Introduction
BRCA1, breast cancer susceptibility gene 1,
maps to 17q21 (1) and encodes a
multifunctional protein involved in DNA repair (2), control of cell-cycle checkpoints
(3), ubiquitinylation and chromatin
remodeling (4). BRCA1 was
originally identified and cloned as a predisposition gene of
familial breast cancer in 1994 (5).
Although a significant fraction of familial types of breast cancer
could be explained by the inherited mutations of BRCA1, a
large proportion of familial and sporadic types of breast cancer
are not associated with mutations in BRCA1 (6–9)
Furthermore, BRCA1 mRNA levels were also found to be reduced
or absent in invasive sporadic types of breast cancer, thus
assigning a role of BRCA1 in these as well (10–12).
This suggests that other mechanisms for loss of functions may
exist.
Breast cancer results from the manifestation of
genetic and epigenetic changes in tumor suppressor genes and
oncogenes (13,14). Although the causal association
remains under debate, increasing evidence has shown that
hypermethylation of promoter CpG islands (15,16),
accompanied by global hypomethylation (17,18),
are common molecular events in cancer cells. Promoter CpG islands,
which frequently locate at the 5′ end regulatory regions of genes,
are subject to epigenetic modification by DNA methylation which is
known to play an important role in regulating gene expression
(16,19). If promoter CpG islands of key genes
were hypermethylated and form a closed repressive chromatin
configuration, the transcription initiation of the corresponding
genes should be affected (20).
There are reports that BRCA1 promoter
methylation status is associated with downregulated mRNA and
protein levels in breast cancerous tissues (21,22)
and cell lines (23). Aberrant
BRCA1 promoter methylation is associated with particular
biological and clinicopathological features (24,25).
However, these studies failed to lead to a conclusive finding. In
the current study, the hypothesis is that the absence of
BRCA1 transcript is associated with promoter methylation in
sporadic types of breast cancer. The present study further
investigates BRCA1 gene expression, methylation status and
their clinical significance in sporadic breast cancer.
Materials and methods
Study cohort and tissue samples
The study was approved by the ethics committee of
Guangxi Medical University (Nanning, China). All patients involved
in the study provided their informed consent. The study cohort
consisted of 49 patients, who were randomly selected from patients
continuously diagnosed with operable breast cancer between
September 2010 and September 2012 in the Department of Breast
Surgery of the Affiliated Tumor Hospital of Guangxi Medical
University. Patients were excluded from participation in the case
of familial types of breast cancer; prior chemotherapy or
radiotherapy for any malignancy; and pregnancy or lactation.
All the studied samples included 49 surgically
resected cancerous tissues and 49 corresponding paired
non-cancerous tissues which were taken >5 cm from the tumor
macroscopically (in cases where such distance was not present, the
non-cancerous sample was taken from the distance furthest from the
tumor sample). These samples were the fresh tissues following
surgical removal, and were immediately put into liquid nitrogen for
10 min and then into a −80°C ultra freezer. All samples were
subsequently reviewed and confirmed by the Department of Pathology
of the Affiliated Tumor Hospital of Guangxi Medical University.
Pathological information was collected from the patient clinical
database, and the information was blinded in another database. The
clinicopathologic characteristics of patients included histological
tumor type, primary tumor size, axillary nodal status, grade of the
disease, estrogen/progesterone receptor (ER/PR) status or HER-2/neu
status.
RNA extraction and quantitative
polymerase chain reaction (PCR)
The RNA isolated from the breast cancerous tissues
and paired non-cancerous tissues were kept using TRIzol®
reagent (Invitrogen Life Technologies, Carlsbad, CA, USA) according
to the manufacturer’s instructions. β-actin mRNA was the reference
gene used as the internal control. The primers of BRCA1 and
β-actin (Invitrogen Life Technologies) are shown in Table I. The PCR cycle conditions used are
95°C for 2 min; 40 cycles at 95°C for 10 sec, 60°C for 30 sec, and
70°C for 30 sec; and final extension at 72°C for 7 min.
