Introduction
As a complex disorder with the whole synovial joint
involved, osteoarthritis (OA) has the highest prevalence in all
forms of arthritis worldwide and contributes to the majority of
disability because of pain (1). The
knee is the most commonly affected joint and OA often takes place
in older adults especially in women (1). It is estimated that there are nearly
27 million adults suffering from OA in 2008 in the USA alone
(2). Consequently, it is necessary
to carry out remarkable efforts to look into the pathogenesis and
pursue better understanding of OA progression and development,
which might result to the development of promising treatment
regimens.
It has been demonstrated that apoptosis of synovial
cells is under control of variable factors, and dysregulation of
apoptosis contributes to the development of OA (3–5). It
has been reported that significant concentrations of nitric oxide
(NO) metabolites, which is believed to promote apoptosis, in human
OA synovial fluids (6,7). Apoptotic cells were reported to be
present in the synovial membrane in OA as well as in RA. As a
result, apoptosis might act as a regulatory event to control
metabolism of synovial cells in OA (8).
Recently, synoviolin 1 (SYVN1) expression is highly
related to rheumatoid arthritis development and has been confirmed
as a rheumatoid regulator. Transgenic SYVN1 mice with high SYVN1
expression had spontaneous arthroplasty, however, mice with reduced
SYVN1 (SYVN1+/− mice) were at lower risk of developing
collagen-induced arthritis (CIA) (9).
As a class of small, non-coding RNAs, microRNA
(miRNA) plays critical roles in development of numerous kinds of
species and frequently participates in numerous genetic diseases,
including cancer (10). Generally,
microRNAs are involved in gene expression downregulation by
targeting mRNAs (11). It is
believed that a common mechanism of microRNA biogenesis is shared
in mammals (12). The primary
microRNAs (pri-microRNAs) are transcribed from the genome, followed
by processed into precursor microRNAs (pre-microRNAs) in the
nucleus by the Microprocessor complex of Drosha (a type of RNase
III enzyme) and its cofactor DGCR8 (also named as Pasha) (13,14). A
typical hairpin structure consisting of approximately 60–70
nucleotides which is characterized by an overhang of approximately
2 nucleotides at the 3′ end exist in pre-microRNAs (15).
Increasing evidence demonstrates that miRNAs play
important roles in controlling numerous basic cell functions.
miRNAs play key roles in all cellular processes including cartilage
remodeling and chondrogenesis (16–18).
Consequently, aberrant expression profiles of miRNA was related to
development of osteoarthritis (19–22).
Previous research has identified a signature of 16 miRNAs which
could discriminate normal osteoarthritic cartilage tissue, with 7
miRNAs downregulated and 9 miRNAs remarkably upregulated in
osteoarthritis tissue by contrast to normal controls (20).
In a recent study, miR-125b-5p was shown to be
upregulated in the Synovial cells collected from OA, and SYVN1 was
found to be involved in the pathogenesis of OA (23–25).
We identified SYVN1 as a virtual target of miR-125b-5p by using
in-silico analysis. In this study, we validated SYVN1 as a
target of miR-125b-5p and tested the role of miR-125b-5p and SYVN1
in the development of OA.
Materials and methods
Samples
Human synovial cells were isolated from synovium
obtained from joint surgery, and synovium was isolated from 36
participants consisting of 12 normal control (K/L, Grade 0: normal
cartilage), 12 mild OA (K/L Grade I and II: low grade OA cartilage)
and 12 severe OA (K/L, Grade III and IV: high grade OA cartilage)
at Department of Orthopedics, The People's Hospital of Linyi
(Linyi, China). Trypsin and collagenase were used to digest the
superficial layer of synovium after dissection. The protocol of the
study was approved by the Ethics Committee of The People's Hospital
of Liny. The patients signed informed consent for participation in
the study after the potential risk was explained. The study was
conducted according to the Declaration of Helsinki.
Synovial cells culture and
transfection
DMEM (Dulbecco's modified Eagle's medium)
(Gibco® Invitrogen, Carlsbad, CA, USA) containing 100
U/ml streptomycin, 100 U/ml penicillin and 10% FBS (fetal bovine
serum) (Hyclone, Logan, UT, USA) was used to culture the synovial
cells under a humidified atmosphere of 5% CO2/95% air at
37°C. Lipofectamine 2000 (Invitrogen) was used to perform the
transfection in accordance with the manufacturer's instructions.
Briefly, when the cells were grown to 80% confluence, the cells
were transfected with miR-125b-5p mimics or inhibitors and scramble
control (RiboBio, Guangzhou, China).
Quantitative PCR
The miRNANeasy Mini kit (Qiagen, Hilden, Germany)
was used to isolate total RNA from synovial cells and tissue
samples in accordance with manufacturer's guideline. Beckman DU-640
spectrophotometer (Beckman Instruments Inc., Fullerton, CA, USA)
was used to determine the concentration of RNA extracted following
the standard protocol by the supplier, and then 1% formaldehyde
agarose gel electrophoresis was used to evaluate quality of RNA.
