formula: Mobility = (Width0 h group - Widthx h
group)/Width0 h group × 100%.
Cell migration
Transwell chambers (Costar, Bethesda, MD, USA) were
used for the cell mobility experiments. The experimental group
which had been transfected with PAR1 siRNA for 24 h, as well as the
positive and negative control groups, were incubated into the upper
compartment of the Transwell chambers, respectively, at a density
of 1×105/ml and 100 μl/well. The cells were incubated at
37°C for 12 h. Cells that did not penetrate the membrane were wiped
off. The membrane was removed, fixed with paraformaldehyde and
stained with 0.1% crystal violet. Five fields of view were randomly
selected and the number of cells that penetrated the membrane was
counted. The mobility inhibition rate was calculated using the
equation: Mobility inhibition rate = (the number of cells in the
control group that penetrated the membrane - the number of cells in
the PAR1 siRNA group that penetrated the membrane)/the number of
cells in the control group that penetrated the membrane × 100%.
Cell invasion
Transwell chambers were used to determine the cell
invasiveness. The membrane at the bottom of the Transwell chamber
was evenly coated with 50 μl diluted Matrigel. Cells from the
experimental group which had been transfected with PAR1 siRNA for
24 h as well as the positive and negative control groups were
inoculated into the upper compartment of the Transwell chambers at
a density of 1×105 cells/ml and 100 μl/well. The cells
were incubated at 37°C for 24 h. Cells that did not penetrate the
polycarbonate membrane were wiped off. The membrane was then fixed
with paraformaldehyde and stained with 0.1% crystal violet. Five
fields of view were randomly selected and the number of cells that
penetrated the membrane was counted. The invasion inhibition rate
was calculated using the formula: Invasion inhibition rate = (the
number of cells in the control group that penetrated the membrane -
the number of cells in the experimental group that penetrated the
membrane)/the number of cells in the control group that penetrated
the membrane × 100%.
Statistical analyses
The PCR and western blot data were normalized to the
GAPDH controls. The results were expressed as the mean ± standard
deviation and the significance of differences was determined using
one-way analysis of variance (ANOVA) followed by Scheffe’s post hoc
test. Differences with P<0.05 were considered to be
statistically significant.
Results
Efficiency of delivery
The A549 cells were seeded in 6-well plates and
incubated overnight. The following day, the cells were transfected
with 12.5 μl negative control-FAM. Six hours following
transfection, the 6-well plates were observed under a fluorescence
microscope to observe green fluorescence resulting from the
negative control-FAM. As shown in Fig.
1, the delivery efficiency to A549 cells, which was 95%, was
sufficiently high to transfect siRNAs into A549 cells in the
present study.
Inhibitory effect of three siRNA duplexes
on PAR1 expression
To examine the silencing effect of siRNAs on PAR1
mRNA and protein, three siRNA duplexes, a positive control and a
negative control at the same final concentration of 100 nM were
used for transfection of A549 cells with 5 μl Lipofectamine RNAiMAX
transfection reagent. Following 24 h of incubation, the cells were
collected for PCR. As shown in Fig.
2A, compared with the control, all three duplexes significantly
decreased PAR1 mRNA levels (P<0.05). However, siRNA2 and siRNA3,
which led to ~89.3 and 91.3% decrease of PAR1 mRNA, respectively,
exerted a greater silencing effect compared with siRNA1 (72.3%,
P<0.05). Following 48 h of incubation post-transfection, the
cells were collected for western blot analysis. As shown in
Fig. 2B, siRNA3, which caused an
~83.6% decrease of PAR1 protein, had the most marked silencing
effect compared with siRNA1 and siRNA2. Transfection of the
negative control did not decrease the mRNA or the protein levels of
the PAR1 gene and transfection of the positive control decreased
both the mRNA and protein levels of the GAPDH gene. From the above
results, siRNA3 was proven to have the most marked silencing effect
among the three siRNAs assessed. Accordingly, siRNA3 was selected
to be used in the present study.