Dissociation curve analyses were used to confirm the specificity of
the SYBR® Green (Invitrogen Life Technologies) signals
in each experiment. Data were analyzed using ABI Prism 7900 SDS
software (Applied Biosystems, Waltham, MA, USA). The mRNA
expression of BRCA1 was analyzed using the 2−ΔΔCt
method (26). Fluorescent data were
converted into RQ measurements, which stand for relative expression
automated by the system software. Thermal dissociation plots were
examined for biphasic melting curves. To ensure experiment
accuracy, quantitative PCR products were randomly selected for
sequencing.
 | Table IPrimer sequences used in the
study. |
Table I
Primer sequences used in the
study.
| Gene/primer | Sequence |
|---|
| BRCA1 |
| Forward |
TGTGAGGCACCTGTGGTGAC |
| Reverse |
GTGGCTGGCTGCAGTCAGTAG |
| β-catenin |
| Forward |
GAAACGGCTTTCAGTTGAGC |
| Reverse |
CTGGCCATATCCACCAGAGT |
| Bisulfite
sequencing primer |
| Forward |
GATTGGGTGGTTAATTTAGAGT |
| Reverse |
AATTATCTAAAAAACCCCACAA |
DNA extraction and sodium bisulfite
modification
Total genomic DNA of the specimens were isolated
from the breast cancerous tissues and paired non-cancerous tissues,
by the DNeasy Tissue AxyPrep DNA extraction kit (Tiangen, Beijing,
China). All procedures were followed according to the
manufacturer’s instructions. Genomic DNA was modified with
bisulfite using MethylCode™ Bisulfite Conversion kit (Invitrogen
Life Technologies) according to the manufacturer’s
instructions.
Bisulfite genomic sequencing
Bisulfite genomic DNA sequencing was carried out as
previously described (27) with
sodium bisulfite modification. The CpG islands of promoter region
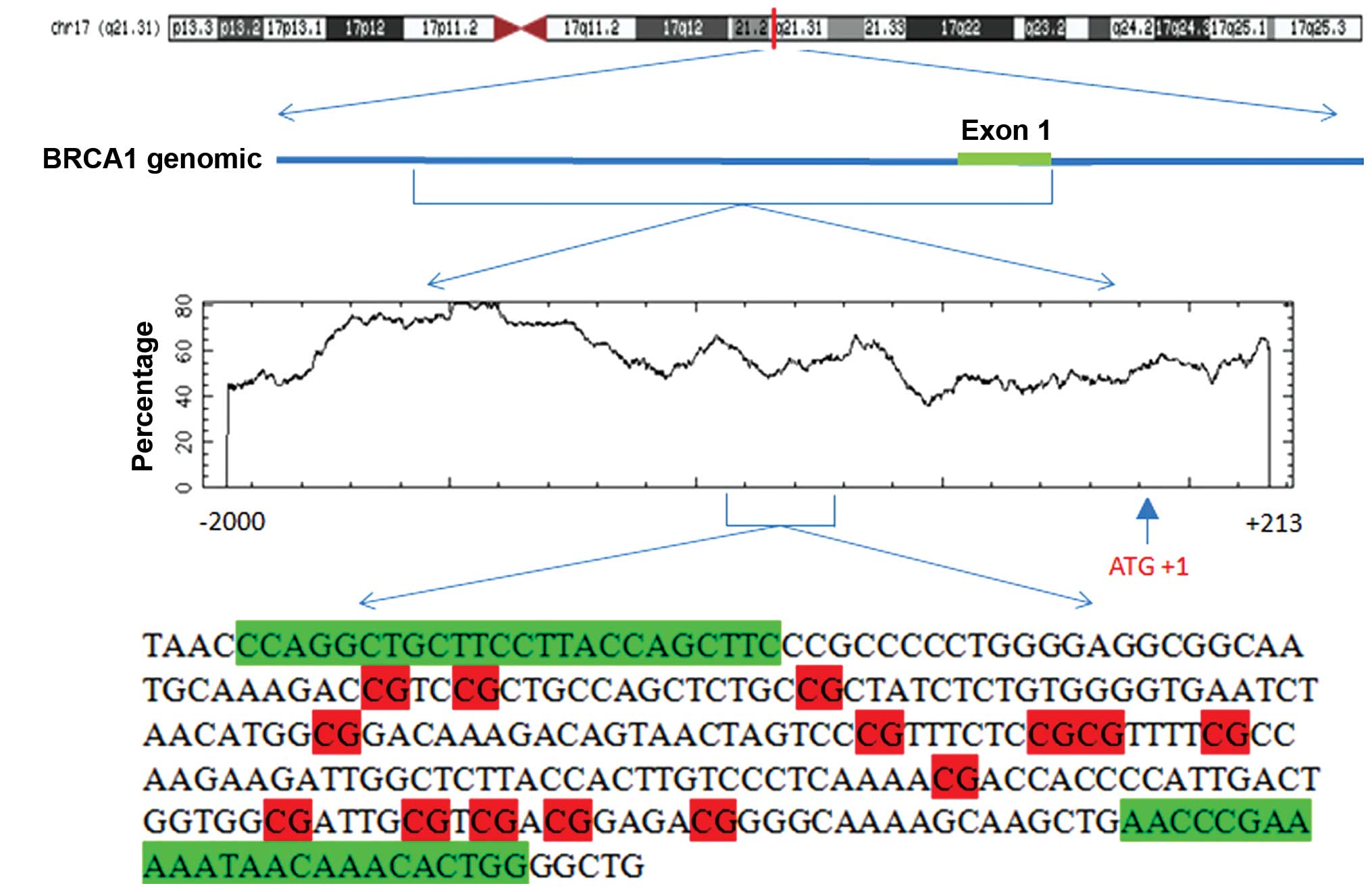
located between −937 and −717 bp (translation start site as 1). The
bisulfite-treated DNA was subjected to PCR in order to amplify the
BRCA1 promoter region. The primers of bisulfite genomic
