Introduction
Cervical cancer is the second most commonly
diagnosed cancer and third leading cause of mortality due to cancer
among women in underdeveloped countries (1). In 2012, there were an estimated 527,600
novel cervical cancer cases and 265,700 mortalities worldwide
(1). Although the majority of
patients can be cured with treatments based on surgery and
radiotherapy, a significant number eventually develop recurrent
disease, with the the risk of recurrence being 10–20% for patients
with stage IB-IIA [International Federation of Gynecology and
Obstetrics (FIGO) stage] and 50–70% for those in stages IIB-IVA2
(2,3).
With advances in the development of molecular-targeted drugs, novel
diagnostic and therapeutic molecular targets may enable the
development of novel therapies for cervical cancer. Therefore, it
is important to study proteins that are differentially expressed or
overexpressed in cervical cancer cells and investigate the
mechanisms regulating the expression of these proteins.
Hypoxia, a reduction in tissue oxygen tension due to
inadequate oxygen supply, is a common phenomenon that occurs in a
majority of solid tumours in humans (4). The presence of hypoxia is associated
with aggressive tumour progression, resistance to chemotherapy and
radiation, and poor prognosis (5,6). Tumour
cells and tissues adapt to a hypoxic microenvironment through the
activation of a number of hypoxia-associated molecules and
pathways, among which the hypoxia-inducible factor (HIF) is the
most predominant factor (7). HIF is a
heterodimeric basic helix-loop-helix PAS (Per-ARNT-Sim)
domain-containing transcription factor that consists of a
constitutively expressed β-subunit (HIF-β/ARNT) and one of the
three oxygen-regulated α-subunits, HIF-1α, HIF-2α and HIF-3α
(8,9).
The α-subunits are constitutively transcribed and translated, but
are regulated at the protein level by oxygen dependent
hydroxylation of specific prolyl residues (10). In cervical cancer, the majority of
studies on HIF have focused on HIF-1α. A number of studies have
revealed significant associations between HIF-1α expression, FIGO
stages, tumour grade and size (11).
Although clinically relevant associations between HIF-1α expression
and lymph node metastasis were not identified (12), HIF-1α was demonstrated to trigger
angiogenesis and tumour invasiveness to surrounding tissues at an
early stage of cervical carcinoma (13), and the inhibition of HIF-1α attenuated
cell migration under normoxia and hypoxia in the uterine cervix
cancer cell line SiHa (14). In
addition, HIF-1α may affect proliferation, apoptosis and cell cycle
(14,15). To date, there are a limited number of
studies on the function of HIF-2α in cervical cancer. To the best
of our knowledge, the only two studies on HIF-2α in cervical cancer
have focused on radiotherapy. One study demonstrated that HIF-2α
expression may have an important role in radioresistance in
patients with locally advanced cervical cancer (16). The other study indicated that the
proportion of HIF-2α-positive cells in tumour infiltrative
macrophages may be a novel predictive indicator for prognosis prior
to radiation therapy for uterine cervical cancer (17).
The difference in the roles of HIF-1α and HIF-2α in
tumorigenesis remains to be clearly delineated. In the present
study, the authors aimed to investigate the effect of HIF-2α on the
characteristics of the cervical cancer cell line, including
proliferation, apoptosis, cell cycle, invasion and cell autophagy.
Furthermore, the authors of the present study investigated the
function of HIF-1α to enable comparison with that of HIF-2α. The
results of the present study indicate that HIF-1α and HIF-2α have
similar effects on the characteristics of a cervical cancer cell
line.
Materials and methods
Cell culture and hypoxic exposure
The human cervical cancer cell line CaSki was
purchased from America Type Culture Collection (ATCC; Manassas, VA,
USA). CaSki cells were grown in RPMI-1640 medium (HyClone; GE
Healthcare Life Sciences, Logan, UT, USA) with 10% fetal bovine
serum (FBS; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA,
USA), supplemented with 100 U/ml penicillin G and 100 µg/ml
streptomycin (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). The
cells were maintained at 37°C in a humidified 5% CO2
incubator. To expose cells to hypoxia, the cells were cultured in a
Billups-Rothenburg chamber with 94% N2, 1% O2 and 5%
CO2 at 37°C.
RNA interference
Small interfering RNA (siRNA) targeting HIF-1α and
HIF-2α were purchased from Shanghai Gene-Pharma Co., Ltd.
(Shanghai, China). The siRNA sequences used were as follows: HIF-1α
siRNA sense, 5′-GCCACUUCGAAGUAGUGCUTT-3′ and antisense,
5′-AGCACUACUUCGAAGUGGCTT-3′; HIF-2α siRNA sense,
5′-GCGACAGCUGGAGUAUGAATT-3′ and antisense,
5′-UUCAUACUCCAGCUGUCGCTT-3′; negative control siRNA sense,
5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense,
5′-ACGUGACACGUUCGGAGAATT-3′. The cells were transfected with siRNAs
by using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's instructions. The knockdown
efficiency was determined by quantitative PCR and western blot
analysis. A total of three independent transfection experiments
were performed.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from each group of CaSki
cells using TRIzol reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's instructions. RNA was reverse
transcribed into cDNA using a PrimeScript RT reagent kit with cDNA
eraser (Takara Biotechnology Co., Ltd., Dalian, China) in a 10 µl
reaction according to the manufacturer's instructions. Equal
amounts of cDNAs were used as templates for RT-qPCR to detect the
levels of HIF-1α and HIF-2α expression relative to that of 18S rRNA
(endogenous control). The expression levels were quantified using
the ABI PRISM 7500 Sequence Detection system with the SYBR Green
qPCR SuperMix (Invitrogen; Thermo Fisher Scientific, Inc.). Primers
used in the present study were as follows: HIF-1α forward,
5′-GTGGATTACCACAGCTGA-3′ and reverse, 5′-GCTCAGTTAACTTGATCCA-3′;
HIF-2α forward, 5′-AATCCGAGCAGTGGAGTCAT-3′ and reverse,
5′-ACGTGCCATCAGACCCTCTT-3′; and 18S rRNA forward,
5′-CCTGGATACCGCAGCTAGGA-3′ and reverse,
5′-GCGGCGCAATACGAATGCCCC-3′. Experiments were performed in
duplicate and were repeated three times. Fold induction of gene
expression was calculated using the 2−ΔΔCq method
(18).
