Introduction
Berberine is the major active component of the
Chinese herbal medicine Huang Lian (1), which has been widely used clinically
as a natural microbial agent in the treatment of diseases like
dysentery and gastroenteritis (2,3). In
recent years, a number of studies have reported that berberine has
antitumor effects on numerous cancer cell line types, including
leukocyte, glioblastoma, esophageal, breast and osteosarcoma
(4–8). The anticancer mechanisms of berberine
appear to be complex: Certain studies have suggested that berberine
may interfere with DNA replication, acting as an inhibitor of
replication and repair enzymes (6), while others have implied it may
induce DNA damage directly (9). In
addition, it has been reported that berberine is capable of
inducing apoptosis through DNA damage-independent signaling
pathways, including fatty acid synthase (Fas) receptor and estrogen
receptor (10,11).
As high doses of berberine provoke a wide variety of
cellular responses, the mechanisms that explain its genotoxicity
remain difficult to define. Rev3, a catalytic subunit of DNA
polymerases ζ (Polζ), participates in translesion DNA synthesis
(TLS) and has been identified as an important component in the
maintenance of chromosomal DNA, in its ability to enhance tolerance
to DNA damage (12). Previously,
we generated Rev3 deficient DNA repair deficient chicken B
lymphocyte (DT40) cells (Rev3−/−) and identified
they were hypersensitive to various DNA damaging agents, including
IR, cisplatin and UV (13,14). Besides their specific sensitivity,
Rev3−/− cells also possess a number of advantages
as DT40 clones. Firstly, the proliferation of DT40 clones with a
cell cycle time of 8 h is more rapid than that of the majority of
mammalian cell lines. Secondly, during logarithmic growth, 70% of
DT40 cells are in S phase, in contrast to the 50% for mammalian
cells, so that exogenous DNA damage may target DNA replication
directly. Finally, due to the absence of functional p53, DT40 cells
lack the G1/S checkpoint. Therefore, DNA repair activity
contributes a greater extent to cell survival, which aids our
observations of the chromosomal breaks in mitotic cells, as cells
carrying double-strand breaks (DSBs) can enter the M phase without
stimulating the apoptosis pathway (15). As a result,
Rev3−/− cells have been characterized as an
excellent tool for studying the DNA damage and repair mechanisms of
different chemicals, including tamoxifen and nitric oxide (NO)
(11,16).
In the present study, we used
Rev3−/− and other DNA repair gene deficient cells
to study the mechanisms of berberine. We identified that
Rev3−/− and several other DNA repair gene
deficient clones including Rev1−/−,
Parp1−/−, Rad18−/−,
Xrcc2−/−, were hypersensitive to the naturally
occurring alkaloid. Cell cycle and chromosomal break analysis
demonstrated that berberine induced a significant increase in G2/M
phase cells and chromosomal breaks in Rev3−/−,
compared with the wild-type (WT) cells. These results suggest that
berberine is able to directly induce DNA damage, and that
Rev3 participates in the processes that aid its repair.
Materials and methods
Chemicals
The berberine chloride and alachlor was obtained
from Sigma-Aldrich (St. Louis, MO, USA). Stock solution of
berberine (20 mM) was prepared in DMSO and stored at −20°C in
aliquots until use.
Cell line and cell culture
The cells were cultured as described previously
(17,18). The cell lines used in this study
are listed in Table I. The
phenotypes of these cells have been previously described (12,19–25).
The WT cell line and the DNA repair-deficient cell lines differ
only in the presence/absence of an endogenous DNA repair gene,
other than this difference, their genetic backgrounds were
identical (15). All cell lines
were cultured in RPMI-1640 medium supplemented with 10%
heat-inactivated fetal bovine serum, 1% chicken serum, 0.1% β
mercaptoethanol and 1% penicillin. The cell lines were maintained
at 39°C under a humidified atmosphere and 5% CO2. The
study was approved by the Ethics Committee of Sichuan University
(Chengdu, China).