sequencing are shown in Table I.
PCR products were purified and cloned into the pMD18-T vector
(Takara, Dalian, China), then transformed into Escherichia
coli strain DH5α (Invitrogen Life Technologies). Five positive
clones for each sample were selected and analyzed using the ABI
3730 DNA Sequencer (Applied Biosystems). The percentage of
methylation for each sample was calculated as the number of
methylated CpG dinucleotides/(5×48) × 100%.
Statistical analysis
Statistical analysis was performed using SPSS
software version 13.0 (SPSS, Inc., Chicago, IL, USA). Gene
expression levels or DNA methylation status of paired samples with
normal distribution were expressed as the mean ± standard
deviation; otherwise, they were expressed as the median with the
first and third interquartile ranges (IQR1 and IQR3). Associations
between BRCA1 mRNA expression or DNA methylation and the
categorical variables were assessed by the Pearson’s χ2
or Mann-Whitney U tests, as appropriate. Correlation coefficients
were assessed by Spearman’s correlation analysis. P<0.05 was
considered to indicate a statistically significant difference, and
all P-values were two-sided.
Results
Expression of BRCA1 in breast cancerous
and paired non-cancerous samples
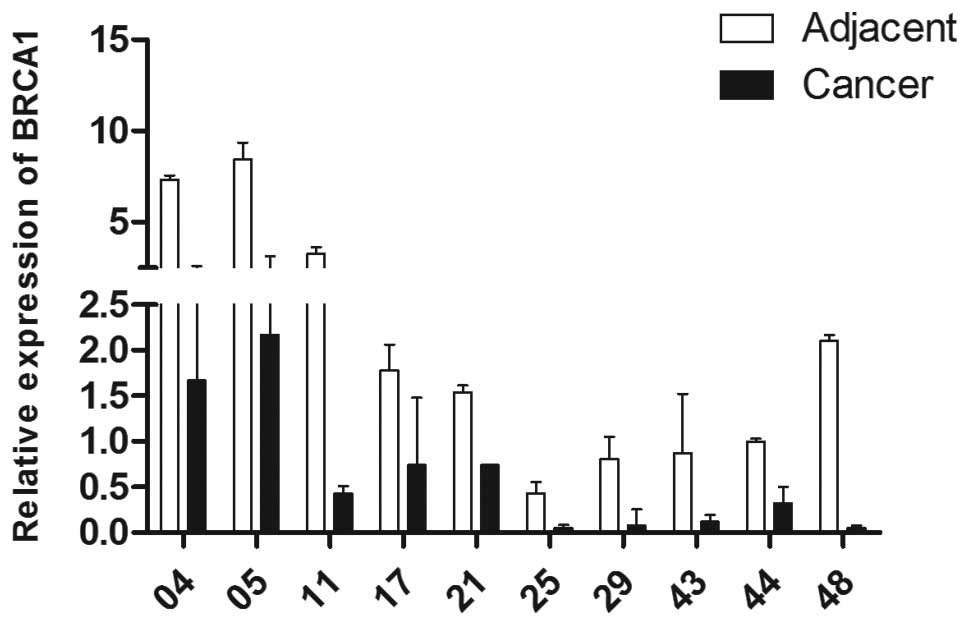
In the present study, the median level of
BRCA1 in non-cancerous samples was set as 1. The median RQs
of BRCA1 mRNA in breast cancerous and paired non-cancerous
samples were 0.33 (IQR1, 0.18; IQR3, 0.95) and 0.94 (IQR1, 0.46;
IQR3, 1.98), respectively. The difference between the two group was
statistically significant (Wilcoxon matched-pairs signed-ranks
test, P=0.001). The representative results of the quantitative PCR
are provided in Fig. 1. The results
indicate that the expression of BRCA1 in breast cancer was
aberrantly decreased at the transcriptional level.