Western blot analysis
CaSki cells in each group were washed twice with
ice-cold phosphate-buffered saline and resuspended in ice-cold RIPA
buffer (Beyotime Institute of Biotechnology, Nantong, China)
containing 1 mmol/l phenylmethanesulfonyl fluoride and a cocktail
of protease inhibitors (1:100; Beyotime Institute of
Biotechnology). The samples were centrifuged at 4°C for 15 min at
800 × g. The supernatants were recovered and the total protein was
quantified using BCA Protein Assay kit (Pierce; Thermo Scientific
Inc.). Protein (30 µg) was loaded and separated on 8–12% SDS
polyacrylamide gels and transferred to polyvinylidene difluoride
membranes (EMD Millipore, Billerica, MA, USA). The membranes were
blocked for 1 h at room temperature with 5% milk in TBS containing
0.05% Tween-20 (TBST) and incubated at 37°C for 1 h with
anti-HIF-1α mouse monoclonal antibody (1:2,000 dilution, cat. no.
ab113642), anti-HIF-2α mouse monoclonal antibody (dilution 1:2,000,
cat. no. ab157249), anti-LC3B rabbit monoclonal antibody (dilution
1:1,000, cat. no. ab192890), to anti-Beclin 1 rabbit polyclonal
antibody (dilution 1:500, cat. no. ab62557) and anti-GAPDH mouse
monoclonal antibody (dilution 1:5,000, cat. no. ab8245) (all
purchased from Abcam, Cambridge, MA, USA). The membranes were
subsequently washed three times with TBST and incubated with a
horseradish peroxidase-conjugated mouse anti-rabbit IgG (1:2,000,
cat no. BM2006) or horseradish peroxidase-conjugated goat
anti-mouse IgG (1:3,000, cat. no. BA1051) (Wuhan Boster Biological
Technology, Ltd., Wuhan, China) for 40 min at 37°C. The membranes
were then washed three times with TBST and visualized using
Immobilon Western Chemiluminescent horseradish peroxidase substrate
(EMD Millipore). GAPDH served as an internal loading control. The
images of the blots were scanned, and densitometric analysis was
performed using Image Pro-Plus 6.0 software (Media Cybernetics,
Inc., Rockville, MD, USA). For quantification of specific bands, a
square of the same size was drawn around each band to measure the
density, and then the value was adjusted according to the density
of the background near that band. The results of densitometric
analysis have been expressed as a relative ratio of the target
protein to reference protein. The relative ratio of the target
protein of control group was arbitrarily presented as 1.
Cell proliferation assays
Proliferation of CaSki cells was monitored using the
Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc.,
Kumamoto, Japan) according to the manufacturer's instructions. A
total of 24 h following transfection, the cells were seeded at
1×104/well in a 96-well plate. The optical density (OD) of each
group was measured at 16, 24, 48 and 72 h. Briefly, 10 µl WST-8 was
added to each well, and after 4 h of incubation at 37°C. Absorbance
at 490 nm was measured using a microplate reader (Multiskan MK3;
Thermo Fisher Scientific, Inc.). The proliferation ratio was
calculated using the formula: Proliferation rate (survival rate) =
(ODtest/ODnegative control) × 100%. All experiments were performed
in triplicate and repeated three times.
Flow cytometric analysis
CaSki cells from each group were digested with
trypsin and centrifuged at 100 × g for 5 min. Following collection,
the cells were washed twice with PBS and centrifuged at 100 × g for
5 min. For cell apoptosis analysis, Annexin V-allophycocyanin (APC)
apoptosis detection kit (Nanjing KeyGen Biotech Co., Ltd., Nanjing,
China) was used to detect the apoptotic rate according to the
manufacturer's instructions (Nanjing KeyGen Biotech Co., Ltd.).
Briefly, the cell pellet (~1-5×105 cells) was
resuspended in 500 µl binding buffer. Then, 1.25 µl Annexin V-APC
and 10 µl 7-aminoactinomycin D were added and mixed at room
temperature (protected from light) for 15 min. Within 1 h, the
cells in each group were detected by flow cytometry (BD
Biosciences, San Jose, CA, USA) and data were interpreted using the
FlowJo software version 8 (Tree Star, Inc., Ashland, OR, USA). For
cell cycle analysis, cell cycle detection kits were used to detect
cell cycle according to the manufacturer's instructions (Nanjing
KeyGen Biotech Co., Ltd.). Briefly, the cells were fixed in 500 µl
70% precooled ethanol at 4°C overnight. An equal amount of PBS was
added twice for washing. Up to 100 µl RNase A was added at 37°C for
30 min, followed by the addition of 100 µl propidium iodide at 4°C
in the dark for 30 min. The different stages of the cell cycle were
detected by flow cytometry (BD Biosciences) and cell cycle
distribution was acquired using the ModFit LT 3.0 program (BD
Biosciences). Each experiment was repeated three times.
Matrigel-Transwell invasion assay
CaSki cells from each group were harvested, and
1×105 cells in 100 µl serum-free medium were placed into
the upper chamber of an insert pre-coated with Matrigel (pore size,
8 µm; BD Biosciences). The lower chamber was filled with 10% FBS
(600 µl, Gibco; Thermo Fisher Scientific, Inc.). After 24 h
incubation and removal of the cells on the upper chamber of the
filter with a cotton swab, the cells on the underside were fixed
with 4% paraformaldehyde for 15 min. The cells were subsequently
stained with 0.1% crystal violet in 20% ethanol and counted in five
randomly selected fields using a phase contrast microscope.
Migrating cells were monitored by imaging them at ×200
magnification with an Olympus microscope (Olympus, Tokyo, Japan) in
six independent fields for each well. The assays were performed in
triplicate.
Transmission electron microscope
At the end of the intervention, CaSki cells from
each group were digested with 0.25% trypsin and collected in
centrifuge tubes, followed by centrifugation at 100 × g for 10 min
at 4°C and then by a second centrifugation at a speed of 80 × g at
4°C for the same duration. The supernatant was discarded, and 2.5%
glutaraldehyde was added to the tubes to fix the cells at 4°C for 2
h. Subsequent to dehydration and embedding, ultra-thin sections (70
nm) were prepared using a microtome and each section was mounted on
a copper grid. Samples were contrasted in 4% aqueous uranyl acetate
(10 min) and then in Reynolds lead citrate (2 min) using normal
methods (19). The autophagosomes
were observed under a transmission electron microscope (JEM-1010;
Matsunaga Manufacturing, Yourou-cho, Japan), and images were
captured. Images were captured in three fields and the assays were
performed in triplicate. The number of autophagosomes was counted
by two independent researchers.
Statistical analysis
All statistical analyses were performed using SPSS
software, version 19.0 (IBM Corp., Armonk, NY, USA). Data are
expressed as the mean ± standard deviation. Statistical comparisons
were performed using one-way analysis of variance, followed by
Scheffe's test. The statistical differences between two groups were
determined using unpaired Student's t-test. P<0.05 was
considered to indicate a statistically significant difference.