 | Figure 2Levels of PAR1 expression following
transfection with siRNAs (100 nmol/l). (A) Column diagram shows the
levels of PAR1 mRNA in A549 cells 24 h following transfection with
the three different siRNA dulplexes examined by PCR. Bars 1,
Control; 2, N.C.; 3, siRNA1; 4, siRNA2; 5, siRNA3. (B) Effects of
the three siRNA duplexes on PAR1 protein expression examined by
western blot analysis 48 h following transfection. The column
diagram shows PAR1 protein levels in A549 cells following
transfection. *P<0.05 compared with untreated control
cells. Experiments were performed at least three times and a
representative experiment is shown in B. The values shown in A and
B are the mean ± standard deviation of three independent
experiments. PAR1, protease-activated receptor 1; mRNA, messenger
ribonucleic acid; siRNA, small interfering ribonucleic acid; PCR,
polymerase chain reaction; N.C., negative control. |
PAR1 siRNA3 suppresses A549 cell
viability
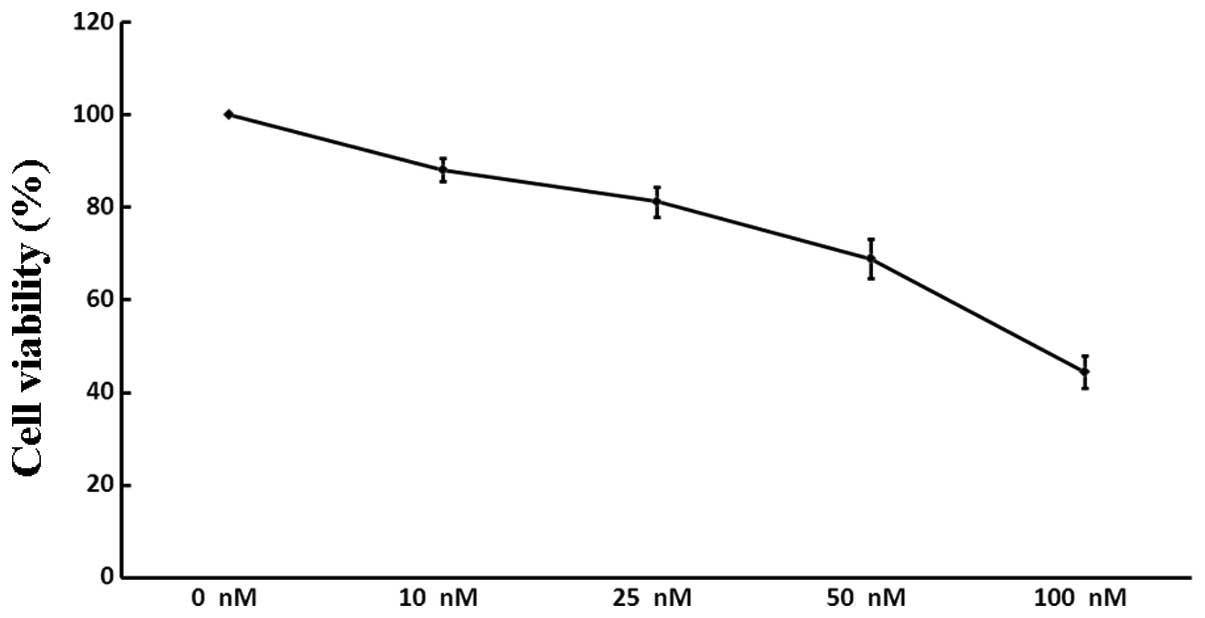
The viability of A549 cells following treatment with
increasing concentrations of siRNA3 (0, 10, 25, 50 and 100 nM) was
assessed. As demonstrated by the WST-8 assay (Fig. 3), siRNA3 decreased the quantity of
viable cells in a dose-dependent manner: Following incubation with
100 nM siRNA, the number of A549 cells was reduced by 55.5%,
whereas siRNA at a lower concentration (10 nM) exerted only a minor
inhibitory effect (11.9%). There was no significant difference
between the negative and positive controls (P>0.05).
In order to investigate the role of PAR1 in the
viability of A549 cells, PAR1 mRNA levels were assessed 48 h
following transfection with siRNA3 at various concentrations. As
shown in Fig. 4, 10 nM siRNA3
decreased PAR1 mRNA levels by 32.5%, while 100 nM siRNA3 led to a
76.1% decrease in PAR1 mRNA levels. Thus, siRNA3 decreased PAR1
mRNA levels in a dose-dependent manner, affecting the viability of
A549 cells.
