Introduction
Gastric cancer (GC) is one of the most common types
of malignant cancer, with poor prognosis and high mortality rates
worldwide (1,2). Therefore, the development of an
effective therapeutic method with minimal side effects is required.
Early diagnosis and curative resection are associated with
increased survival in patients with GC. However, currently the
majority of GC cases are diagnosed at later stages. The generation
of differential expression profiles from pathological samples can
markedly improve cancer biomarker discovery (2). In a previous study, it was demonstrated
that Sox7 and β-catenin expression significantly correlated with
the depth of invasion, lymph node metastasis, distant metastasis
and the TNM (tumor, node and metastasis) stage in GC (3). Overexpression of zinc finger, DHHC-type
containing 14 (ZDHHC14) promotes the migration and invasion of
scirrhous GC (4). Caspase-associated
recruitment domain-containing protein (CARP) is a potential tumor
suppressor in GC, and a single-nucleotide polymorphism in the CARP
gene may increase the risk of GC (5).
Ras association domain family member 10 (RASSF10) is an
epigenetically silenced tumor suppressor in GC (6). It has previously been demonstrated that
the inactivation of tumor suppressor genes and activation of
oncogenes serve a significant role in carcinogenesis (3–6). However,
the etiology of GC remains poorly understood.
Lactotransferrin (LTF; also termed lactoferrin) is
an iron-binding glycoprotein involved in a large array of
protective processes in mammals, resulting in antibacterial,
antioxidant and anticarcinogenic effects for the host (7–9). LTF is
produced in the exocrine glands and is secreted in numerous
external fluids as a first line of defense (9). LTF has the capacity to induce apoptosis
and inhibit proliferation in cancer cells, and restore white and
red blood cell levels following chemotherapy (9). The tumor suppressor function of LTF has
been reported in a variety of tumors (10–15),
however the main functions of LTF are within innate immunity and
nutrition (7,8). In a previous study, LTF was observed to
inhibit the cell migration of three gastrointestinal cell lines
(Caco-2, AGS and IEC-18) in vitro (16). Other studies have examined the
association between LTF and GC, but the underlying mechanism
remains unclear.
The MAPK pathway is an important signaling pathway
in many malignancies. We previously found that lactotransferrin, a
candidate tumor suppressor, exhibited deficient expression in human
nasopharyngeal carcinoma and inhibited nasopharyngeal carcinoma
cell proliferation by modulating the mitogen-activated protein
kinase pathway (17) Therefore, in
the present study, the expression levels of LTF and the key
molecular intermediates of the MAPK signaling pathway [p38,
c-Jun N-terminal kinase (JNK) and c-Jun] were
investigated in GC tissues. In addition, the effects of
overexpressing LTF were studied in the GC cell line, SGC7901.
Materials and methods
Cell culture
The human SGC7901 GC cell line was cultured in RPMI
1640 (Gibco Life Technologies, Carlsbad, CA, USA) supplemented with
10% fetal bovine serum (Gibco Life Technologies), 100 U/ml
penicillin and 100 μg/ml streptomycin (GE Healthcare Life Sciences,
Logan, UT, USA) at 37°C in the presence of 5% CO2.
Patient samples
A total of 8 participants were recruited at Xiangya
Hospital, Central South University (Changsha, China). Consent forms
were obtained from individual patients and experimental protocols
were approved by the Institutional Review Board of Xiangya Hospital
(Changsha, China). All subjects enrolled in the study were Chinese.
All clinical and biological data available for the samples are
listed in Table I. GC and
corresponding non-tumor tissues were collected and each biopsy
sample was divided into two sections; one was submitted for routine
histological diagnosis and the remaining section was assessed by
quantitative polymerase chain reaction (qPCR) and western blot
analysis.
 | Table I.Characteristics of patients with
gastric cancer. |
Table I.