Results
Expression profile of HIF-1α and
HIF-2α under hypoxic exposure
To investigate the function of HIF-2α in cervical
cancer, the authors of the present study monitored the protein
expression levels of HIF-2α under hypoxic exposure at different
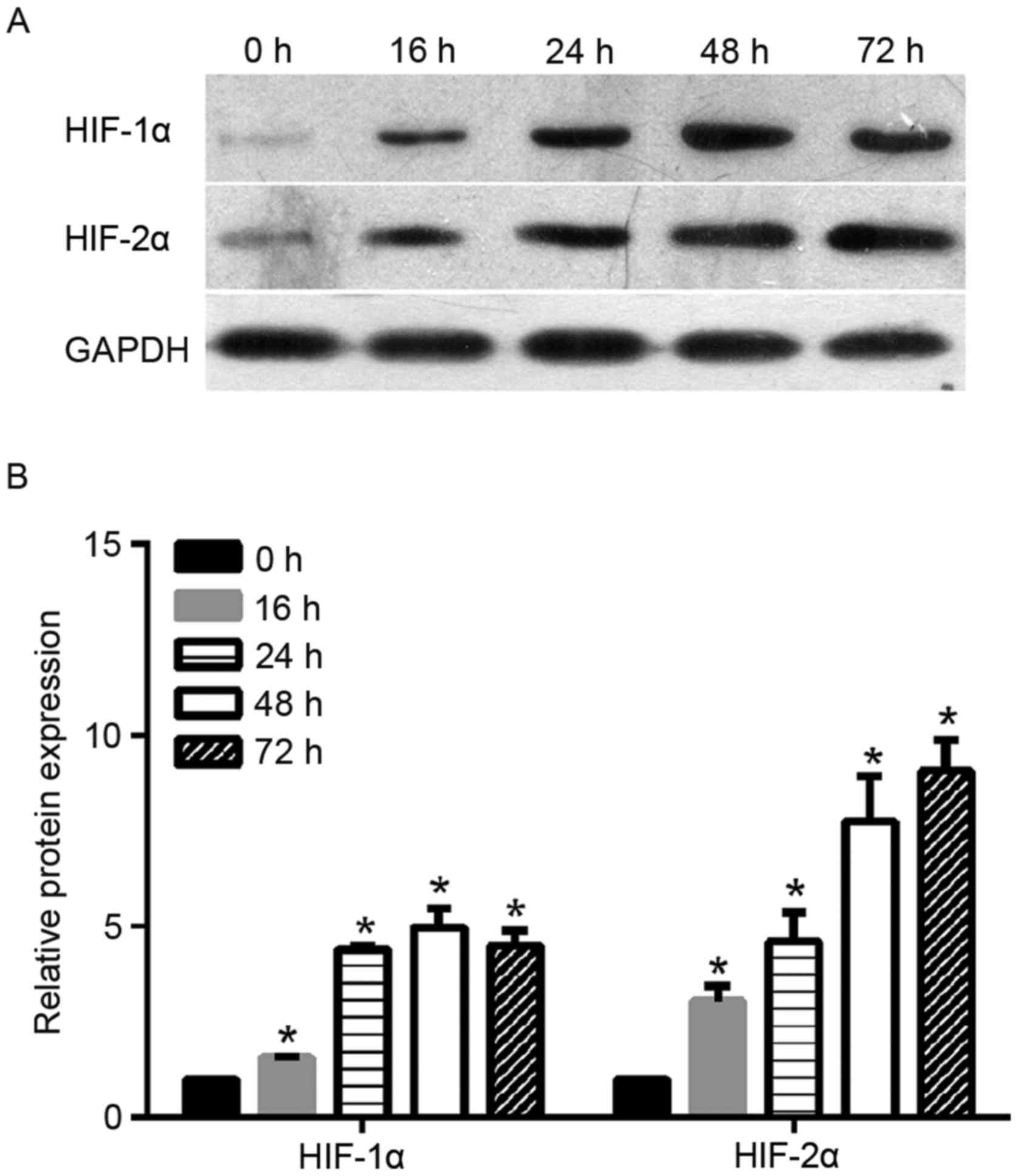
time-points (0, 16, 24, 48 and 72 h). As shown in Fig. 1, the levels of HIF-2α protein
increased steadily in the cervical cancer cells under hypoxic
exposure from 16 to 72 h. The increase in HIF-2α protein expression
in cells under hypoxic exposure was statistically significant when
compared with the expression in cells under normoxia (0 h). As a
control, the authors also monitored the protein expression level of
HIF-1α under hypoxic exposure. As shown in Fig. 1, HIF-1α protein level increased
steadily under hypoxic exposure from 16 to 24 h and peaked at 48 h.
Similar to HIF-2α, the increase of level HIF-1α protein expression
in cells under hypoxic exposure was statistically significant when
compared with cells under normoxia (0 h).