The quantification of miR-125b and SYVN1 mRNA expression was
performed using Applied BiosystemsTaqMan MicroRNA Reverse
Transcription kit (Thermo Fisher Scientific, Waltham, MA, USA),
TaqMan MicroRNA Assay kits was used to perform the reverse
transcription of complementary DNA in accordance with
manufacturer's recommendations. Applied Biosystems StepOne
Real-Time PCR System (Thermo Fisher Scientific) was used to carry
out qRT-PCR (quantitative polymerase chain reaction) following the
guideline by the supplier. The protocol of the reaction was carried
out at 40 cycles of 95°C for 15 sec, 15 sec at 60°C, and 30 sec at
70°C. The Comparative CT (2-∆∆Ct) method was used to calculate the
expression of miR-125b and SYNV1 mRNA. Three independent
experiments were performed.
Luciferase assay
PCR (polymerase chain reaction) was used to amplify
the human SYVN1 cDNA containing a putative target site for
miR-125b-5p. Then the PCR products were inserted into the pGL3
control vector (Promega, Madison, WI, USA) immediately downstream
of the stop codon of the firefly luciferase gene (pGL3-SYVN1
−3′UTR). The Quik Change II Site-Directed Mutagenesis kit
(Stratagene, La Jolla, CA, USA) was used to generate the mutated
version of the 3′UTR including a 7-bp mutation (3′ mutant).
Lipofectamine 2000 (Invitrogen) was used to transiently
co-transfect miR-125 mimics and the mutant or wild-type reporter
plasmid with the 3′UTR of SYVN1. The dual-luciferase assay kit
(Promega) was used to measure the activities of both the Renilla
luciferases and firefly 24–48 h after transfection. Three
independent experiments were repeated.
Cell proliferation assay
MTT
[3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide]
assay was used to perform the cell proliferation. The synovial
cells were cultured in 48-well plates, and then 20 µl of MTT (5
mg/ml) was added into each well 1, 2, 3 days after trancfection,
and the supernatant was removed. DMSO (150 µl) was used to dissolve
the remaining crystals. A microplate reader was employed to measure
the optical density of the cells based on the absorption at 490 nm.
Each test was repeated in triplicate.
Western blot analysis
Synovial cells were harvested and washed twice with
cold PBS. Radio immune precipitation assay lysis buffer
(Invitrogen) was used to lyse the cells according to the protocol,
and then centrifuged the cell lysates at 12,000 × g at 4°C for 10
min to obtain the supernatants contained in the whole-cell protein
extracts. DC protein assay (Bio-Rad, Berkeley, CA, USA) was used to
determine the concentration of proteins. The protein was treated
with boiling water with loading buffer to obtain heat-denatured
protein samples, and 10% SDS-PAGE (poly-acrylamide gel
electrophoresis) was used to separate the protein, and then
transferred to an Immobilon-P membrane (Millipore, Bedford, MA,
USA) for 2 h (120 V). PBS containing 0.1% Tween-20 and 5% no-fat
dry milk was used to incubate the membrane for 1 h to avoid
non-specific binding. The primary antibody anti-SYVN1 (1:1000
dilution, Alomone labs Ltd., Jerusalem, Israel) anti-β-actin
(1:8000, Sigma-Aldrich Co., LLC, St. Louis, MO, USA) were used to
incubate the membrane at room temperature for 60 min, and washed
twice with PBS including 0.1% Tween-20, and then a secondary
antibody (1:15000 dilution, Invitrogen) was used to treat the
membrane for another 60 min, and finally washed twice with PBS
including 0.1% Tween-20. Enhanced chemiluminescence detection
reagents (Amersham Biosciences) was used to detect the bound
antibody in accordance with manufacturer's protocol.
Apoptosis analysis
BioVision Annexin V-FITC reagent kit (Sigma-Aldrich)
and flow cytometry were used to perform apoptosis analysis in
accordance with the manufacturer's instruction. At 48 h after
transfection, the synovial cells were trypsinised, and PBS was used
to wash the cells, and then the cells were collected by
centrifugation of 2000 rpm/min, 5 min, then 5 µl propidium iodide,
5 µl Annexin VFITC and 500 µl binding buffer was added into each
well, and incubated for 5–15 min. Finally, flow cytometry was used
to estimate the apoptosis of the cells. Three independent tests
were performed.
Statistical analysis
Statistical Package of Social Science Software
program (SPSS), version 21 (SPSS Inc., Chicago, IL, USA) was used
to statistically analyze the data. The data are shown as the means
± SD (standard deviation) unless otherwise indicated. Independent
sample t-test was used to analyze the quantitative variables
comparison between groups, and Pearson's Chi-square test was used
to analyze qualitative variables comparison between groups. P-value
<0.05 was considered to indicate a statistically significant
difference.