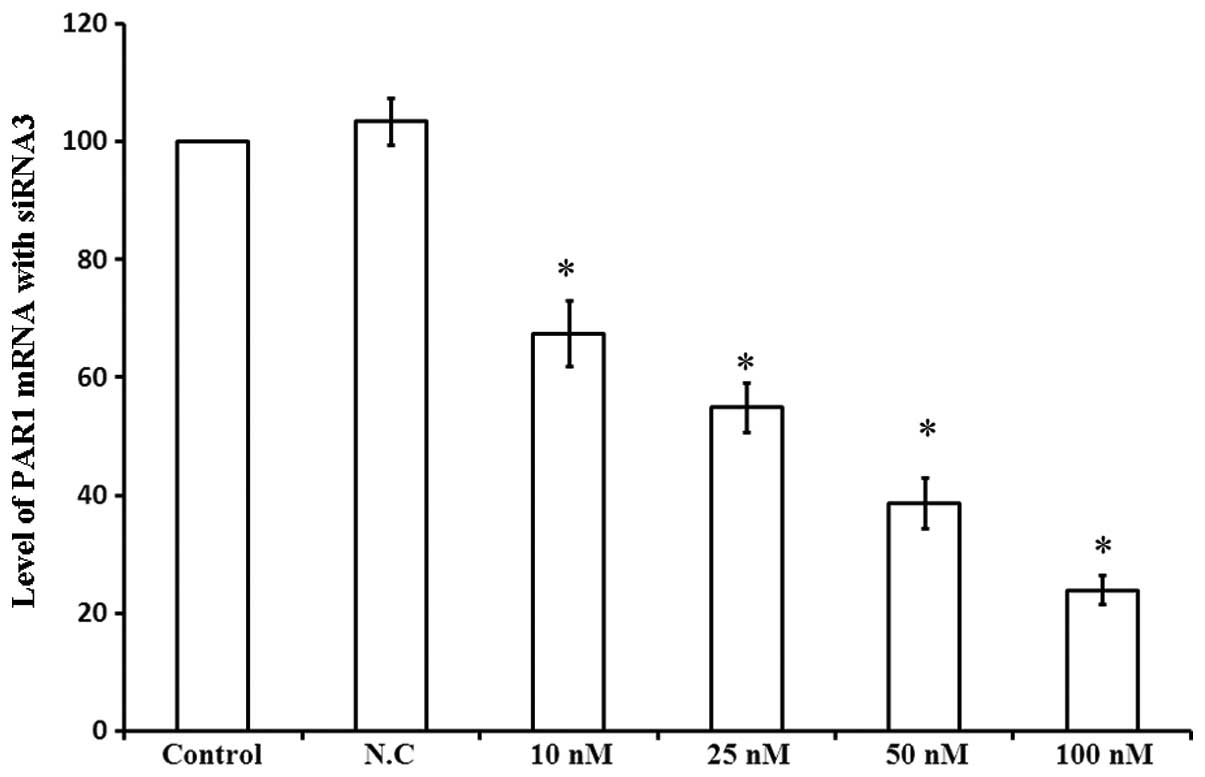 | Figure 4Levels of PAR1 mRNA following
transfection with different concentrations of siRNA3. Column
diagram shows the levels of PAR1 mRNA in A549 cells 48 h following
transfection with four different concentrations of siRNA3 examined
by Bars PCR. 1, Control; 2, N.C.; 3, 10 nM; 4, 25 nM; 5, 50 nM; 6,
100 nM. *P<0.05 compared with control cells. PAR1,
protease-activated receptor 1; mRNA, messenger ribonucleic acid;
siRNA, small interfering ribonucleic acid PCR, polymerase chain
reaction; N.C., negative control. |
PAR1 siRNA inhibits the migration of A549
cells
As demonstrated in the wound healing experiment
(Fig. 5A), PAR1 siRNA inhibited
the migration of A549 cells within 72 h post-perforation of the
cell layer at 100 nM. In the Transwell chamber experiment (Fig. 5B), a significant decrease in
migration (63.6%) was observed between the cells penetrated from
the control and the treated groups.