siHIF1α and siHIF2α may suppress the
expression of HIF-1α and HIF-2α under hypoxic exposure
To attenuate the expression of HIF-1α and HIF-2α
under hypoxic exposure, siHIF1α and siHIF2α were synthesized and
transiently transfected into CaSki cells. The transfected CaSki
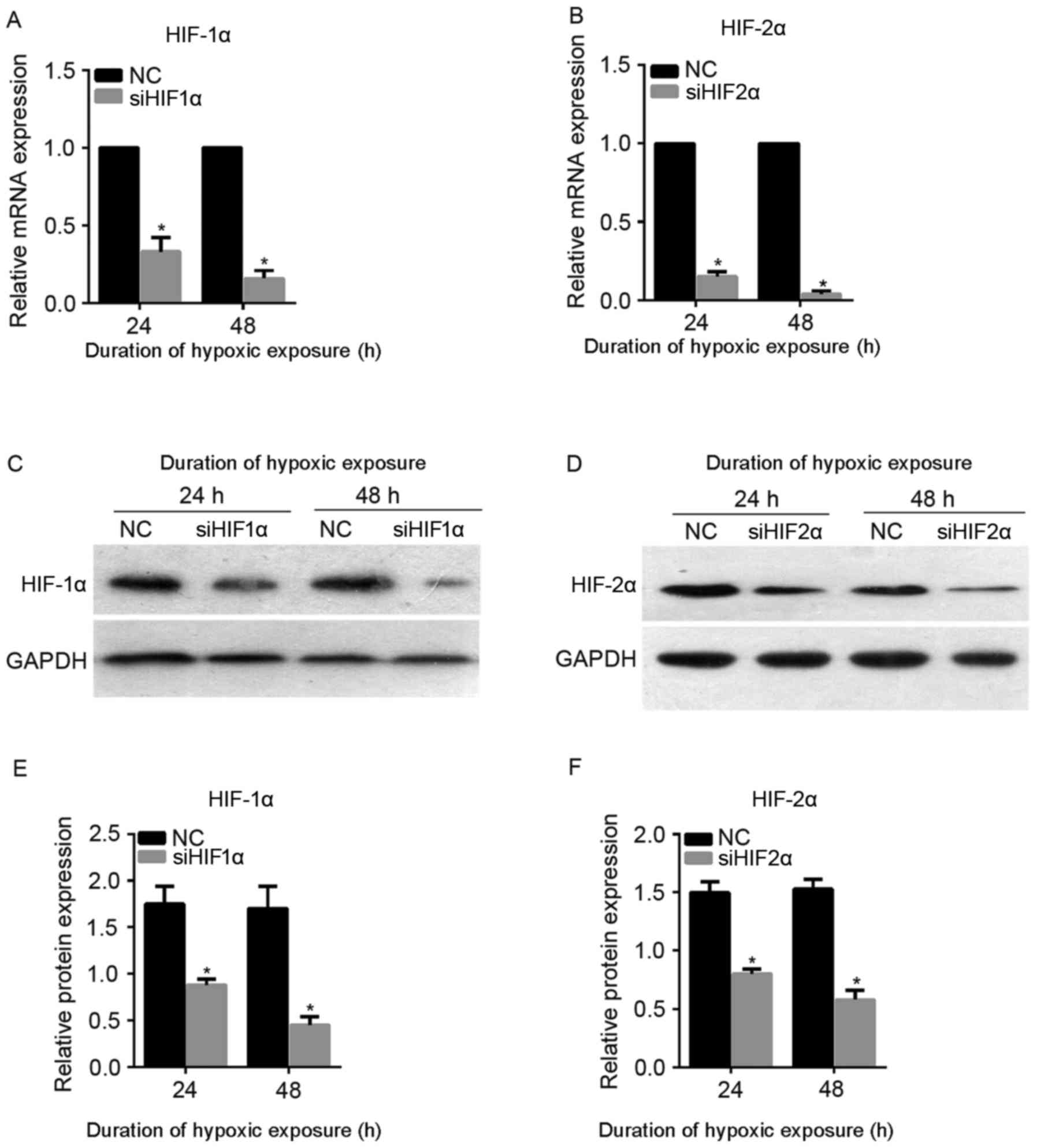
cells were cultured under hypoxic exposure for 24 or 48 h. As shown
in Fig. 2A and B, the mRNA expression
level of HIF-1α and HIF-2α was successfully downregulated following
siHIF1α or siHIF2α transfection at 24 and 48 h under hypoxic
exposure. Furthermore, the level of protein expression was also
successfully downregulated following siHIF1α or siHIF2α
transfection at 24 and 48 h under hypoxic exposure (Fig. 2C-F). Therefore, transfection with
siHIF1α or siHIF2α was able to successfully suppress the expression
of HIF-1α and HIF-2α, respectively, under hypoxic exposure, and
siHIF1α and siHIF2α were used for subsequent assays.
Effect of HIF-1α and HIF-2α silencing
on cell proliferation in cervical cancer under hypoxic
exposure
To determine the suitable duration of hypoxic
exposure for the assays performed during the present study, CaSki
cells were cultured under hypoxic conditions for 0, 16, 24, 48 and
72 h. CCK-8 assay was performed to detect the viability of each
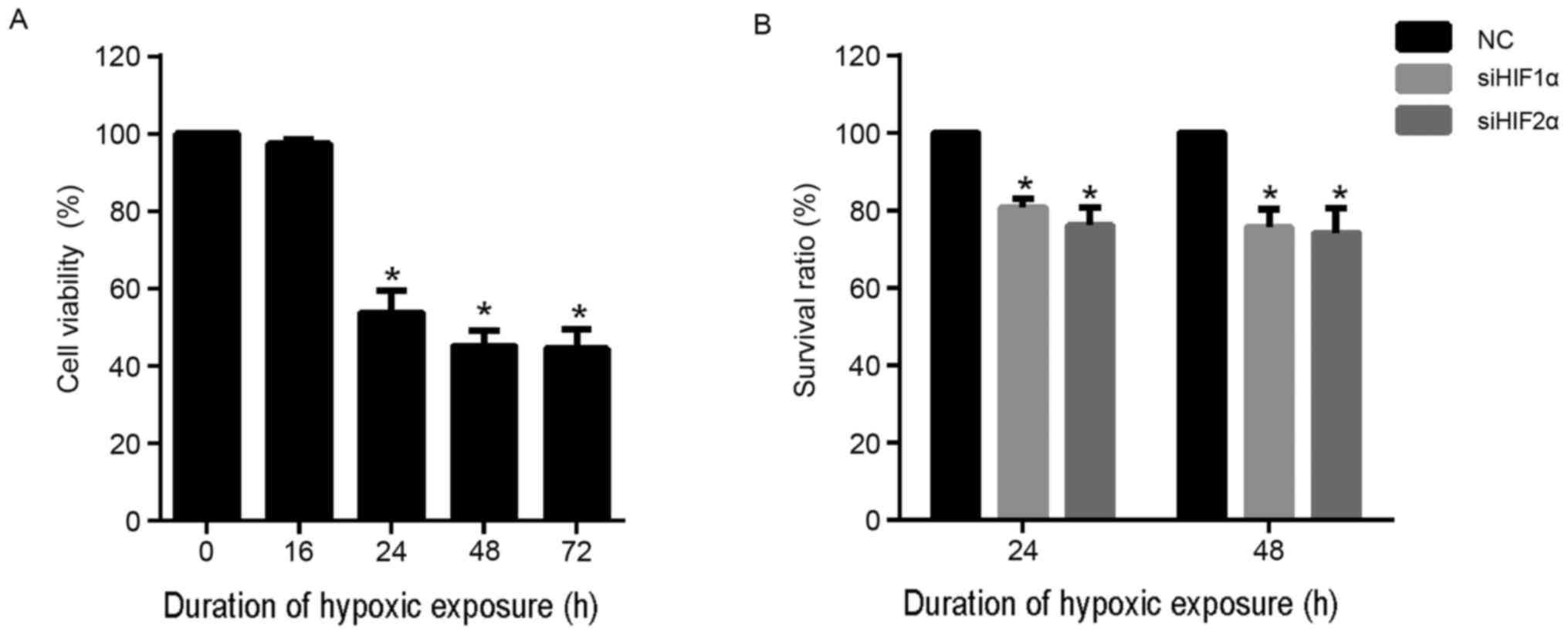
group of cells. As shown in Fig. 3A,
there was no evident effect on cell viability when the duration of
hypoxic exposure was 16 h. However, hypoxic exposure for 24, 48 and
72 h was able to significantly inhibit viability compared with
normoxia (0 h). In addition, the effect of hypoxic exposure for 48
and 72 h was similar. Based on these results, duration of hypoxic
exposure of 24 and 48 h were selected for further assays.