Characteristics of patients with
gastric cancer.
| Samples | Gender | Age | Histological
diagnosis |
|---|
| a | Male | 69 | Well
differentiated |
| b | Female | 63 | Intermediately
differentiated |
| c | Male | 49 | Poorly
differentiated |
| d | Male | 60 | Intermediately
differentiated |
| e | Male | 73 | Poorly
differentiated |
| f | Male | 62 | Poorly
differentiated |
| g | Female | 61 | Intermediately
differentiated |
| h | Male | 58 | Poorly
differentiated |
RNA isolation and reverse
transcription (RT)-qPCR analysis
Total RNA was extracted using a TRIzol reagent
(Invitrogen Life Technologies, Carlsbad, CA, USA). The quality of
RNA samples was determined by 1% agarose (Invitrogen Life
Technologies) gel electrophoresis at 80 V for 40 min followed by 60
min at 120 V, and staining with SYBR Safe DNA Gel Stain kit
(Invitrogen Life Technologies). Bands of 18S and 28S RNA were
visualized under UV light (multi-purpose ultraviolet analyzer,
model: WD-9403F; Beijing Liuyi Instrument Factory, Beijing, China).
The optical density 260/280 ratios of the samples were demonstrated
to be in the range of 1.8–2.0. The chromosomal DNA was removed from
the total RNA preparation by using RQ1 RNase-Free DNase (Promega
Corporation, Beijing, China). For detection of LTF mRNA expression
levels, cDNA was synthesized from total RNA using a Reverse
Transcription System (Promega Corporation) according to the
manufacturer's instructions; GAPDH was used as an internal control.
The sequences of the primers used for the qPCR were as follows:
LTF, F 5′-GCA TGG GCT AAG GAT TTGAA-3′ and R 5′-TCC CAA ATT TAG CCT
GTTGG-3′; GAPDH, F 5′-CGA CCA CTT TGT CAA GCT CA-3′ and R 5′-ACT
GAG TGT GGC AGG GACTC-3′. The expression levels of mRNA were
assessed by evaluating the relative threshold cycle (Ct) values.
The Ct values were normalized against the expression levels of
GAPDH, and the relative amount of mRNA specific to each of the
target genes was calculated using the ΔΔCT method (17–19).
Construction of the pIRES-LTF
vector
The full-length LTF coding sequence entirely lacking
the 3′-UTR (GeneCopoeia, Rockville, MD, USA) was subcloned into the
eukaryotic expression vector pIRES (Clontech Laboratories, Inc.,
Mountainview, CA, USA) as previously described (20). The fusion sequences were verified by
DNA sequencing using a Sanger ABI 3730×1 DNA sequencer (GATC
Biotech AG, Konstanz, Germany). The empty pIRES vector was used as
a negative control.
Cell transfection
Cell transfection was performed using Lipofectamine
(Invitrogen Life Technologies), according to the manufacturer's
protocol. Cells were seeded into each well of a 6-well plate, at a
density of 2×105 cells/well, 24 h prior to transfection.
For transfection, 2 μg each of the pIRES-LTF and pIRES plasmids
were transfected into the SGC7901 cells. The plasmids were diluted
with 100 μl serum-free media and 4 μl Lipofectamine was added into
100 μl serum-free media. The two solutions were combined, mixed
gently and incubated at room temperature for 30 min. Next, the 200
μl mixture and a further 200 μl serum-free media were added into
each well. The cells were then incubated at 37°C for 24 h, followed
by replacing the transfection media with fresh complete culture
media. Following an additional 48-h culture, the cells were
harvested for the following western blot analysis.
Western blot analysis
The GC tissues, corresponding non-tumor tissues and
SGC7901 cells were lysed in RIPA buffer (Beijing Comwin Biotech
Co., Ltd, Beijing, China) and the total protein concentration was
determined using a Pierce BCA Protein Assay Kit (Thermo Fisher
Scientific, Inc., Rockford, IL USA). Extracts containing 50 μg
protein were electrophoretically separated on 10% SDS-PAGE gels and
electroblotted onto nitrocellulose membranes (GE Healthcare Life
Sciences, Logan, UT, USA). The membranes were blocked using
Tris-buffered saline/Tween 20 (25 mM Tris-HCl, 150 mM NaCl, pH 7.5
and 0.05% Tween 20) containing 5% non-fat milk followed by an
overnight incubation at 4°C with the following primary antibodies:
Rabbit anti-human p38 antibody (1:200; catalog no. B7178; Anbo
Biotechnology Company, Changzhou, China); rabbit anti-human JNK2
antibody (catalog no. sc-827) and rabbit anti-human c-Jun
antibody (1:500; catalog no. sc-1694; Santa Cruz Biotechnology,
Inc., Santa Cruz, CA, USA); and rabbit anti-human p53 antibody
(1:200; catalog no. PB0076; Wuhan Boster Biological Technology,
Ltd., Wuhan, China). Following three washes with phosphate buffered
saline with Tween (Beyotime Institute of Biotechnology, Hangzhou,
China), the membranes were incubated with horseradish
peroxidase-conjugated secondary antibodies (Santa Cruz
Biotechnology, Inc.) and the specific signals were visualized using
an ECL detection system [MultiSciences (Lianke) Biotech Co., Ltd.