According to the median RQ of the paired
non-cancerous tissues which was 0.94, the tumor tissues were
divided into three groups: overexpression (>0.94), normal
expression (=0.94) and reduced expression (<0.94). Due to the
limited number of tissues in the over and normal expression groups,
these two groups were combined into one group, named the unreduced
expression group. The correlation between BRCA1 mRNA and the
main clinicopathologic characteristics was also analyzed. The
associations between them are shown in Table II. No significant correlation was
observed between BRCA1 mRNA and the various parameters.
 | Table IICorrelations between BRCA1
mRNA expression and the main clinicopathologic characteristics. |
Table II
Correlations between BRCA1
mRNA expression and the main clinicopathologic characteristics.
| | BRCA1 mRNA
expression | |
|---|
| |
| |
|---|
| Variable | n | Reduced | % | Overexpression | % | P-value |
|---|
| Age (years) | | | | | | 0.304a |
| <50 | 28 | 19 | 67.9 | 9 | 32.1 | |
| ≥50 | 21 | 17 | 81.0 | 4 | 19.0 | |
| Menopause | | | | | | 0.129a |
| Pre | 29 | 19 | 65.5 | 10 | 34.5 | |
| Post | 20 | 17 | 85.0 | 3 | 15.0 | |
| TNM stage | | | | | | 0.078b |
| I | 9 | 5 | 55.6 | 4 | 44.4 | |
| II | 24 | 17 | 70.8 | 7 | 29.2 | |
| III | 16 | 14 | 87.5 | 2 | 12.5 | |
| ER | | | | | | 0.219a |
| Negative | 14 | 12 | 85.7 | 2 | 14.3 | |
| Positive | 35 | 24 | 68.6 | 11 | 31.4 | |
| PR | | | | | | 0.232a |
| Negative | 22 | 18 | 81.9 | 4 | 18.2 | |
| Positive | 27 | 18 | 66.7 | 9 | 33.3 | |
| HER-2/neu | | | | | | 0.156a |
| Negative | 34 | 27 | 79.4 | 7 | 20.6 | |
| Positive | 15 | 9 | 60.0 | 6 | 40.0 | |
| Ki-67 | | | | | | 0.492a |
| <0.15 | 15 | 12 | 80.0 | 3 | 20.0 | |
| ≥0.15 | 34 | 24 | 70.6 | 10 | 29.4 | |
| Axillary nodes | | | | | | 0.682a |
| Negative | 24 | 17 | 70.8 | 7 | 29.2 | |
| Positive | 25 | 19 | 76.0 | 6 | 24.0 | |
| Tumor stage | | | | | | 0.140b |
| T1 | 6 | 3 | 50.0 | 3 | 50.0 | |
| T2 | 25 | 18 | 72.0 | 7 | 28.0 | |
| T3 | 12 | 10 | 83.3 | 2 | 16.7 | |
| T4 | 6 | 5 | 83.3 | 1 | 16.7 | |
Correlation of BRCA1 expression and
methylation in breast cancerous and paired non-cancerous
samples
Analysis was carried out using the Methyl Primer
Express version 1.0 (Applied Biosystems) to analyze the CpG islands
of the region between −2,000 and +1,000 bp, including the
translational initiation codon (ATG) in detail. In the 5′ end of
the BRCA1 gene, two CpG islands were revealed, a 244 bp
(between −1,279 and −1,036 bp) and a 221 bp (between −937 and −717
bp) segment (Fig. 2).