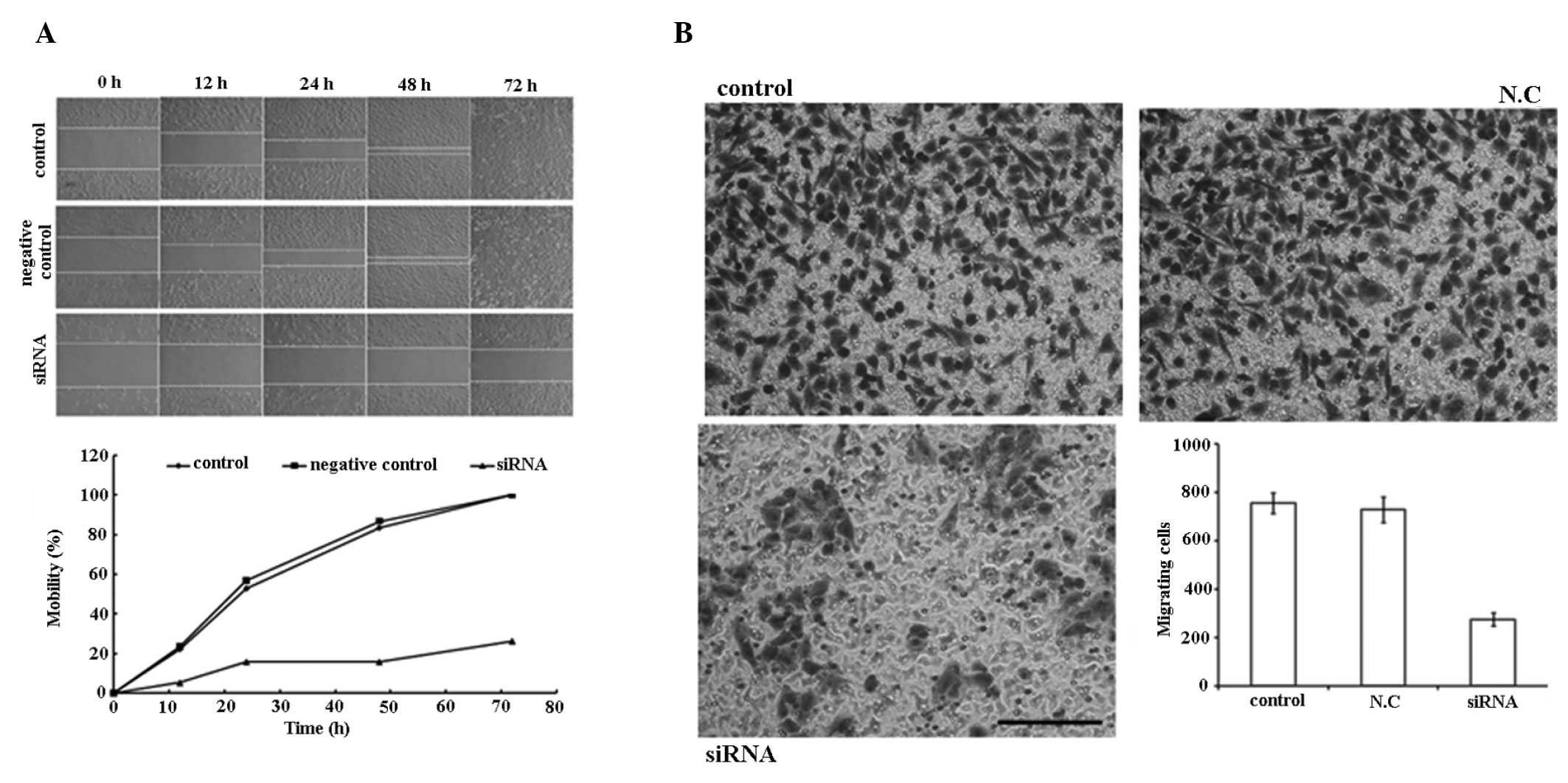 | Figure 5siRNA (100 nM) inhibits the mobility
of A549 cells. (A) Migration ability of the control group, negative
control group and siRNA transfection group 0, 12, 24, 48 and 72 h
following perforation of the A549 cell monolayer with a sterile 10
μl plastic pipette tip. The graph shows the mobility of cells in
three groups at different time-points following perforation. (B)
Cell migration evaluated using Transwell chambers. Cells that
migrated through the pores to the lower surface of the membrane
were fixed, stained and counted. Representative images of the
membrane surface in the three groups (control, N.C.,
siRNA-transfected). Column diagram shows the quantification of the
cell migration results. Each bar represents the mean ± standard
deviation of the counts from three independent experiments. Values
for siRNA-treated cells versus control show significant
differences. Scale bars, 100 μm. siRNA, small interfering
ribonucleic acid; N.C, negative control. |
PAR1 siRNA inhibits the invasiveness of
A549 cells
As shown in Fig. 6,
PAR1 siRNA visibly inhibited the invasiveness of the cells by
67.6%, at 100 nM compared with the control group. These results
suggest that PAR1 has a role in promoting the invasive phenotype of
A549 adenocarcinoma cells. A significant difference (P<0.05) was
observed between the control and treatment groups.