(Hangzhou, China]. Anti-human GAPDH antibody (1:3,000; Santa Cruz
Biotechnology, Inc.) was used as a loading control.
Statistical analysis
Data are expressed as the mean ± standard deviation.
Differences of the quantitative variables between groups were
analyzed by Student's t-test using SPSS software, version 11.0
(SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to
indicate a statistically significant difference.
Results
Detection of mRNA expression levels of
LTF in GC
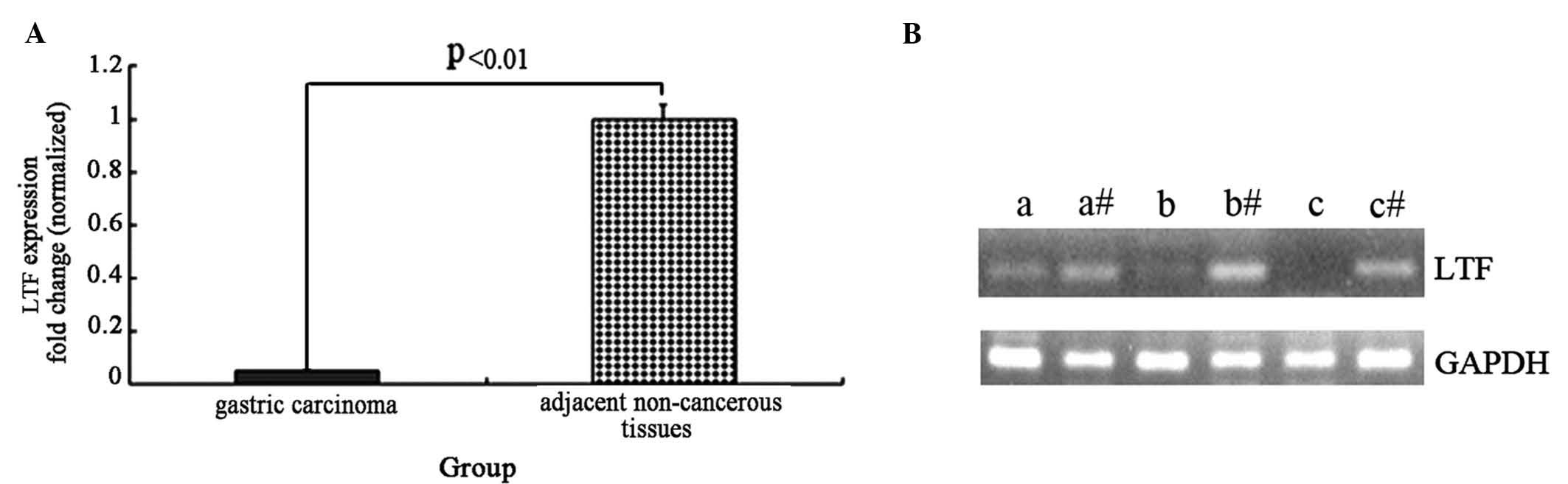
To detect the mRNA expression levels of the LTF gene
in GC and adjacent non-cancerous tissues, eight samples of each
were collected and subjected to RT-qPCR to examine the LTF gene.
The data were analyzed using the ΔΔCT method and the fold change in
the expression of LTF was calculated relative to the internal
control gene, GAPDH. The expression of the LTF gene was lower in
the GC samples compared with the adjacent non-cancerous tissues,
and the normalized LTF gene expression in gastric cancer tissue
samples was reduced by ~20-fold (t=4.56, P<0.01; Fig. 1A). Agarose gel electrophoresis was
performed following RT-qPCR to assess the LTF and GAPDH genes in
three GC tissues and adjacent non-cancerous tissues (Fig. 1B). The results from the
electrophoresis further demonstrate that LTF expression is
deficient in GC tissues compared with adjacent non-cancerous
tissues.