To determine whether epigenetic silencing of the
BRCA1 gene also occurs in primary breast cancer, the
BRCA1 methylation status in 49 paired breast cancer and
corresponding non-cancerous tissues was examined (Fig. 3). Aberrant hypermethylation was
detected in 24 of 49 (49%) tumors, which was more frequent than
that in the paired no-cancerous tissues (11 of 49, 22.4%; Wilcoxon
matched-pairs signed-ranks test, P=0.007). In 24 cases of
hypermethylation of cancerous tissues, 20 (83.3%) showed a lower
BRCA1 mRNA expression. Furthermore, it was revealed that the
association between BRCA1 mRNA expression level and
methylation status was a negative correlation (r=-0.311, P=0.029),
which indicated a correlation between CpG island hypermethylation
and transcriptional silencing.
Association between BRCA1 methylation
level in breast cancer and the main clinicopathological
parameters
To ascertain the potential clinical significance of
the epigenetic event, analysis was conducted on the main
clinicopathological characters and methylation status of
BRCA1 in the 49 cases. The associations between BRCA1
methylation status and various clinicopathological parameters are
shown in Table III. No
significant correlation was observed between BRCA1
hypermethylation and main parameters such as age at diagnosis,
menopausal status, tumor, node and metastasis (TNM) stage, primary
tumor size, axillary nodal status, ER/PR status or HER-2/neu
status.
 | Table IIICorrelations between BRCA1
methylation status and the main clinicopathological
characteristics. |
Table III
Correlations between BRCA1
methylation status and the main clinicopathological
characteristics.
| | BRCA1
methylation status | |
|---|
| |
| |
|---|
| Variable | n | Reduced | % | Overexpression | % | P-value |
|---|
| Age | | | | | | 0.458a |
| <50 | 28 | 13 | 46.4 | 15 | 53.6 | |
| ≥50 | 21 | 12 | 57.1 | 9 | 42.9 | |
| Menopause | | | | | | 0.644a |
| Pre | 29 | 14 | 48.3 | 15 | 51.7 | |
| Post | 20 | 11 | 55.0 | 9 | 45.0 | |
| TNM stage | | | | | | 0.465b |
| I stage | 9 | 4 | 44.4 | 5 | 55.6 | |
| II stage | 24 | 15 | 62.5 | 9 | 37.5 | |
| III stage | 16 | 6 | 37.5 | 10 | 62.5 | |
| ER | | | | | | 0.470a |
| Negative | 14 | 6 | 42.9 | 8 | 57.1 | |
| Positive | 35 | 19 | 54.3 | 16 | 45.7 | |
| PR | | | | | | 0.201a |
| Negative | 22 | 9 | 40.9 | 13 | 59.1 | |
| Positive | 27 | 16 | 59.3 | 11 | 40.7 | |
| HER-2/neu | | | | | | 0.686a |
| Negative | 34 | 18 | 52.9 | 16 | 47.1 | |
| Positive | 15 | 7 | 46.7 | 8 | 53.3 | |
| Ki-67 | | | | | | 0.086a |
| <0.15 | 15 | 6 | 40.0 | 9 | 60.0 | |
| ≥0.15 | 34 | 19 | 55.9 | 15 | 44.1 | |
| Axillary nodes | | | | | | 0.666a |
| Negative | 24 | 13 | 54.2 | 11 | 45.9 | |
| Positive | 25 | 12 | 48.0 | 13 | 52.0 | |
| Tumor stage | | | | | | 0.508b |
| T1 | 6 | 2 | 33.3 | 4 | 66.7 | |
| T2 | 25 | 11 | 44.0 | 14 | 56.0 | |
| T3 | 12 | 9 | 75.0 | 3 | 25.0 | |
| T4 | 6 | 3 | 50.0 | 3 | 50.0 | |
Discussion
BRCA1 is a well-established breast cancer
susceptibility gene, and is involved in maintaining genome
integrity through pathways including participation in DNA damage
repair, the control of cell cycle checkpoints and apoptosis
(2–4). In these functions, BRCA1 is
implicated in the repair of double strand DNA breaks by homologous
chromosomal recombination (28,29).