 | Table IDNA repair genes mutated in the
analyzed DT40 clones. |
Table I
DNA repair genes mutated in the
analyzed DT40 clones.
| Gene | Function | Reference |
|---|
| Rev3 | TLS, catalytic
subunit of Polζ | 12 |
| Rev1 | TLS, deoxycytidyl
transferase activity | 19 |
| Rad18 | TLS | 20 |
| PARP1 | Poly(ADP)
ribosylation, related to single-strand and base excision
repair | 21 |
| XPA | An initial step of
nucleotide excision repair | 20 |
|
DNA-PKcs | Nonhomologous
end-joining-dependent double-strand break repair, the catalytic
subunit of DNA-dependent protein kinase | 22 |
| Ku70 | Initial step of
nonhomologous end-joining-dependent double-strand break repair,
associated with DNA-PK catalytic subunit | 19 |
| Rad52 | Homologous
recombination | 23 |
| Xrcc2 | Rad51 paralog,
homologous recombination, promotion of Rad51 assembly | 24 |
| Brca1 | Homologous
recombination | 23 |
| Brca2 | Homologous
recombination | 25 |
Measurement of cell proliferation by ATP
assay
To assess cell numbers following treatment with
berberine, we measured the amount of ATP in the whole cell lysate
(26). Cells
(1.5×103–1.5×104) were incubated in 1 ml
culture medium per well, containing various concentrations of
berberine. At 72 h, the ATP in the cellular lysates was measured to
assess the number of live cells. At least three independent
experiments were conducted. Sensitivity was calculated by dividing
the number of drug-treated cells, by the number of untreated cells
(13).
Cell-cycle analysis
Following treatment with 5 μM berberine for 16 h,
cells were labeled for 10 min with 20 μM Bromodeoxyuridine (BrdU)
and subsequently harvested. Harvested cells were fixed and analyzed
as previously described (19): (i)
In 4N HCl, 0.5% Triton X-100 for 30 min at room temperature; (ii)
in FITC-conjugated anti-BrdU antibody for 1 h at room temperature;
(iii) in 5 μg/ml propidium iodide (PI) in PBS. Following
incubation, cells were washed with PBS containing 2% FCS and 0.1%
sodium azide. Subsequent flow cytometric analysis was performed on
a FACScan. Fluorescence data were displayed as dot plots using the
Cell Quests software.
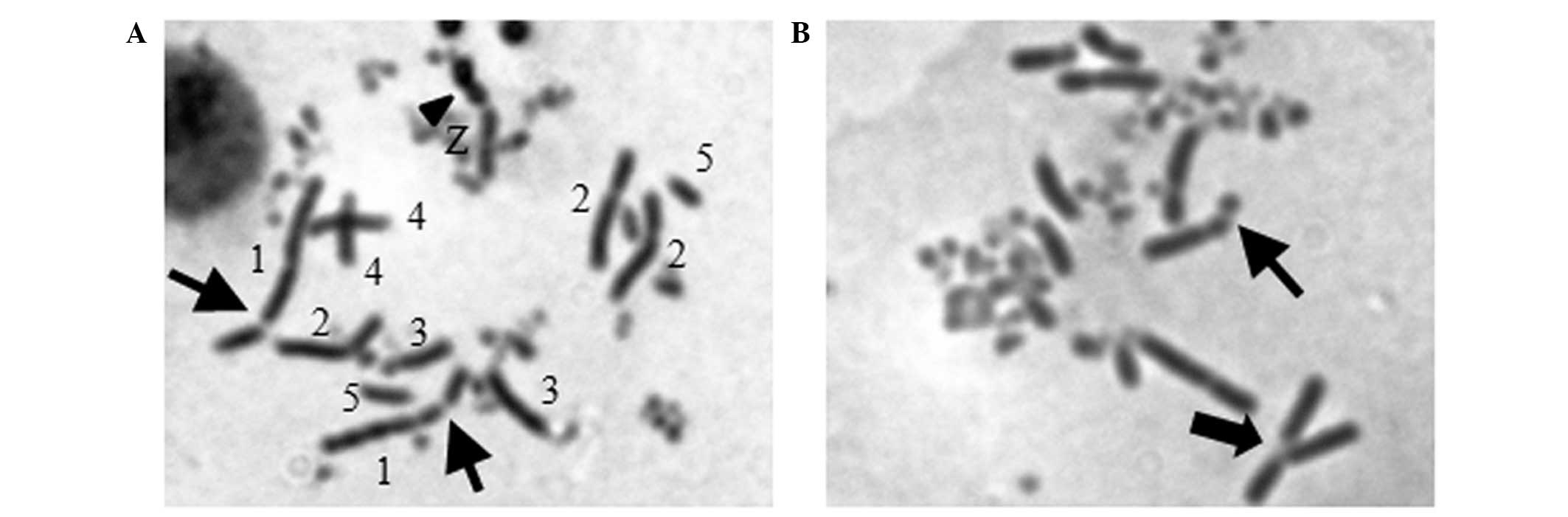
Chromosomal aberration (CA) analysis
Measurement of CA was performed as previously
described (12). The chicken
karyotype consisted of 80 chromosomes, including 11 major autosomal
macrochromosomes, the ZW sex chromosomes, and 67 microchromosomes
(15). In the present study, only
the 11 major autosomal macrochromosomes were measured. For CA
analysis, cells (3.0×106/3 ml) were treated with the
test doses of 2.5 and 5 μM for berberine and 5 μM for alachlor for