Western blot analysis of protein
levels of LTF in GC
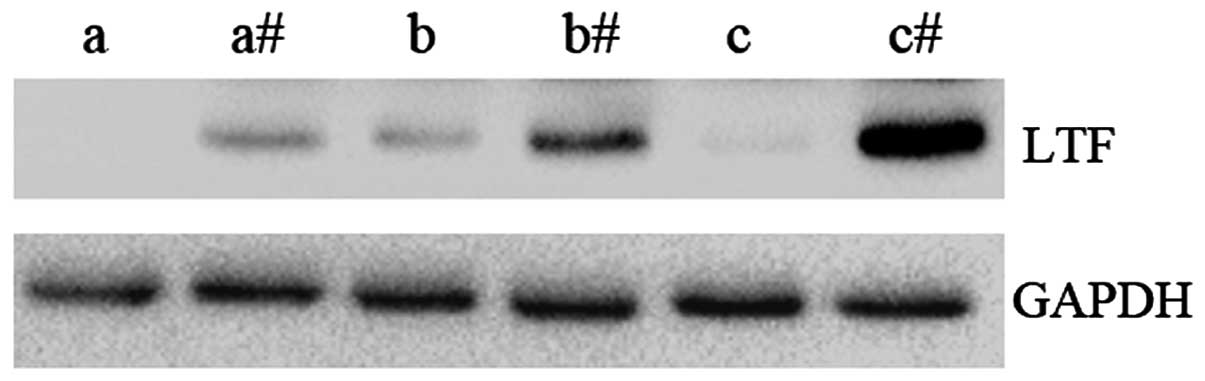
The protein expression levels of LTF were examined
in GC samples and compared to adjacent non-cancerous tissues
(Fig. 2). In agreement with the qPCR
results, the expression level of LTF protein was observed to be
lower in GC tissues compared with the adjacent non-cancerous
tissues. These results further demonstrate that LTF expression
levels are downregulated in GC.
LTF correlates with dysregulation of
the MAPK signaling pathway in GC tissues
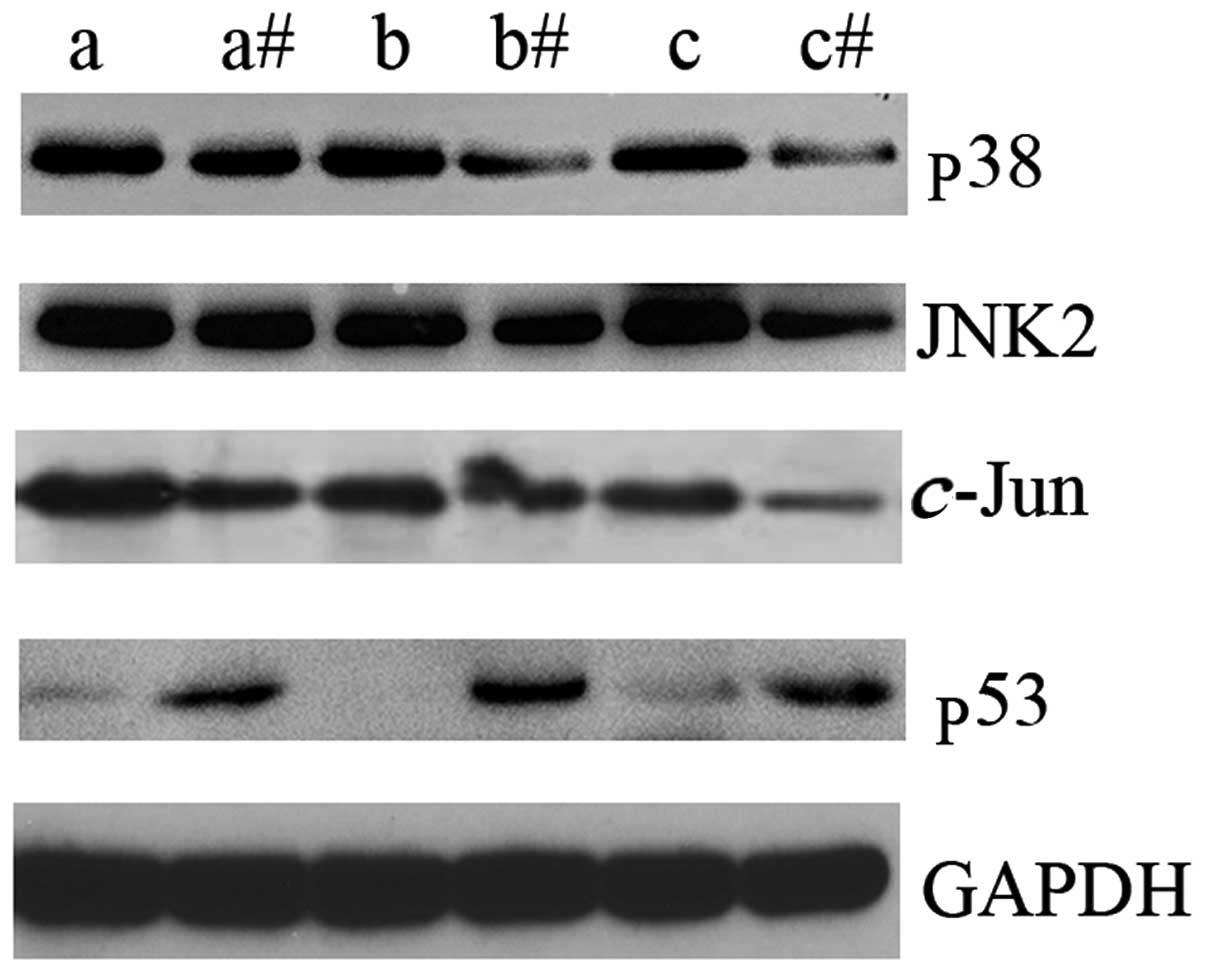
In order to identify the possible underlying
mechanism of LTF action in GC, the expression levels of key
intermediate molecules in the MAPK signaling pathway were assessed
by western blot analysis. p38, JNK2 and c-Jun were
upregulated in GC tissues compared with the adjacent non-cancerous
tissues (Fig. 3). In addition, the
expression levels of p53 protein were markedly downregulated in GC
tissues compared with the adjacent non-cancerous tissues (Fig. 3). The results of the present study
indicate that LTF may be associated with the dysregulation of the
MAPK signaling pathway in GC tissues. These results may indicate an
association between reduced LTF expression levels and the
upregulation of certain key molecular intermediates of the MAPK
signaling pathway in GC.
LTF overexpression affects the
expression of p38, JNK2 and c-Jun in vitro
To confirm whether LTF affected the expression of
p38, JNK2 and c-Jun in vitro, pIRES-LTF and pIRES
were transfected into the SGC7901 GC cell line. The cells were
harvested 48 h following transfection and the levels of the p38,
JNK2, c-Jun and p53 proteins were determined. The results