Discussion
PAR1, the prototype of the PAR family, was
originally recognized to transmit cellular responses to thrombin,
the main effector protease of the coagulation cascade (3). Subsequently, PAR1 was identified to
be involved in tumor progression. Bar-Shavit et al (13) reported that in epithelial
malignancies, by recruiting the dishevelled homolog (DVL), an
upstream signaling partner of the canonical wingless type mouse
mammary tumor virus integration site (Wnt) signaling pathway, PAR1,
is able to eventually cause β-catenin stabilization, a core event
in both tumorigenesis and developmental processes. Tantivejkul
et al (14) proved that
PAR1 is able to activate the necrosis factor κB (NF-κB) signaling
pathway, which finally results in the growth of prostate cancer
cells. Additionally, PAR1 is a regulator of several genes and
molecules involved in tumor growth and metastatic progression,
including the vascular endothelial growth factor (VEGF),
interleukin 8 (IL-8), and matrix metalloproteinases (MMPs)
(15,16).
Although the aforementioned studies support the fact
that PAR1 is an important tumor-associated gene, the precise
mechanism of its contribution to tumor progression remains to be
elucidated. Recently, MMP-1 was reported to proteolytically
activate PAR1 (17). In addition,
MMP-1 has been identified as one of the most upregulated proteins
in various types of cancer, including breast, esophageal and
colorectal carcinomas (18–20).
Silencing of MMP1-PAR1 signaling may improve the outcome of
Taxotere treatment in advanced, metastatic breast cancer (21).
All of these findings suggest that the inhibition of
PAR1 is beneficial to patients with tumors. RNAi is a tool which is
able to silence genes in a sequence-specific manner. Following the
finding that RNAi is mediated by long, double-stranded RNA in
Caenorhabditis elegans in 1998 and the revelation of
synthetic siRNAs being able to silence target genes in mammalian
systems in 2001, there has been a large number of reports on
therapeutic applications harnessing RNAi. Numerous cancer targets
for RNAi therapies have been found in previous studies and by using
RNAi, cancer therapy or its outcome may be improved (22–26).
In the present study, siRNA3 decreased PAR1 mRNA
levels by 91.3% as determined by PCR and PAR1 protein levels were
decreased by 83.6% as determined by western blot analysis.
Furthermore, the present study provided substantial evidence for
the role of PAR1 in survival, invasiveness and the metastatic
capabilities of the A549 lung adenocarcinoma cell line. By
silencing PAR1 with RNAi, the migration ability of A549 cells was
inhibited by 63.6%, invasion was decreased by 67.6% and viability
was only 44.5% of the control group.
The diffusion of the tumor cells from the primary
site and the infiltration of the extracellular matrix (ECM) were
two significant steps in tumor invasion and metastasis, which are
hallmarks of malignant tumors and are the major causes of mortality
of patients with cancer. Besides PAR1, MMP and urokinase-type
plasminogen activator (uPA) also participate in basement membrane
destruction. RNA-interfering technology which targets these
proteins in these pathways may contribute to favourable cancer
prognosis.
The principal advantage of RNAi is that all targets,
are theoretically druggable with RNAi, since any transcript that
encodes a protein that causes or contributes to a disease is able
to be targeted by RNAi (27,28).
This includes ‘undruggable’ targets which are, due to their
structure and location, not accessible by other therapeutics.