16 h. Then cells were treated for 30 min with 0.1 μg/ml colcemid
and went on harvesting for 2.5 h. We then suspended the cells with
75 mM KCL and incubated them for 15 min at room temperature, adding
5 ml of Carnoy’s solution (mixture of acetic acid and methanol,
1:3) and left for at least 30 min at room temperature. The cells
were then dropped onto ethanol-cleaned slides and dried by a flame.
The dried slide was then dipped into 5% Giemsa solution, left for 8
min at room temperature and rinsed carefully with water and
finally, they were air-dried. The cells were observed under a light
microscope (magnification, ×1,000).
Statistical analysis
Survival data were log-transformed giving
approximate normality. Analysis of covariance (ANCOVA) was used to
test for differences in the linear dose-response curves between WT
and a series of mutant cells. Viability of the DT40 cells was
estimated using regression curves. Regression-curve equations were
used to calculate LC50 (lethal concentration, 50%) values. Relative
LC50 values were normalized according to the LC50 value of the
parental WT cells.
Results
Rev3−/− cells are sensitive to
berberine
As Rev3−/− are specifically
sensitive to a variety of DNA damaging agents, in the present study
we assessed whether Rev3−/− cells exhibit a
higher sensitivity to berberine. We treated WT and
Rev3−/− cells with varying concentrations of
berberine for 72 h, and measured the viability of cells using an
ATP assay. As summarized in Fig.
1, Rev3−/− cells exhibited a significant
increase in sensitivity to berberine when compared with WT cells.
This observation indicated that berberine may induce DNA damage,
and that Rev3 associated DNA repair is an important
regulatory process in cellular tolerance to berberine-induced
genotoxicity. To study whether other DNA repair pathways were also
involved in tolerance to berberine, we assessed the sensitivity of
berberine in a panel of DT40 clones, which were separately
deficient in different DNA repair pathways (Table I). The DNA repair pathway mutants
demonstrated sensitivity to berberine in the following order: Rev3
> Rev1 > Parp1 = Xrcc2 = Rad18 > WT (parental DT40
cells)
Berberine induces enhanced arrest of cell
cycle in G2/M phase
As DT40 cells are deficient in p53, and agents
causing DNA damage would lead to cell arrest in G2/M phase, we
assessed if the increased sensitivity of Rev3−/−
to berberine was also correlated with G2/M arrest. Data from the
pulse labeled with BrdU indicated that berberine induced an
increase in the accumulation of Rev3−/− cells in
the G2/M phase compared with that in the WT cells (Fig. 2).