demonstrated that p38, JNK2 and c-Jun proteins were
expressed at lower levels in the SGC7901 cells transfected with
pIRES-LTF compared with the SGC7901 cells transfected with pIRES
(Fig. 4). However, the p53 protein
expression levels displayed the opposite pattern. p53 was highly
expressed in the SGC7901 cells transfected with pIRES-LTF compared
with the SGC7901 cells transfected with pIRES (Fig. 4). These results demonstrate that LTF
overexpression affects the expression of p38, JNK2, c-Jun
and p53 in vitro.
In conclusion, the overexpression of LTF
downregulates the expression of p38, JNK2 and c-Jun.
However, the mechanism by which LTF affects the MAPK signaling
pathway requires further investigation.
Discussion
It has previously been demonstrated in numerous
studies that LTF possesses antitumor action (10,15,21,22).
In the current study, it was demonstrated that the mRNA and protein
expression levels of LTF in GC were 20-fold lower compared with the
adjacent non-cancerous tissues. Sousa et al (2) observed significantly lower levels of LTF
in GC samples with more advanced disease progression and worse
prognosis. In a previous study, quantitative analysis of LTF mRNA
expression revealed a marked downregulation in prostate cancer
(23). In addition, the LTF gene is
inactivated by genetic and epigenetic mechanisms in lung cancer
(24). Previous studies have also
indicated that LTF may have a direct effect on tumor cell growth,
as suggested by the fact that LTF and an LTF splice variant are
downregulated or absent in certain types of cancer (25–28). The
results of the present study indicate that LTF may also serve as an
important tumor suppressor gene in GC.