Efficient delivery to targeted tissues is the main
issue in developing RNAi as therapeutics. Both the non-viral
delivery of siRNAs and viral delivery of shRNAs are being advanced
as potential RNAi-based therapeutic approaches. Viral delivery
approaches include retroviral, lentiviral, adenoviral and
adeno-associated viral vectors. With regard to non-viral delivery,
liposomes, lipid complexes or conjugates with small molecules
(polymers, proteins and antibodies), electroporation and
hydrodynamic gene transfer have all been used to facilitate the
delivery of siRNAs to target cells.
Electroporation (EP) has been extensively used for
drugs and plasmid delivery in a large number of organs and tissues
(25,29–31).
By selecting appropriate electrical parameters and electrodes, gene
transfer may be optimized and tissue injury minimized. However,
electroporation is often limited to tumors that are accessible and
it is not possible to use it for the treatment of deep tumors,
currently, only electrodes for the treatment of cutaneous and
subcutaneous tumours have been designed and produced, including
needle electrodes and plate electrodes. It is aspired that in the
future, the development of technologies including microelectrodes
may be beneficial for cancer therapy.
In conclusion, in the present study PAR1 was proven
to be a significant target for clinical cancer therapy and
additionally provides a novel target in small-molecular drug
design. With the rapid progression of research and development of
applications, RNAi may remain a significant class of therapeutics
in the foreseeable future.
References
|
1
|
Wang Y, Yang H, Liu H, Huang J and Song X:
Effect of staurosporine on the mobility and invasiveness of lung
adenocarcinoma A549 cells: an in vitro study. BMC Cancer.
9:1742009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Arora P, Ricks TK and Trejo J:
Protease-activated receptor signalling, endocytic sorting and
dysregulation in cancer. J Cell Sci. 120:921–928. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rasmussen UB, Vouret-Craviari V, Jallat S,
Schlesinger Y, Pagès G, Pavirani A, Lecocq JP, Pouysségur J and Van
Obberghen-Schilling E: cDNA cloning and expression of a hamster
alpha-thrombin receptor coupled to Ca2+ mobilization.
FEBS Lett. 288:123–128. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Vu TK, Hung DT, Wheaton VI and Coughlin
SR: Molecular cloning of a functional thrombin receptor reveals a
novel proteolytic mechanism of receptor activation. Cell.
64:1057–1068. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Villares GJ, Zigler M, Wang H, et al:
Targeting melanoma growth and metastasis with systemic delivery of
liposome-incorporated protease-activated receptor-1 small
interfering RNA. Cancer Res. 68:9078–9086. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tellez C and Bar-Eli M: Role and
regulation of the thrombin receptor (PAR-1) in human melanoma.
Oncogene. 22:3130–3137. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chay CH, Cooper CR, Gendernalik JD,
Dhanasekaran SM, Chinnaiyan AM, Rubin MA, Schmaier AH and Pienta
KJ: A functional thrombin receptor (PAR1) is expressed on
bone-derived prostate cancer cell lines. Urology. 60:760–765. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Even-Ram S, Uziely B, Cohen P,
Grisaru-Granovsky S, Maoz M, Ginzburg Y, Reich R, Vlodavsky I and
Bar-Shavit R: Thrombin receptor overexpression in malignant and
physiological invasion processes. Nat Med. 4:909–914. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Darmoul D, Gratio V, Devaud H, Lehy T and
Laburthe M: Aberrant expression and activation of the thrombin
receptor protease-activated receptor-1 induces cell proliferation
and motility in human colon cancer cells. Am J Pathol.
162:1503–1513. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang Y, Zhan H, Xu W, Yuan Z, Lu P, Zhan
L and Li Q: Upregulation of matrix metalloproteinase-1 and
proteinase-activated receptor-1 promotes the progression of human
gliomas. Pathol Res Pract. 207:24–29. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hernández NA, Correa E, Avila EP, Vela TA
and Pérez VM: PAR1 is selectively over expressed in high grade
breast cancer patients: a cohort study. J Transl Med.