Berberine induces an increase of
chromosome breaks in Rev3−/− cells
To examine if the higher sensitivity of
Rev3−/− to berberine was specifically associated
with DNA damage, we measured the number of chromosome breaks in
mitotic cells. Fifteen CAs were detectable in 50 mitotic
Rev3−/− cells when the concentration of berberine
was 2.5 μM, while only 4 CAs were identified in WT cells. As the
concentration elevated to 5 μM, the CAs in
Rev3−/− cells markedly increased to 23, by
contrast, WT cells showed fewer aberrations of only 7 (Table II and Fig. 3). This result was consistent with
the positive control, alachlor. The data suggest that the
hypersensitivity of Rev3−/− to berberine is
correlated with a significant increase of berberine-induced
CAs.
 | Table IIFrequencies of chromosomal
aberrations in WT and Rev3 deficient DT40 cells. |
Table II
Frequencies of chromosomal
aberrations in WT and Rev3 deficient DT40 cells.
| | Chromatid | Isochromatid | | |
|---|
| |
|
| | |
|---|
| Cell | Treatment | Gaps | Breaks | Gaps | Breaks | Exchange | Total |
|---|
| WT | - | 0 | 2 | 0 | 0 | 0 | 2 |
| WT | Berberine (2.5
μm) | 0 | 2 | 0 | 1 | 1 | 4 |
| WT | Berberine (5
μm) | 1 | 3 | 1 | 1 | 1 | 7 |
| WT | Alachlor (5
μm) | 0 | 13 | 1 | 2 | 2 | 18 |
|
Rev3−/− | - | 1 | 1 | 1 | 1 | 0 | 4 |
|
Rev3−/− | Berberine (2.5
μm) | 1 | 9 | 1 | 2 | 2 | 15 |
|
Rev3−/− | Berberine (5
μm) | 1 | 17 | 2 | 2 | 1 | 23 |
|
Rev3−/− | Alachlor (5
μm) | 0 | 35 | 4 | 2 | 5 | 46 |
Discussion
In the present study, we utilized a panel of DNA
repair deficient DT40 cells to study the genotoxicity mechanisms of
the natural alkaloid, berberine. Our results demonstrated that
cells deficient in Rev3 exhibited the highest sensitivity to
berberine. The cell cycle and chromosome breaks assay provided
evidence that more G2/M arrest and chromosome breaks occurred in
Rev3−/− cells than in WT cells. While the
Rev−/− cells were the most sensitive, the other
DNA repair gene deficient cells, including
Rev1−/−, Parp1−/−,
Rad18−/− and Xrcc2−/− cells
were also sensitive to berberine, but polβ−/−,
XPA−/−, DNA-PK−/− cells were
not. Two important homologous recombination (HR) enzymes, Brca1 and
Brca2, were slightly resistant to berberine. These results provided
clear evidence that berberine is able to induce DNA damages, and
that Rev3 as well as Rev1, Rad18, Parp1
and Xrcc2 are important in processes that repair these
damages.
Rev3 is a catalytic subunit of Polζ. Studies
in yeast have revealed that Polζ cells are able to efficiently
bypass abasic sites, through extending from nucleotides inserted
opposite the lesion, by other TLS DNA polymerases. These data
suggest Polζ may be involved in the extension process of TLS in
higher eukaryotes (27). One
reverse genetics study in DT40 cells identified that Rev3
participated in not only TLS but also HR (12). Rev1 and Rad18 appear to have a
similar function to Rev3 in maintaining genomic stability by TLS
and HR (19). The high sensitivity
of Rev3−/−, Rev1−/− and
Rad18−/− to berberine implies that TLS or HR may
participate in berberine-induced DNA damage. TLS and HR are two
major pathways that promote survival following post-replication DNA
damage (28,29). HR accurately repairs DSBs that
arise during the mitotic cell cycle or those induced by
radiotherapy (30,31). This process also releases
replication forks that stall at damaged template DNA strands, by
using intact sister chromatids as a template (32,33).