To explore the possible mechanism of LTF in GC, the
expression levels of key signaling intermediates (p38, JNK2 and
c-Jun) in the MAPK signaling pathway were assessed. Compared
with the adjacent non-cancerous tissues, p38, JNK2 and c-Jun
were upregulated in GC samples. This result may reflect a negative
association between the expression of LTF and the key molecular
intermediates of the MAPK signaling pathway in GC tissues. In
addition, the effect of overexpression of LTF on levels of p38,
JNK2 and c-Jun was determined in vitro. The results
demonstrated that overexpression of LTF downregulated the
expression of p38, JNK2 and c-Jun in the SGC7901 cells.
MAPKs transduce extracellular signals, promoting a variety of
cellular processes, including cell proliferation, survival, death
and differentiation (29). JNK and
p38 MAPK signaling are associated with various types of cancer in
humans and mice (30). JNK is a
family of protein kinases that are activated by stress stimuli and
regulate various cellular processes, including proliferation,
apoptosis and survival (31). Yuan
et al (32) observed that the
overexpression of human DNA polymerase ι (pol ι) is positively
correlated with clinical tumor grade in bladder cancer samples and
may contribute to hypermutagenesis. Dysregulation of pol ι by
JNK/c-Jun is involved in carcinogenesis and offers a novel
understanding of the role of pol ι or c-Jun in mutagenesis
(32). In A549 human non-small cell
lung cancer cells, JNK may be specifically required in vivo
for the maintenance of the tumor-initiating population of tumor
cells rather than for the proliferation and survival of the entire
cell population (33). Taken
together, the results of the present study and previous studies
indicate that a lack of LTF expression in GC may increase the
expression of p38, JNK2 and c-Jun.
In addition, the present study demonstrated that p53
was downregulated in GC, and that the overexpression of LTF
increased the levels of p53 protein in vivo. p53 is a
well-documented tumor suppressor gene (34). The functions of p53 include responding
to cellular stress, apoptosis and cell cycle arrest. The p53
pathway is inactivated in the majority of types of human cancer
(34–37). MicroRNA hsa-miR-125a-3p activates the
p53 protein and induces lung cancer cell apoptosis (35). p53 acts as a safeguard of protein
synthesis by regulating the rRNA methyltransferase fibrillarin and
the subsequent quality and intrinsic activity of ribosomes
(36). A previous study demonstrated
that the loss of p53 promotes the invasion and metastasis
capability of prostate cancer cells through the FAK-Src signaling
pathway (37). The association
between LTF, p53 and GC requires further investigation.
In conclusion, the present study demonstrates that
LTF expression is downregulated in GC, and that this may affect the
MAPK signaling pathway. However, the details of the underlying
mechanism require further study.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81272975 and
81172302); the Key Project of Hunan Provincial Natural Science
Foundation (grant no. 12JJ2044); the Project of Hunan Provincial
Natural Science Foundation (grant no. 12JJ3121); and the Project of
Hunan Provincial Development and Reform Commission.
Glossary
Abbreviations
Abbreviations:
|
GC
|
gastric cancer
|
|
LTF
|
lactotransferrin
|
|
JNK
|
c-Jun N-terminal kinase
|
|
MAPK
|
mitogen-activated protein kinase
|
|
TP53
|
tumor protein p53
|
|
GADPH
|
glyceraldehyde-3-phosphate
dehydrogenase
|
References
|
1
|
Cho JY: Molecular diagnosis for
personalized target therapy in gastric cancer. J Gastric Cancer.
13:129–135. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sousa JF, Ham AJ, Whitwell C, Nam KT, Lee
HJ, Yang HK, et al: Proteomic profiling of paraffin-embedded
samples identifies metaplasia-specific and early-stage gastric
cancer biomarkers. Am J Pathol. 181:1560–1572. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cui J, Xi H, Cai A, Bian S, Wei B and Chen
L: Decreased expression of Sox7 correlates with the upregulation of
the Wnt/β-catenin signaling pathway and the poor survival of
gastric cancer patients. Int J Mol Med. 34:197–204. 2014.PubMed/NCBI
|
|
4
|
Oo HZ, Sentani K, Sakamoto N, Anami K,
Naito Y, Uraoka N, Oshima T, Yanagihara K, Oue N and Yasui W:
Overexpression of ZDHHC14 promotes migration and invasion of
scirrhous type gastric cancer. Oncol Rep. 32:403–410.