Deficiencies in homology-directed DNA repair cause high levels of
genomic instability that increases the risk of tumorigenesis
(30). BRCA1 that impairs
such function leads to increased proliferation and chromosomal
instability. It has been proved that BRCA1 mutation is one
of the main genetic events in the hereditary type of breast cancer
(6), but no or limited somatic
mutations in BRCA1 have been found in the sporadic form of
breast cancer. On the another hand, a growing number of studies
have demonstrated loss of heterozygosity and a reduced level or
absence of BRCA1 expression in sporadic breast cancer
(31,32). These two factors suggest that
transcriptional and/or posttranscriptional repression of
BRCA1 may participate in the development of sporadic breast
cancer. One of the common mechanisms of functional inactivation of
tumor suppressor genes in cancer cells is the aberrant DNA
hypermethylation of CpG islands in the promoter region of the gene
that is associated with the loss of gene expression.
Firstly, BRCA1 expression at the mRNA level
was detected in paired cancerous and non-cancerous tissue of
sporadic breast cancer. BRCA1 expression of breast cancerous
tissues showed a relatively lower level as compared with those of
the paired non-cancerous tissues. The difference between them was
statistically significant. The data indicated that the expression
of BRCA1 in breast cancer was aberrantly reduced.
Subsequently, the present study demonstrated that the low
expression of BRCA1 was significantly correlated with the
hypermethylation in its promoter region. In the present study,
BRCA1 hypermethylation was detected in 49% of the cases,
which was consistent with other previous reports (9.1~59%)
(33–35). The differences in the frequency of
hypermethylation among the studies may be accounted for by several
factors including: Methodology, study cohort, adjacent
non-cancerous tissues contaminated by cancer cells and population
differences due to exposure to specific environmental factors.
Furthermore, the correlation between BRCA1
hypermethylation and the main clinicopathological characters was
analyzed. Ever since BRCA1 hypermethylation was proved to be
involved in sporadic breast cancer, some studies were dedicated to
explore the correlation between its aberrant methylation and the
disease characteristics. BRCA1 promoter methylation status
displayed various disease characteristic phenotypes in different
studies; however, the majority of studies demonstrated that
BRCA1 hypermethylation correlated with lack of estrogen and
progesterone receptor expression in younger females (<50 years).
Nevertheless, the present study did not discover a significant
association between BRCA1 hypermethylation and ER/PR status.
This result was similar to that reported in a previous study by Xu
et al (36). Furthermore, in
the study by Matros et al (37), they even found that BRCA1
hypermethylation is correlated with progesterone receptor positive
expression, suggesting a more complex phenotypic association.
In addition, two interesting details were revealed
which may be associated with favorable clinical prognosis, though
there was no association between BRCA1 hypermethylation and
the main clinicopathological characters including age at diagnosis,
menopausal status, TNM stage, primary tumor size, axillary nodal
status, ER/PR status or HER-2/neu status in sporadic breast cancer.
Firstly, the BRCA1 hypermethylation exhibited a higher
percentage of the smaller size primary tumor (T1 and T2, tumor size
≤5 cm) compared to the BRCA1 non-methylation ( 58.1% vs.
49.1%). The result seemed to display a trend that BRCA1
hypermethylation tumors tended to be the smaller tumor size. The
larger size of tumor is one of the most important indicators for
poor prognosis. Secondly, there was more BRCA1
hypermethylation of low Ki-67 index (<15%) cancerous tissues
compared with the BRCA1 non-methylation (60% vs. 40%). The
high Ki-67 index (≥15%), which is one of the important parameters
for luminal phenotype, has been proven to correlate with a greater
carcinogenic aggressiveness and worse prognosis. The reasons
underlying the phenomenon of BRCA1 methylation were not
elucidated, but some evidence was found correlating BRCA1
hypermethylation and favorable disease characteristics in a study
by Li et al (38). On the
basis of a smaller sample the study demonstrated high survival
rates associated with BRCA1 hypermethylation. Krasteva et
al (39) also reported that
breast cancer with BRCA1 hypermethylation was associated
with improved overall survival rates. Those evidences may partly
explain the present findings. Following cautious consideration, the
findings from the present study do not appear to be contradictory
to previous studies. By contrast, the present study results once
again manifested that breast cancer was a type of heterogeneous
disease from one aspect.