To investigate the effect of inhibition of HIF-2α
expression on cell proliferation, CaSki cells were transfected with
siHIF2α or negative control (NC) and cultured under hypoxia for 24
and 48 h. The results for the CCK-8 assay demonstrated that
suppression of HIF-2α expression significantly suppressed the
viability of CaSki cells at 24 and 48 h compared with the NC group
under hypoxia (Fig. 3B). Similarly,
HIF-1α suppression also significantly suppressed the viability of
CaSki cells at 24 and 48 h compared with the NC group under hypoxia
(Fig. 3B).
Effect of HIF-1α suppression and
HIF-2α expression on cell cycle in cervical cancer under hypoxic
exposure
To uncover the underlying mechanism of suppression
of cell proliferation by downregulation of HIF-1α and HIF-2α
expression, flow cytometry was used to observe the distribution of
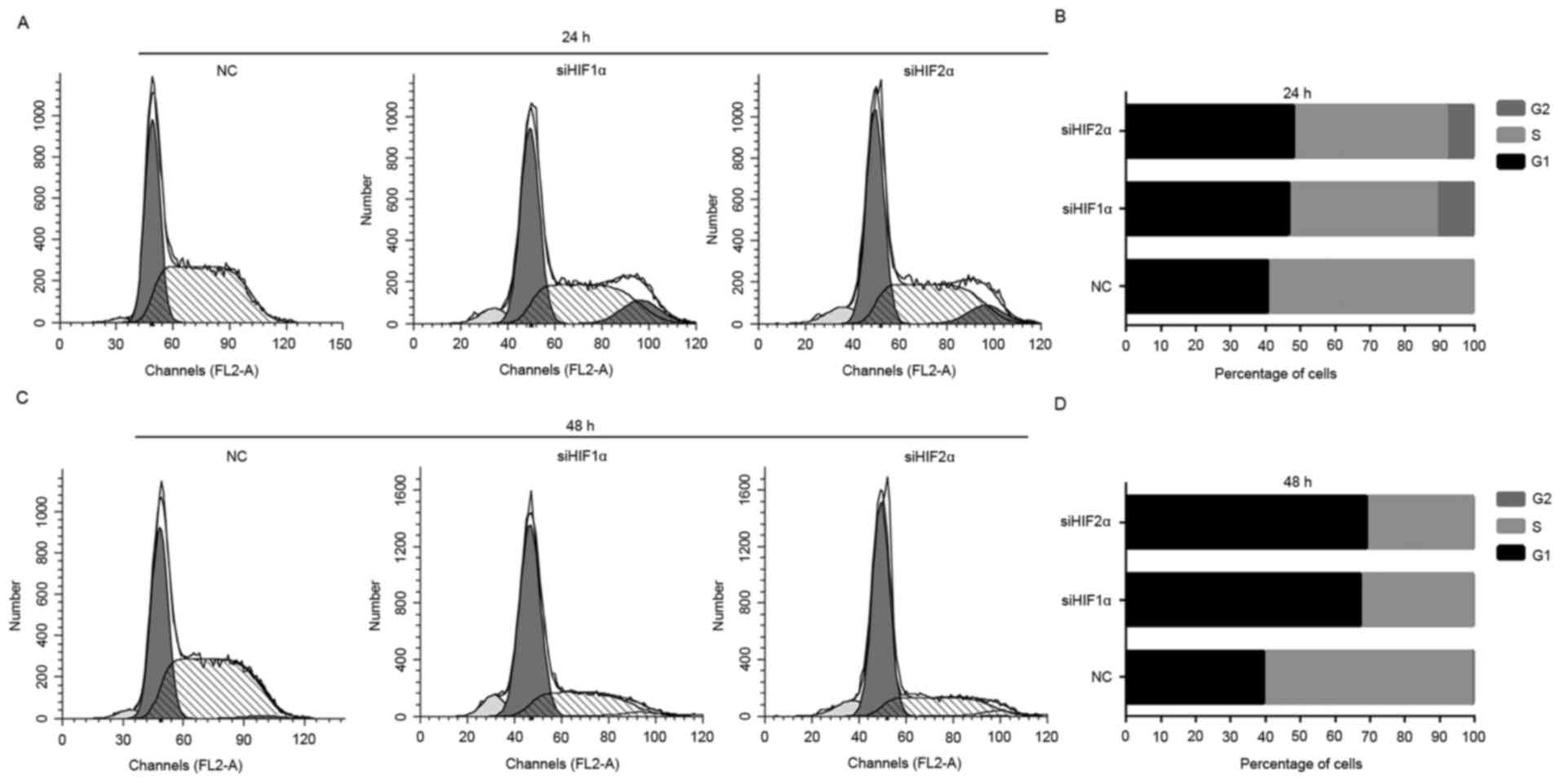
cells at different stages of the cell cycle. The downregulation of
HIF-1α and HIF-2α expression induced a significant G1-phase and
G2-phase arrest in CaSki cells, and the percentage of CaSki cells
in S phase decreased significantly when placed for 24 h under
hypoxic exposure (Fig. 4A and B).
Downregulation of HIF-1α and HIF-2α expression induced a
significant G1 phase arrest of CaSki cells, and the percentage of
cells in S phase decreased markedly when placed for 48 h under
hypoxic exposure compared with the NC-transfected group (Fig. 4C and D).
Effect of HIF-1α and HIF-2α
suppression on cell apoptosis in cervical cancer under hypoxic
exposure
To investigate the effect of HIF-1α and HIF-2α
suppression on apoptosis in cervical cancer under hypoxic exposure,
the authors of the present study monitored cell apoptosis by flow
cytometry. As shown in Fig. 5A and B,
when all the cells were exposed to hypoxia for 24 h, the percentage
of late apoptotic cells increased when the expression of HIF-1α and
HIF-2α was suppressed, while there was no effect on the percentage
of early apoptotic cells in the siHIF1α and siHIF2α-transfected
group compared with the NC-transfected group. The effect of HIF-1α
and HIF-2α suppression on cell apoptosis at 48 h under hypoxic
exposure was similar to the effect elicited by culturing the cells
for 24 h under hypoxic exposure, as shown in Fig. 5C and D.