2014.PubMed/NCBI
|
|
5
|
Lu F, Xue JX, Hu YC, Gan L, Shi Y, Yang HS
and Wei YQ: CARP is a potential tumor suppressor in gastric
carcinoma and a single-nucleotide polymorphism in CARP gene might
increase the risk of gastric carcinoma. PLoS One. 9:e977432014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li Z, Chang X, Dai D, Deng P and Sun Q:
RASSF10 is an epigenetically silenced tumor suppressor in gastric
cancer. Oncol Rep. 31:1661–1668. 2014.PubMed/NCBI
|
|
7
|
Legrand D: Lactoferrin, a key molecule in
immune and inflammatory processes. Biochem Cell Biol. 90:252–268.
2012. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Legrand D, Pierce A, Elass E, Carpentier
M, Mariller C and Mazurier J: Lactoferrin structure and functions.
Adv Exp Med Biol. 606:163–194. 2008.PubMed/NCBI
|
|
9
|
Gibbons JA, Kanwar RK and Kanwar JR:
Lactoferrin and cancer in different cancer models. Front Biosci
(Schol Ed). 3:1080–1088. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bezault J, Bhimani R, Wiprovnick J and
Furmanski P: Human lactoferrin inhibits growth of solid tumors and
development of experimental metastases in mice. Cancer Res.
54:2310–2312. 1994.PubMed/NCBI
|
|
11
|
Varadhachary A, Wolf JS, Petrak K,
O'Malley BW Jr, Spadaro M, Curcio C, Forni G and Pericle F: Oral
lactoferrin inhibits growth of established tumors and potentiates
conventional chemotherapy. Int J Cancer. 111:398–403. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tsuda H, Sekine K, Nakamura J, Ushida Y,
Kuhara T, et al: Inhibition of azoxymethane initiated colon tumor
and aberrant crypt foci development by bovine lactoferrin
administration in F344 rats. Adv Exp Med Biol. 443:273–284.
1998.PubMed/NCBI
|
|
13
|
Sekine K, Watanabe E, Nakamura J, Takasuka
N, et al: Inhibition of azoxymethane-initiated colon tumor by
bovine lactoferrin administration in F344 rats. Jpn J Cancer Res.
88:523–526. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Matsuda Y, Saoo K, Hosokawa K, Yamakawa K,
Yokohira M, Zeng Y, Takeuchi H and Imaida K: Post-initiation
chemopreventive effects of dietary bovine lactoferrin on
4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone-induced lung
tumorigenesis in female A/J mice. Cancer Lett. 246:41–46. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li WY, Li QW, Han ZS, Jiang ZL, Yang H, Li
J and Zhang XB: Growth suppression effects of recombinant
adenovirus expressing human lactoferrin on cervical cancer in vitro
and in vivo. Cancer Biother Radiopharm. 26:477–483. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nakajima M, Shinoda I, Samejima Y,
Miyauchi H, Fukuwatari Y and Hayasawa H: Lactoferrin as a
suppressor of cell migration of gastrointestinal cell lines. J Cell
Physiol. 170:101–105. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhou Y, Zeng Z, Zhang W, Xiong W, Wu M,
Tan Y, Yi W, Xiao L, Li X, Huang C, et al: Lactotransferrin: a
candidate tumor suppressor - Deficient expression in human
nasopharyngeal carcinoma and inhibition of NPC cell proliferation
by modulating the mitogen-activated protein kinase pathway. Int J
Cancer. 123:2065–2072. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pfaffl MW: A new mathematical model for
relative quantification in real-time RT-PCR. Nucleic Acids Res.