In conclusion, the present study revealed that
BRCA1 expression was expressed at low levels in the majority
of sporadic breast cancerous tissues, and DNA promoter
hypermethylation may be the potential mechanism accounting for
BRCA1 expression silence. Secondly, the reduced BRCA1
expression and BRCA1 hypermethylation did not correlate with
any clinicopathological features. Finally, partial sporadic breast
cancer with BRCA1 hypermethylation may exhibit favorable
clinicopathological status. It is thus reasonable to explore
BRCA1 epigenetic inactive mechanism and identify a subset of
sporadic breast cancer with a specific epigenetic phenotype.
Further studies to observe whether a specific BRCA1-related
sporadic breast cancer can indicate a favorable prognosis would be
beneficial.
Acknowledgements
This study was supported by funds from the Natural
Scientific Foundation of Guangxi (no. 2011GXNSFB018102).
References
|
1
|
Hall JM, Lee MK, Newman B, et al: Linkage
of early-onset familial breast cancer to chromosome 17q21. Science.
250:1684–1689. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Deng CX and Wang RH: Roles of BRCA1 in DNA
damage repair: a link between development and cancer. Hum Mol
Genet. 12(Spec No 1): R113–R123. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Xu B, St K and Kastan MB: Involvement of
BRCA1 in S-phase and G(2)-phase checkpoints after ionizing
irradiation. Mol Cell Biol. 21:3445–3450. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ralhan R, Kaur J, Kreienberg R and
Wiesmüller L: Links between DNA double strand break repair and
breast cancer: accumulating evidence from both familial and
nonfamilial cases. Cancer Lett. 248:1–17. 2007. View Article : Google Scholar
|
|
5
|
Miki Y, Swensen J, Shattuck-Eidens D, et
al: A strong candidate for the breast and ovarian cancer
susceptibility gene BRCA1. Science. 266:66–71. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Easton DF, Bishop DT, Ford D and Crockford
GP: Genetic linkage analysis in familial breast and ovarian cancer:
results from 214 families. The Breast Cancer Linkage Consortium. Am
J Hum Genet. 52:678–701. 1993.PubMed/NCBI
|
|
7
|
Narod SA, Ford D, Devilee P, et al: An
evaluation of genetic heterogeneity in 145 breast-ovarian cancer
families. Breast Cancer Linkage Consortium. Am J Hum Genet.
56:254–264. 1995.PubMed/NCBI
|
|
8
|
Ford D, Easton DF, Stratton M, et al:
Genetic heterogeneity and penetrance analysis of the BRCA1 and
BRCA2 genes in breast cancer families. The Breast Cancer Linkage
Consortium. Am J Hum Genet. 62:676–689. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hedenfalk I, Ringner M, Ben-Dor A, et al:
Molecular classification of familial non-BRCA1/BRCA2 breast cancer.
Proc Natl Acad Sci USA. 100:2532–2537. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Thompson ME, Jensen RA, Obermiller PS,
Page DL and Holt JT: Decreased expression of BRCA1 accelerates
growth and is often present during sporadic breast cancer
progression. Nat Genet. 9:444–450. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Magdinier F, Ribieras S, Lenoir GM,
Frappart L and Dante R: Down-regulation of BRCA1 in human sporadic
breast cancer; analysis of DNA methylation patterns of the putative
promoter region. Oncogene. 17:3169–3176. 1998. View Article : Google Scholar
|
|
12
|
Bianco T, Chenevix-Trench G, Walsh DC,
Cooper JE and Dobrovic A: Tumour-specific distribution of BRCA1
promoter region methylation supports a pathogenetic role in breast
and ovarian cancer. Carcinogenesis. 21:147–151. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jones PA and Baylin SB: The fundamental
role of epigenetic events in cancer. Nat Rev Genet. 3:415–428.
2002.PubMed/NCBI
|
|
14
|
Franco R, Schoneveld O, Georgakilas AG and
Panayiotidis MI: Oxidative stress, DNA methylation and
carcinogenesis. Cancer Lett. 266:6–11. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Herman JG and Baylin SB: Gene silencing in
cancer in association with promoter hypermethylation. N Engl J Med.