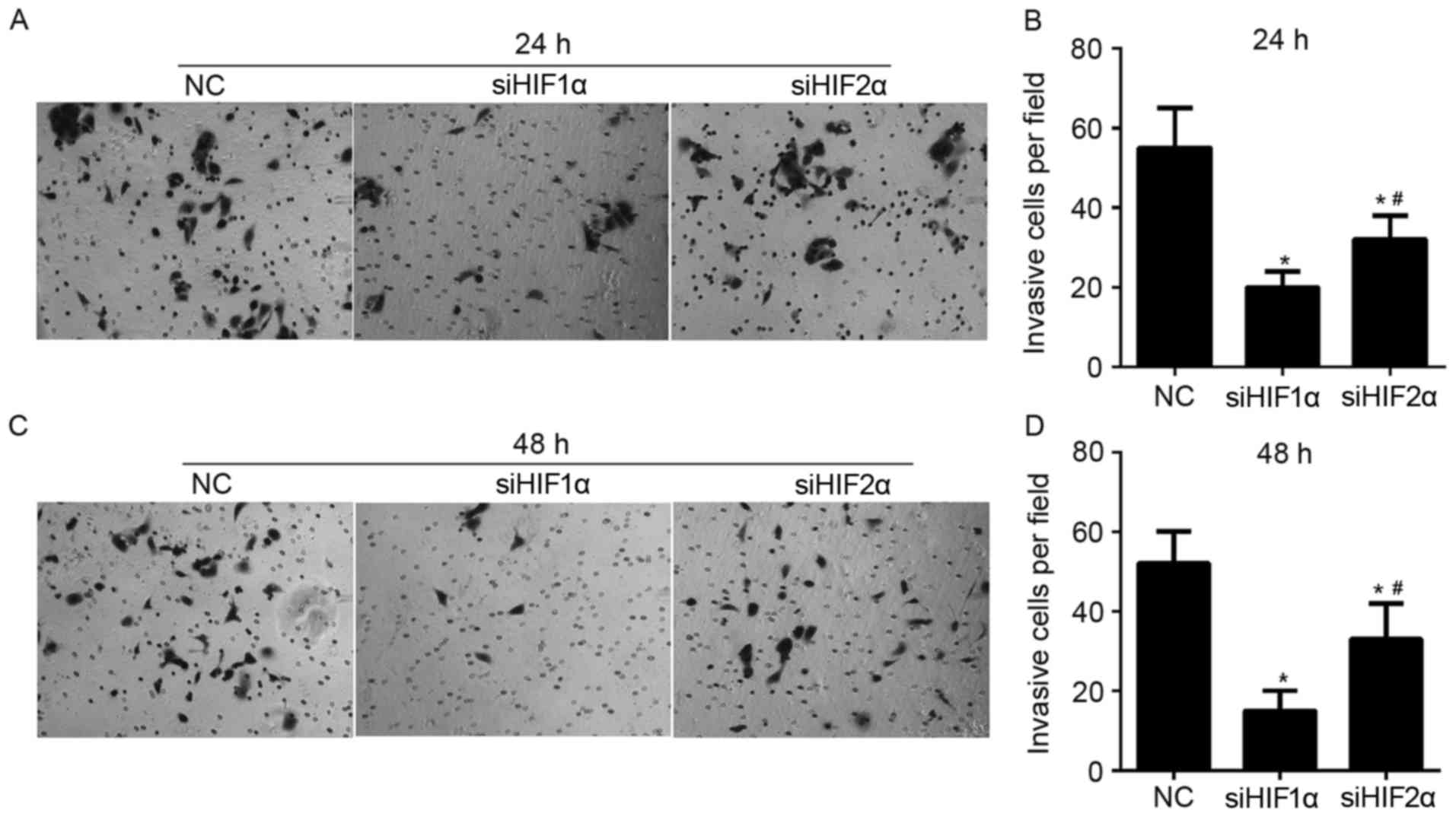
Effect of HIF-1α and HIF-2α
suppression on cell invasion in cervical cancer under hypoxic
exposure
To demonstrate the role of HIF-1α and HIF-2α in
regulating invasion of CaSki cells, Matrigel-Transwell assays were
performed following siHIF1α, siHIF2α or NC transfection under
hypoxic exposure. The number of cells that passed through the
Matrigel-coated membrane into the lower chamber was significantly
lower in the siHIF1α or siHIF2α-transfected cells compared with the
NC-transfected cells that were cultured for 24 h under hypoxic
exposure (Fig. 6A and B). The number
of cells that passed through the Matrigel-coated membrane onto the
lower chamber was significantly higher in siHIF2α-transfected cells
compared with the siHIF1α-transfected cells cultured for 24 h under
hypoxic exposure (Fig. 6A and B). The
effect of HIF-1α and HIF-2α suppression on invasion in cells
cultured for 48 h under hypoxic exposure was similar to the effect
on cells cultured for 24 h under hypoxic exposure, as shown in
Fig. 6C and D.
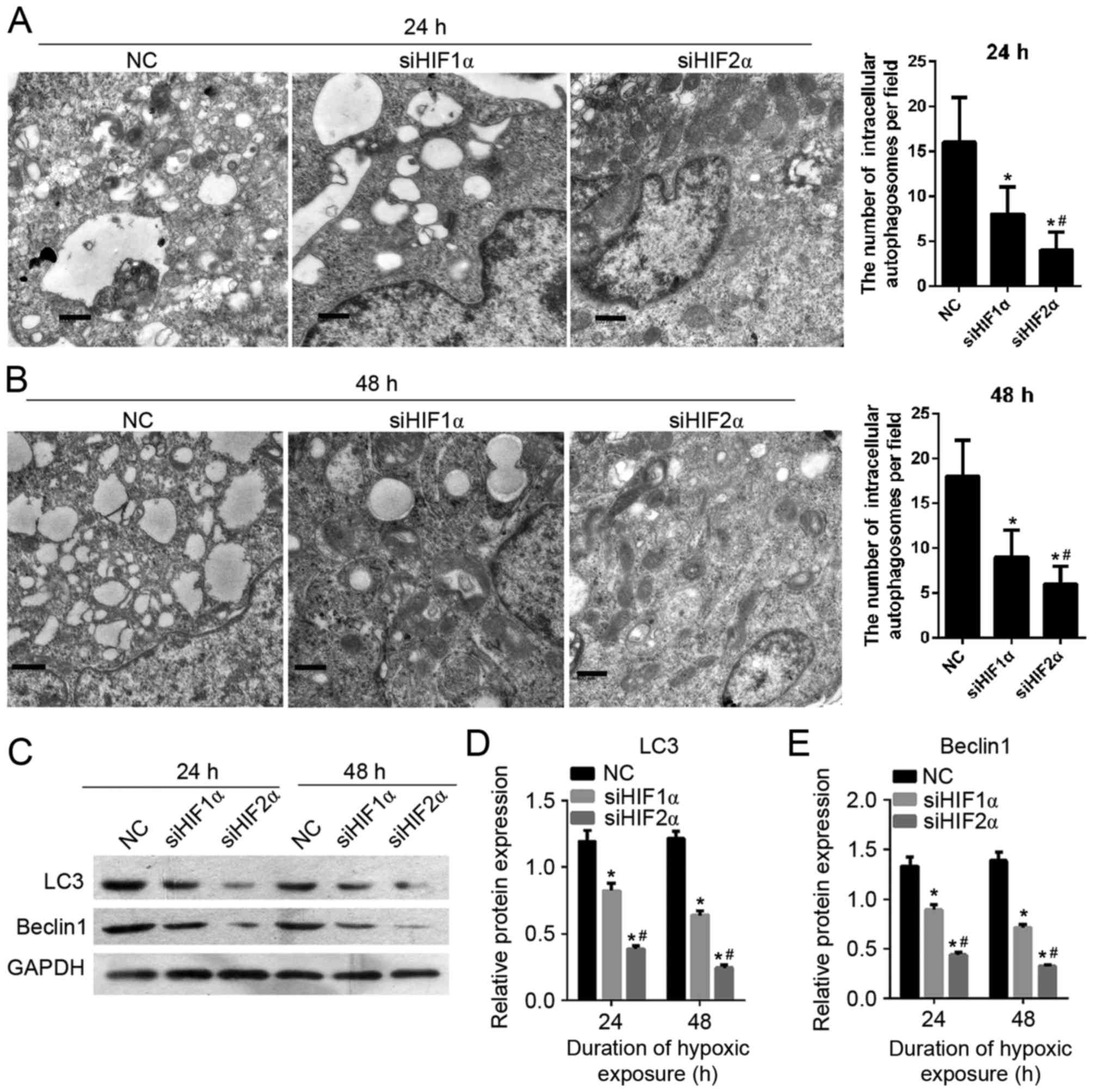
Effect of HIF-1α and HIF-2α
suppression on cell autophagy in cervical cancer under hypoxic
exposure
To detect the effect of HIF-1α and HIF-2α
suppression on CaSki cell autophagy, the formation of
autophagosomes was observed using transmission electron microscopy.
As shown in Fig. 7A and B, the number
of intracellular autophagosomes following siHIF1α or siHIF2α
transfection decreased compared with the cells in the
NC-transfected group which were cultured under hypoxic exposure for
24 h. The cells in the siHIF2α and siHIF1α-transfected groups were
cultured under hypoxia for 24 h. There was a lower number of
intracellular autophagosomes in the siHIF2α-transfected group
compared with the siHIF-1α-transfected group. Furthermore in order
to determine the effect of HIF-1α and HIF-2α suppression on cell
autophagy, the expression of autophagy-related gene LC3 and beclin
1 was examined using western blotting. The level of protein
expression of LC3 and beclin 1 following siHIF1α or siHIF2α
transfection was decreased compared with the NC-transfected group
under hypoxia for 24 h (Fig. 7C-E).
The level of LC3 and beclin1 protein expression in cells (cultured
under hypoxia for 24 h) in the siHIF2α-transfected group was lower
compared with the expression in the NC-transfected group (Fig. 7C-E). The effect of HIF-1α and HIF-2α
suppression on cell autophagy on cells cultured for 48 h under
hypoxia was similar to the effect on cells cultured for 24 h under
hypoxia, as shown in Fig. 7C-E.
Discussion
Cancer cells adapt to hypoxia by stabilising HIF-α
isoforms, which increase the transcription of several genes
(10). Among the genes regulated by
HIF are enzymes that have a role in proliferation, invasion,
metastasis and metabolism of tumour cells (20). To date, numerous cancer studies have
investigated HIF-1α as an important cancer drug target (21). In cervical cancer, HIF-1α is able to
affect proliferation, apoptosis, cell cycle and invasion (14,15).
However, less is known about HIF-2α, another HIF-α isoform.