29:e452001. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Deng M, Zhang W, Tang H, Ye Q, Liao Q,
Zhou Y, Wu M, Xiong W, Zheng Y, Guo X, et al: Lactotransferrin acts
as a tumor suppressor in nasopharyngeal carcinoma by repressing AKT
through multiple mechanisms. Oncogene. 32:4273–4283. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ward PP, Paz E and Conneely OM:
Multifunctional roles of lactoferrin: a critical overview. Cell Mol
Life Sci. 62:2540–2548. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Rodrigues L, Teixeira J, Schmitt F,
Paulsson M and Månsson HL: Lactoferrin and cancer disease
prevention. Crit Rev Food Sci Nutr. 49:203–217. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shaheduzzaman S, Vishwanath A, Furusato B,
Cullen J, Chen Y, Bañez L, Nau M, Ravindranath L, Kim KH, Mohammed
A, et al: Silencing of Lactotransferrin expression by methylation
in prostate cancer progression. Cancer Biol Ther. 6:1088–1095.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Iijima H, Tomizawa Y, Iwasaki Y, Sato K,
Sunaga N, Dobashi K, Saito R, Nakajima T, Minna JD and Mori M:
Genetic and epigenetic inactivation of LTF gene at 3p21.3 in lung
cancers. Int J Cancer. 118:797–801. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yi HM, Li H, Peng D, Zhang HJ, Wang L,
Zhao M, Yao KT and Ren CP: Genetic and epigenetic alterations of
LTF at 3p21.3 in nasopharyngeal carcinoma. Oncol Res. 16:261–272.
2006.PubMed/NCBI
|
|
26
|
Campbell T, Skilton RA, Coombes RC,
Shousha S, Graham MD and Luqmani YA: Isolation of a lactoferrin
cDNA clone and its expression in human breast cancer. Br J Cancer.
65:19–26. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kholodnyuk ID, Kozireva S, Kost-Alimova M,
Kashuba V, Klein G and Imreh S: Down regulation of 3p genes, LTF,
SLC38A3 and DRR1, upon growth of human chromosome 3-mouse
fibrosarcoma hybrids in severe combined immunodeficiency mice. Int
J Cancer. 119:99–107. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yang Y, Li J, Szeles A, Imreh MP,
Kost-Alimova M, Kiss H, Kholodnyuk I, Fedorova L, Darai E, Klein G
and Imreh S: Consistent downregulation of human lactoferrin gene,
in the common eliminated region 1 on 3p21.3, following tumor growth
in severe combined immunodeficient (SCID) mice. Cancer Lett.
191:155–164. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Huang P, Han J and Hui L: MAPK signaling
in inflammation-associated cancer development. Protein Cell.
1:218–226. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wagner EF and Nebreda AR: Signal
integration by JNK and p38 MAPK pathways in cancer development. Nat
Rev Cancer. 9:537–549. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
You H, Lei P and Andreadis ST: JNK is a
novel regulator of intercellular adhesion. Tissue Barriers.
1:e268452013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yuan F, Xu Z, Yang M, Wei Q, Zhang Y, Yu
J, Zhi Y, Liu Y, Chen Z and Yang J: Overexpressed DNA polymerase
iota regulated by JNK/c-Jun contributes to hypermutagenesis in
bladder cancer. PLoS One. 8:e693172013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Okada M, Shibuya K, Sato A, Seino S,
Watanabe E, Suzuki S, Seino M and Kitanaka C: Specific role of JNK
in the maintenance of the tumor-initiating capacity of A549 human
non-small cell lung cancer cells. Oncol Rep. 30:1957–1964.
2013.PubMed/NCBI
|
|
34
|
Suzuki K and Matsubara H: Recent advances
in p53 research and cancer treatment. J Biomed Biotechnol.
2011:9783122011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jiang L, Chang J, Zhang Q, Sun L and Qiu
X: MicroRNA hsa-miR-125a-3p activates p53 and induces apoptosis in
lung cancer cells. Cancer Invest. 31:538–544. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Marcel V, Ghayad SE, Belin S, Therizols G,
Morel AP, Solano-Gonzàlez E, Vendrell JA, Hacot S, Mertani HC,
Albaret MA, et al: p53 acts as a safeguard of translational control
by regulating fibrillarin and rRNA methylation in cancer. Cancer
Cell. 24:318–330. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang Y, Zhang YX, Kong CZ, Zhang Z and Zhu
YY: Loss of P53 facilitates invasion and metastasis of prostate
cancer cells. Mol Cell Biochem. 384:121–127. 2013. View Article : Google Scholar : PubMed/NCBI
|