The function of TLS is to release stalled replication forks by
filling gaps on daughter strands that remain, following DNA
replication caused by damage on the mother strand (34). Although cells deficient in
Brca1 or Brca2, were not sensitive but slightly
resistant to berberine, cells deficient in Xrcc2, a RAD51
paralog in HR (35), did exhibit
sensitivity to berberine. Parp1 regulates several processes
of DNA repair and also participates in HR. The sensitivity of
Parp1−/− and Xrcc2−/− cells to
berberine suggested that HR may participate in berberine-induced
DNA repair. The importance of HR in repairing berberine-induced DNA
damage has also been proved in yeast (9). Earlier studies have demonstrated that
Brca1−/− and Brca2−/− cells
were highly sensitive to several chemotherapeutic agents including
cisplatin, camptothecin and olaparib, which separately stalled DNA
replicative polymerases, via the formation of intra- and
inter-strand crosslinks, inhibition of topoisomerase I and
inhibition of Parp1 (36).
In addition, previous studies have shown that
Rad18−/− rather than Rev3−/−
was more sensitive to camptothecin (37). These differences imply that the
anticancer mechanisms of berberine are different from the
aforementioned chemotherapeutic agents, although all of them are
able to induce DSBs and HR repair. These results present that there
are different DNA damage mechanisms between berberine and these
agents, although certain studies considered that berberine may
function as a topoisomerase I inhibitor like camptothecin (38).
Acknowledgements
This study was mainly supported by the
Administration of Traditional Chinese Medicine of Sichuan Province,
China.
References
|
1
|
Yin J, Ye JP and Jia WP: Effects and
mechanisms of berberine in diabetes treatment. Acta Pharmaceutica
Sinica Sin B. 2:327–334. 2012. View Article : Google Scholar
|
|
2
|
Takase H, Yamamoto K, Ito K and Yumioka E:
Pharmacological studies on antidiarrheal effects of berberine and
geranii herba. Nihon Yakurigaku Zasshi. 102:101–112. 1993.(In
Japanese).
|
|
3
|
Lin JP, Yang JS, Lee JH, Hsieh WT and
Chung JG: Berberine induces cell cycle arrest and apoptosis in
human gastric carcinoma SNU-5 cell line. World J Gastroenterol.
12:21–28. 2006.PubMed/NCBI
|
|
4
|
Lin CC, Kao ST, Chen GW, Ho HC and Chung
JG: Apoptosis of human leukemia HL-60 cells and murine leukemia
WEHI-3 cells induced by berberine through the activation of
caspase-3. Anticancer Res. 26:227–242. 2006.PubMed/NCBI
|
|
5
|
Sanders MM, Liu AA, Li TK, Wu HY, Desai
SD, Mao Y, Rubin EH, LaVoie EJ, Makhey D and Liu LF: Selective
cytotoxicity of topoisomerase-directed protoberberines against
glioblastoma cells. Biochem Pharmacol. 56:1157–1166. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu Q, Jiang H, Liu Z, Wang Y, Zhao M, Hao
C, Feng S, Guo H, Xu B, Yang Q, et al: Berberine radiosensitizes
human esophageal cancer cells by downregulating homologous
recombination repair protein RAD51. PLoS ONE. 6:e234272011.
View Article : Google Scholar
|
|
7
|
Wang J, Liu Q and Yang QF:
Radiosensitization effects of berberine on human breast cancer
cells. Int J Mol Med. 30:1166–1172. 2012.PubMed/NCBI
|
|
8
|
Liu ZJ, Liu Q, Xu B, Wu JJ, Guo C, Zhu FL,
Yang QZ, Gao GM, Gong YQ and Shao CS: Berberine induces
p53-dependent cell cycle arrest and apoptosis of human osteosarcoma
cells by inflicting DNA damage. Mutat Res. 662:75–83. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pasqual MS, Lauer CP, Moyna P and
Henriques JAP: Genotoxicity of the isoquinoline alkaloid berberine
in prokaryotic and eukaryotic organisms. Mutat Res. 286:243–252.