Furthermore, one previous study demonstrated that the HIF isoforms
(HIF-1α and HIF-2α) have divergent effects on invasion, metastasis,
metabolism and formation of lipid droplets in human breast cancer
cells (10). Therefore, the present
study comprehensively investigated the function of HIF-2α in
cervical cancer cell line CaSki and compared the function of HIF-1α
and HIF-2α on proliferation, cell cycle, apoptosis, cell invasion
and cell autophagy. To the best our knowledge, this is the first
study where two isoforms of HIFα have been investigated
simultaneously in cervical cancer. The effect of HIF-1α and HIF-2α
on cell autophagy has also been investigated in the present
study.
In the present study, the authors investigated the
function of HIF-1α and HIF-2α on proliferation, cell cycle,
apoptosis, invasion and cell autophagy. The results indicated that
HIF-1α and HIF-2α have similar effects on proliferation, cell cycle
and apoptosis. HIF-1α or HIF-2α suppression may suppress
proliferation, induce G1 phase arrest and promote apoptosis in
cervical cancer cell line CaSki. However, the effect of HIF-1α and
HIF-2α on invasion and cell autophagy was different. The inhibitory
effect of HIF-1α on invasion was greater compared with the effect
of HIF-2α, while the inhibitory effect of HIF-1α on cell autophagy
was weaker compared with HIF-2α. The results revealed that HIF-1α
and HIF-2α have similar effects on the characteristics of the
cervical cancer cell line. The only difference is that the degree
of effect on cell invasion and autophagy. This similarity in the
roles of HIF-1α and HIF-2α in cervical cancer was supported by the
studies of HIF expression under hypoxia in the present study. It
was demonstrated that the level of HIF-1α and HIF-2α expression was
increased under hypoxia compared with normoxia. The major
difference was that the time-point at which the expression of these
proteins peaked. These results show that HIF-1α and HIF-2α may have
different roles at different stages of tumour progression, and
hence at different stages of hypoxia.
The results of the present study vary from a number
of previous studies (10,22,23). In
colon cancer, HIF-1α and HIF-2α have divergent roles in cell
proliferation and migration (22).
Specifically, HIF-1α-deficient cells exhibit lower rates of
proliferation and migration. However, HIF-2α-deficient cells
exhibit increased anchorage independent growth in a soft agar assay
(22). In human breast cancer cells,
suppression of only one of the HIF-1α or HIF-2α protein expression
was not sufficient to attenuate invasiveness, and cells with
suppression of expression of both proteins show significant
reduction in invasive characteristics (10). In hepatocellular carcinoma, the
knockdown of HIF-2α increased the autophagic activity of the tumour
cells, and attenuated apoptosis by increasing HIF-1α expression
(23). These results show that the
functions of HIF-1α and HIF-2α in cancer cells may differ with the
type of cancer, and this may be due to differences in the
downstream genes regulated by them.