7:472009.PubMed/NCBI
|
|
12
|
Du X, Wang S, Lu J, et al: Correlation
between MMP1-PAR1 axis and clinical outcome of primary gallbladder
carcinoma. Jpn J Clin Oncol. 41:1086–1093. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bar-Shavit R, Turm H, Salah Z, Maoz M,
Cohen I, Weiss E, Uziely B and Grisaru-Granovsky S: PAR1 plays a
role in epithelial malignancies: transcriptional regulation and
novel signaling pathway. IUBMB Life. 63:397–402. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tantivejkul K, Loberg RD, Mawocha SC, Day
LL, John LS, Pienta BA, Rubin MA and Pienta KJ: PAR1-mediated
NFkappaB activation promotes survival of prostate cancer cells
through a Bcl-xL-dependent mechanism. J Cell Biochem. 96:641–652.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shimizu S, Gabazza EC, Hayashi T, Ido M,
Adachi Y and Suzuki K: Thrombin stimulates the expression of PDGF
in lung epithelial cells. Am J Physiol Lung Cell Mol Physiol.
279:L503–L510. 2000.PubMed/NCBI
|
|
16
|
Huang YQ, Li JJ, Hu L, Lee M and Karpatkin
S: Thrombin induces increased expression and secretion of VEGF from
human FS4 fibroblasts, DU145 prostate cells and CHRF
megakaryocytes. Thromb Haemost. 86:1094–1098. 2001.PubMed/NCBI
|
|
17
|
Boire A, Covic L, Agarwal A, Jacques S,
Sherifi S and Kuliopulos A: PAR1 is a matrix metalloprotease-1
receptor that promotes invasion and tumorigenesis of breast cancer
cells. Cell. 120:303–313. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Poola I, DeWitty RL, Marshalleck JJ,
Bhatnagar R, Abraham J and Leffall LD: Identification of MMP-1 as a
putative breast cancer predictive marker by global gene expression
analysis. Nat Med. 11:481–483. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Murray GI, Duncan ME, O’Neil P, McKay JA,
Melvin WT and Fothergill JE: Matrix metalloproteinase-1 is
associated with poor prognosis in oesophageal cancer. J Pathol.
185:256–261. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Migita T, Sato E, Saito K, Mizoi T, Shiiba
K, Matsuno S, Nagura H and Ohtani H: Differing expression of MMPs-1
and -9 and urokinase receptor between diffuse- and intestinal-type
gastric carcinoma. Int J Cancer. 84:74–79. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yang E, Boire A, Agarwal A, Nguyen N,
O’Callaghan K, Tu P, Kuliopulos A and Covic L: Blockade of PAR1
signaling with cell-penetrating pepducins inhibits Akt survival
pathways in breast cancer cells and suppresses tumor survival and
metastasis. Cancer Res. 69:6223–6231. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gartel AL and Kandel ES: RNA interference
in cancer. Biomol Eng. 23:17–34. 2006. View Article : Google Scholar
|
|
23
|
Kim DH and Rossi JJ: Overview of gene
silencing by RNA interference. Curr Protoc Nucleic Acid Chem.
Beaucage SL: 16(Unit 16.1)Wiley; New York, NY: 2009, View Article : Google Scholar
|
|
24
|
Takeshita F and Ochiya T: Therapeutic
potential of RNA interference against cancer. Cancer Sci.
97:689–696. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wu Z, Li X, Zeng Y, Zhuang X, Shen H, Zhu
H, Liu H and Xiao H: In vitro and in vivo inhibition of MRP gene
expression and reversal of multidrug resistance by siRNA. Basic
Clin Pharmacol Toxicol. 108:177–184. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xiao H, Wu Z, Shen H, Luo AL, Yang YF, Li
XB and Zhu DY: In vivo reversal of P-glycoprotein-mediated
multidrug resistance by efficient delivery of stealth RNAi. Basic
Clin Pharmacol Toxicol. 103:342–348. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Perrimon N, Ni JQ and Perkins L: In vivo
RNAi: today and tomorrow. Cold Spring Harb Perspect Biol.
2:a0036402010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Seyhan AA: RNAi: a potential new class of
therapeutic for human genetic disease. Hum Genet. 130:583–605.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Heller LC, Ugen K and Heller R:
Electroporation for targeted gene transfer. Expert Opin Drug Deliv.
2:255–268. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li S: Electroporation gene therapy: new
developments in vivo and in vitro. Curr Gene Ther. 4:309–316. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wells DJ: Gene therapy progress and
prospects: electroporation and other physical methods. Gene Ther.
11:1363–1369. 2004. View Article : Google Scholar : PubMed/NCBI
|