1993. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ho YT, Lu CC, Yang JS, Chiang JH, Li TC,
Ip SW, Hsia TC, Liao CL, Lin JG, Wood WG and Chung JG: Berberine
induced apoptosis via promoting the expression of caspase-8, −9 and
−3, apoptosis-inducing factor and endonuclease G in SCC-4 human
tongue squamous carcinoma cancer cells. Anticancer Res.
29:4063–4070. 2009.PubMed/NCBI
|
|
11
|
Mizutani A, Okada T, Shibutani S, Sonoda
E, Hochegger H, Nishigori C, Miyachi Y, Takeda S and Yamazoe M:
Extensive chromosomal breaks are induced by tamoxifen and estrogen
in DNA repair-deficient cells. Cancer Res. 64:3144–3147. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sonoda E, Okada T, Zhao GY, Tateishi S,
Araki K, Yamaizumi M, Yagi T, Verkaik NS, van Gent DC, Takata M and
Takeda S: Multiple roles of Rev3, the catalytic subunit of polζ in
maintaining genome stability in vertebrates. EMBO J. 22:3188–3197.
2003.
|
|
13
|
Nojima K, Hochegger H, Saberi A, Fukushima
T, Kikuchi K, Yoshimura M, Orelli BJ, Bishop DK, Hirano S, Ohzeki
M, Ishiai M, et al: Multiple repair pathways mediate tolerance to
chemotherapeutic cross-linking agents in vertebrate cells. Cancer
Res. 65:11704–11711. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Takezawa J, Aiba N, Kajiwara K and Yamada
K: Caffeine abolishes the ultraviolet-induced REV3 translesion
replication pathway in mouse cells. Int J Mol Sci. 12:8513–8529.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yamamoto KN, Hirota K, Kono K, Takeda S,
Sakamuru S, Xia M, Huang R, Austin CP, Witt KL and Tice RR:
Characterization of environmental chemicals with potential for DNA
damage using isogenic DNA repair-deficient chicken DT40 cell lines.
Environ Mol Mutagen. 52:547–561. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wu XH, Takenaka K, Sonoda E, Hochegger H,
Kawanishi S, Kawamoto T, Takeda S and Yamazoe M: Critical roles for
polymeraseζ in cellular tolerance to nitric oxide-induced DNA
damage. Cancer Res. 66:748–754. 2006.
|
|
17
|
Sonoda E, Sasaki MS, Buerstedde JM,
Bezzubova O, Shinohara A, Ogawa H, Takata M, Yamaguchi-Iwai Y and
Takeda S: Rad51-deficient vertebrate cells accumulate chromosomal
breaks prior to cell death. EMBO J. 17:598–608. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Buerstedde JM and Takeda S: Increased
ratio of targeted to random integration after transfection of
chicken B cell lines. Cell. 67:179–188. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Takata M, Sasaki MS, Sonoda E, Morrison C,
Hashimoto M, Utsumi H, Yamaguchi-Iwai Y, Shinohara A and Takeda S:
Homologous recombination and non-homologous end-joining pathways of
DNA double-strand break repair have overlapping roles in the
maintenance of chromosomal integrity in vertebrate cells. EMBO J.
17:5497–5508. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Okada T, Sonoda E, Yamashita YM, Koyoshi
S, Tateishi S, Yamaizumi M, Takata M, Ogawa O and Takeda S:
Involvement of vertebrate polkappa in Rad18-independent
postreplication repair of UV damage. J Biol Chem. 277:48690–48695.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hossain MB, Ji P, Anish R, Jacobson RH and
Takada S: Poly(ADP-ribose) Polymerase 1 interacts with nuclear
respiratory factor 1 (NRF-1) and plays a role in NRF-1
transcriptional regulation. J Biol Chem. 284:8621–8632. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fukushima T: Genetic analysis of the
DNA-dependent protein kinase reveals an inhibitory role of Ku in
late S-G2 phase DNA Double-strand break repair. J Biol Chem.