In conclusion, the expression of HIF-1α and HIF-2α
increased under hypoxic exposure. The suppression of HIF-1α or
HIF-2α was able to inhibit proliferation, migration and autophagy,
induce G1 phase arrest and promote apoptosis. Therefore, HIF-1α and
HIF-2α exert a similar role in altering the biological
characteristics of cervical cancer cell line CaSki.
Acknowledgements
The present study was supported by grants from the
National Natural Science Foundation of China (grant no. 30760280),
the Natural Science Foundation of Jiangxi, China (grant nos.
2011ZBAB204019 and 20132BAB205101), and Science and Technology
Support Program of Jiangxi Province, China (grant nos. 2007BS12802
and 20142BBG70044). Funding was also provided by the Educational
Department Foundation of Jiangxi Province, China (grant nos.
GJJ08126, GJJ10060 and GJJ13682), and the Public Health Department
of Jiangxi Province, China (grant nos. 20072016, 20092025 and
20113071).
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kawano M, Mabuchi S, Matsumoto Y, Sasano
T, Takahashi R, Kuroda H, Kozasa K, Hashimoto K, Isobe A, Sawada K,
et al: The significance of G-CSF expression and myeloid-derived
suppressor cells in the chemoresistance of uterine cervical cancer.
Sci Rep. 5:182172015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Eifel PJ, Winter K, Morris M, Levenback C,
Grigsby PW, Cooper J, Rotman M, Gershenson D and Mutch DG: Pelvic
irradiation with concurrent chemotherapy versus pelvic and
para-aortic irradiation for high-risk cervical cancer: An update of
radiation therapy oncology group trial (RTOG) 90–01. J Clin Oncol.
22:872–880. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li SH, Shin DH, Chun YS, Lee MK, Kim MS
and Park JW: A novel mode of action of YC-1 in HIF inhibition:
Stimulation of FIH-dependent p300 dissociation from HIF-1{alpha}.
Mol Cancer Ther. 7:3729–3738. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lou JJ, Chua YL, Chew EH, Gao J, Bushell M
and Hagen T: Inhibition of hypoxia-inducible factor-1alpha
(HIF-1alpha) protein synthesis by DNA damage inducing agents. PLoS
One. 5:00105222010. View Article : Google Scholar
|
|
6
|
Zhang J, Cao J, Weng Q, Wu R, Yan Y, Jing
H, Zhu H, He Q and Yang B: Suppression of hypoxia-inducible factor
1α (HIF-1α) by tirapazamine is dependent on eIF2α phosphorylation
rather than the mTORC1/4E-BP1 pathway. PLoS One. 5:00139102010.
View Article : Google Scholar
|
|
7
|
Rankin EB and Giaccia AJ: The role of
hypoxia-inducible factors in tumorigenesis. Cell Death Differ.
15:678–685. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hu CJ, Sataur A, Wang L, Chen H and Simon
MC: The N-terminal transactivation domain confers target gene
specificity of hypoxia-inducible factors HIF-1alpha and HIF-2alpha.
Mol Biol Cell. 18:4528–4542. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tian H, McKnight SL and Russell DW:
Endothelial PAS domain protein 1 (EPAS1), a transcription factor
selectively expressed in endothelial cells. Genes Dev. 11:72–82.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shah T, Krishnamachary B, Wildes F,
Mironchik Y, Kakkad SM, Jacob D, Artemov D and Bhujwalla ZM: HIF
isoforms have divergent effects on invasion, metastasis, metabolism
and formation of lipid droplets. Oncotarget. 6:28104–28119. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Huang M, Chen Q, Xiao J, Yao T, Bian L,
Liu C and Lin Z: Overexpression of hypoxia-inducible factor-1α is a
predictor of poor prognosis in cervical cancer: A clinicopathologic
study and a meta-analysis. Int J Gynecol Cancer. 24:1054–1064.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ellingsen C, Andersen LM, Galappathi K and
Rofstad EK: Hypoxia biomarkers in squamous cell carcinoma of the
uterine cervix. BMC Cancer. 15:8052015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kim NS, Kang YJ, Jo JO, Kim HY, Oh YR, Kim
YO, Jung MH, Ock MS and Cha HJ: Elevated expression of thymosin β4,
vascular endothelial growth factor (VEGF) and hypoxia inducible
factor (HIF)-1α in early-stage cervical cancers. Pathol Oncol Res.
17:493–502. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tang B, Qu Y, Zhao F, Mao M, Tang J, Li X,
Ferriero D and Mu D: In vitro effects of hypoxia-inducible factor
1alpha on the biological characteristics of the SiHa uterine cervix
cancer cell line. Int J Gynecol Cancer. 19:898–904. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Schwock J, Geddie WR and Hedley DW:
Analysis of hypoxia-inducible factor-1alpha accumulation and cell
cycle in geldanamycin-treated human cervical carcinoma cells by
laser scanning cytometry. Cytometry A. 68:59–70. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kim MK, Kim TJ, Sung CO, Choi CH, Lee JW,
Bae DS and Kim BG: Clinical significance of HIF-2α immunostaining
area in radioresistant cervical cancer. J Gynecol Oncol. 22:44–48.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kawanaka T, Kubo A, Ikushima H, Sano T,
Takegawa Y and Nishitani H: Prognostic significance of HIF-2alpha
expression on tumor infiltrating macrophages in patients with
uterine cervical cancer undergoing radiotherapy. J Med Invest.
55:78–86. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lin Q, Wang Y, Chen D, Sheng X, Liu J and
Xiong H: Cisplatin regulates cell autophagy in endometrial cancer
cells via the PI3K/AKT/mTOR signalling pathway. Oncol Lett.
13:3567–3571. 2017.PubMed/NCBI
|
|
20
|
Lin D and Wu J: Hypoxia inducible factor
in hepatocellular carcinoma: A therapeutic target. World J
Gastroenterol. 21:12171–12178. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Masoud GN and Li W: HIF-1alpha pathway:
Role, regulation and intervention for cancer therapy. Acta Pharm
Sin B. 5:378–389. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Imamura T, Kikuchi H, Herraiz MT, Park DY,
Mizukami Y, Mino-Kenduson M, Lynch MP, Rueda BR, Benita Y, Xavier
RJ and Chung DC: HIF-1alpha and HIF-2alpha have divergent roles in
colon cancer. Int J Cancer. 124:763–771. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Menrad H, Werno C, Schmid T, Copanaki E,
Deller T, Dehne N and Brüne B: Roles of hypoxia-inducible
factor-1alpha (HIF-1alpha) versus HIF-2alpha in the survival of
hepatocellular tumor spheroids. Hepatology. 51:2183–2192. 2010.
View Article : Google Scholar : PubMed/NCBI
|