349:2042–2054. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Das PM and Singal R: DNA methylation and
cancer. J Clin Oncol. 22:4632–4642. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Feinberg AP and Vogelstein B:
Hypomethylation distinguishes genes of some human cancers from
their normal counterparts. Nature. 301:89–92. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Choi JY, James SR, Link PA, et al:
Association between global DNA hypomethylation in leukocytes and
risk of breast cancer. Carcinogenesis. 30:1889–1897. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Robertson KD: DNA methylation and
chromatin-unraveling the tangled web. Oncogene. 21:5361–5379. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Baylin SB and Ohm JE: Epigenetic gene
silencing in cancer-a mechanism for early oncogenic pathway
addiction. Nat Rev Cancer. 6:107–116. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Esteller M, Silva JM, Dominguez G, et al:
Promoter hypermethylation and BRCA1 inactivation in sporadic breast
and ovarian tumors. J Natl Cancer Inst. 92:564–569. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Birgisdottir V, Stefansson OA,
Bodvarsdottir SK, Hilmarsdottir H, Jonasson JG and Eyfjord JE:
Epigenetic silencing and deletion of the BRCA1 gene in sporadic
breast cancer. Breast Cancer Res. 8:R382006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rice JC, Massey-Brown KS and Futscher BW:
Aberrant methylation of the BRCA1 CpG island promoter is associated
with decreased BRCA1 mRNA in sporadic breast cancer cells.
Oncogene. 17:1807–1812. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Catteau A, Harris WH, Xu CF and Solomon E:
Methylation of the BRCA1 promoter region in sporadic breast and
ovarian cancer: correlation with disease characteristics. Oncogene.
18:1957–1965. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Turner N, Tutt A and Ashworth A: Hallmarks
of ‘BRCAness’ in sporadic cancers. Nat Rev Cancer. 4:814–819. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
27
|
Chu D, Zhang Z, Li Y, et al: Prediction of
colorectal cancer relapse and prognosis by tissue mRNA levels of
NDRG2. Mol Cancer Ther. 10:47–56. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang J and Powell SN: The role of the
BRCA1 tumor suppressor in DNA double-strand break repair. Mol
Cancer Res. 3:531–539. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ting NS and Lee WH: The DNA double-strand
break response pathway: becoming more BRCAish than ever. DNA Repair
(Amst). 3:935–944. 2004. View Article : Google Scholar
|
|
30
|
De Vargas Roditi L and Michor F:
Evolutionary dynamics of BRCA1 alterations in breast tumorigenesis.
J Theor Biol. 273:207–215. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Valentin MD, da Silva SD, Privat M,
Alaoui-Jamali M and Bignon YJ: Molecular insights on basal-like
breast cancer. Breast Cancer Res Treat. 134:21–30. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Warmoes M, Jaspers JE, Pham TV, et al:
Proteomics of mouse BRCA1-deficient mammary tumors identifies DNA
repair proteins with potential diagnostic and prognostic value in
human breast cancer. Mol Cell Proteomics. 11:M111.0133342012.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Esteller M, Silva JM, Dominguez G, et al:
Promoter hypermethylation and BRCA1 inactivation in sporadic breast
and ovarian tumors. J Natl Cancer Inst. 92:564–569. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Rice JC, Ozcelik H, Maxeiner P, Andrulis I
and Futscher BW: Methylation of the BRCA1 promoter is associated
with decreased BRCA1 mRNA levels in clinical breast cancer
specimens. Carcinogenesis. 21:1761–1765. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Krasteva ME, Bozhanov SS, Antov GG,
Gospodinova ZI and Angelov SG: Breast cancer patients with
hypermethylation in the promoter of BRCA1 gene exhibit favorable
clinical status. Neoplasma. 59:85–91. 2012. View Article : Google Scholar
|
|
36
|
Xu X, Gammon MD, Zhang Y, et al: BRCA1
promoter methylation is associated with increased mortality among
women with breast cancer. Breast Cancer Res Treat. 115:397–404.
2009. View Article : Google Scholar :
|
|
37
|
Matros E, Wang ZC, Lodeiro G, Miron A,
Iglehart JD and Richardson AL: BRCA1 promoter methylation in
sporadic breast tumors: relationship to gene expression profiles.
Breast Cancer Res Treat. 91:179–186. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li S, Rong M and Iacopetta B: DNA
hypermethylation in breast cancer and its association with
clinicopathological features. Cancer Lett. 237:272–280. 2006.
View Article : Google Scholar
|
|
39
|
Krasteva ME, Bozhanov SS, Antov GG,
Gospodinova ZI and Angelov SG: Breast cancer patients with
hypermethylation in the promoter of BRCA1 gene exhibit favorable
clinical status. Neoplasma. 59:85–91. 2012. View Article : Google Scholar
|