276:44413–44418. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Qing Y, Yamazoe M, Hirota K, Dejsuphong D,
Sakai W, Yamamoto KN, Bishop DK, Wu X and Takeda S: The epistatic
relationship between BRCA2 and the other RAD51 mediators in
homologous recombination. PLoS Genet. 7:e10021482011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Danoy P, Sonoda E, Lathrop M, Takeda S and
Matsuda F: A naturally occurring genetic variant of human XRCC2
(R188H) confers increased resistance to cisplatin-induced DNA
damage. Biochem Biophys Res Commun. 352:763–768. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hatanaka A, Yamazoe M, Sale JE, Takata M,
Yamamoto K, Kitao H, Sonoda E, Kikuchi K, Yonetani Y and Takeda S:
Similar effects of Brca2 truncation and Rad51 paralog deficiency on
immunoglobulin V Gene diversification in DT40 cells support an
early role for Rad51 paralogs in homologous recombination. Mol Cell
Biol. 25:1124–1134. 2005. View Article : Google Scholar
|
|
26
|
Ji K, Kogame T, Choi K, Wang X, Lee JY,
Taniguchi Y and Takeda S: A novel approach using
DNA-repair-defcient chicken DT40 cell lines for screening and
characterizing the genotoxicity of environmental contaminants.
Environ Health Perspect. 117:1737–1744. 2009.PubMed/NCBI
|
|
27
|
Haracska L, Unk I, Johnson RE, Johansson
E, Burgers PM, Prakash S and Prakash L: Roles of yeast DNA
polymerasesδ and ζ and of Rev1 in the bypass of abasic sites. Genes
Dev. 15:945–954. 2001.
|
|
28
|
Hochegger H, Sonoda E and Takeda S:
Post-replication repair in DT40 cells: translesion polymerases
versus recombinases. Bioessays. 26:151–158. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
van Gent DC, Hoeijmakers JH and Kanaar R:
Chromosomal stability and the DNA double-stranded break connection.
Nat Rev Genet. 2:196–206. 2001.
|
|
30
|
Sharma S, Hicks JK, Chute CL, Brennan JR,
Ahn JY, Glover TW and Canman CE: REV1 and polymeraseζ facilitate
homologous recombination repair. Nucleic Acids Res. 40:682–691.
2012.
|
|
31
|
Venkitaraman AR: Linking the cellular
functions of BRCA genes to cancer pathogenesis and treatment. Annu
Rev Pathol. 4:461–487. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yamazoe M, Sonoda E, Hochegger H and
Takeda S: Reverse genetic studies of the DNA damage response in the
chicken B lymphocyte line DT40. DNA Repair (Amst). 3:1175–1185.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Branzei D, Vanoli F and Foiani M:
SUMOylation regulates Rad18-mediated template switch. Nature.
456:915–920. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Friedberg E, Lehmann A and Fuchs R:
Trading places: How do DNA polymerases switch during translesion
DNA synthesis? Mol Cell. 18:499–505. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Murakawa Y, Sonoda E, Barber LJ, Zeng W,
Yokomori K, Kimura H, Niimi A, Lehmann A, Zhao GY, Hochegger H, et
al: Inhibitors of the proteasome suppress homologous DNA
recombination in mammalian cells. Cancer Res. 67:8536–8543. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bhat A, Andersen PL, Qin Z and Xiao W:
Rev3, the catalytic subunit of Polζ, is required for maintaining
fragile site stability in human cells. Nucleic Acids Res.
41:2328–2339. 2013.
|
|
37
|
Yoshimura A, Nishino K, Takezawa J, Tada
S, Kobayashi T, Sonoda E, Kawamoto T, Takeda S, Ishii Y, Yamada K,
Enomoto T and Seki M: A novel Rad18 function involved in protection
of the vertebrate genome after exposure to camptothecin. DNA Repair
(Amst). 5:1307–1316. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Tomicic MT and Kaina B: Topoisomerase
degradation, DSB repair, p53 and IAPs in cancer cell resistance to
camptothecin-like topoisomerase I inhibitors. Biochim Biophys Acta.
1835:11–27. 2013.PubMed/NCBI
|