Results
Expression of miR-125b-5p in normal
control, mild OA and severe OA groups
A total of 36 participants were enrolled which
consist of 12 normal control (K/L, Grade 0: normal cartilage), 12
mild OA (K/L Grade I and II: low grade OA cartilage) and 12 severe
OA (K/L, Grade III and IV: high grade OA cartilage) according to a
Kellgren/Lawrence Criterion, cells were cultured, followed by the
detection of the gene level using quantitative PCR (qPCR)
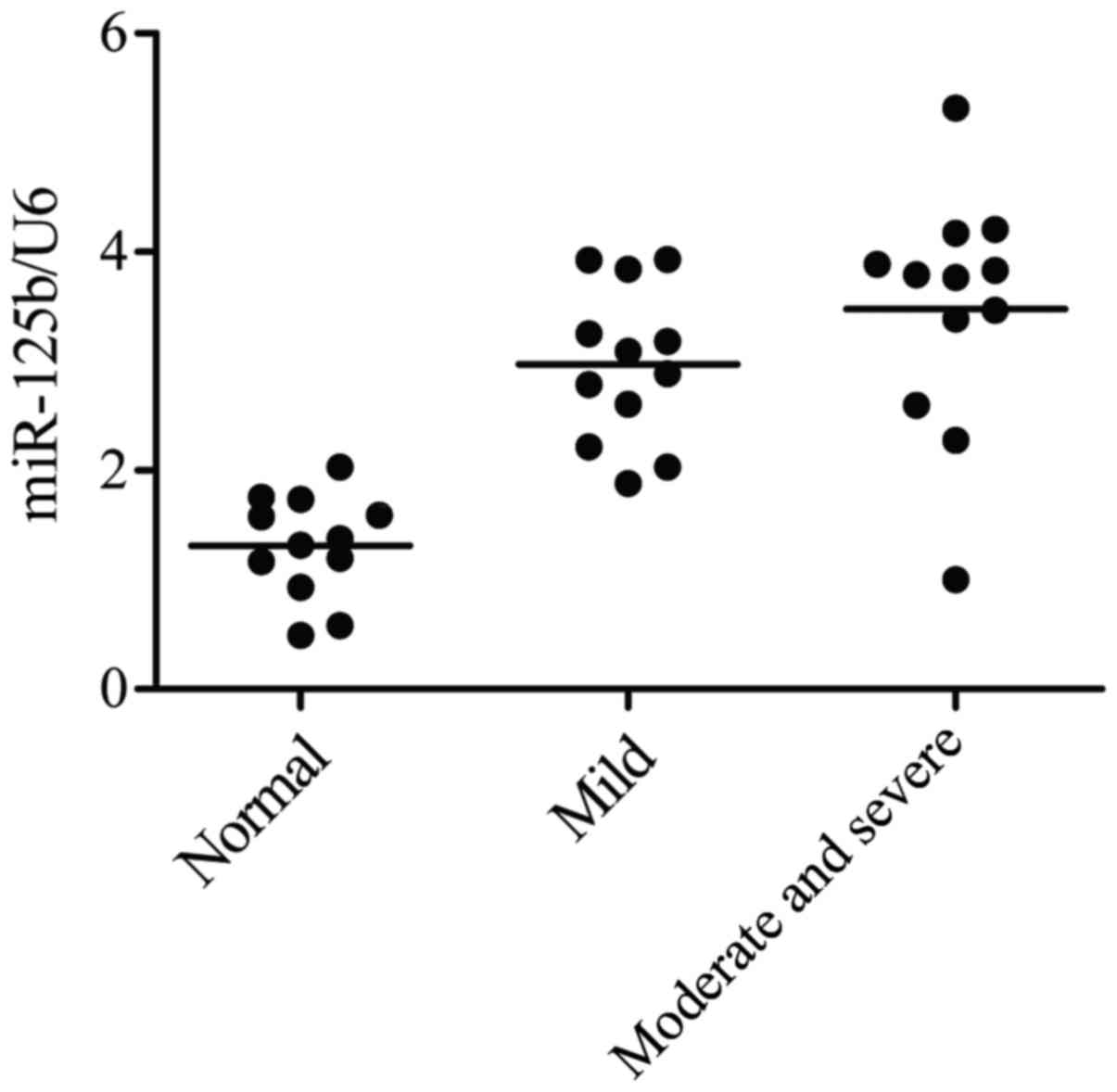
technique. As shown in Fig. 1, the
gene level of miR-125b-5p in severe OA group was the highest in the
three groups, while the gene level of miR-125b-5p in normal group
was the lowest in the three groups. The data thus indicated that
the overexpression level of miR-125b-5p induce the development of
OA, and increased at high grade OA groups, while it decreased at
low grade OA group, compared with normal control groups.
Expression of SYVN1 in normal control,
mild OA and severe OA groups
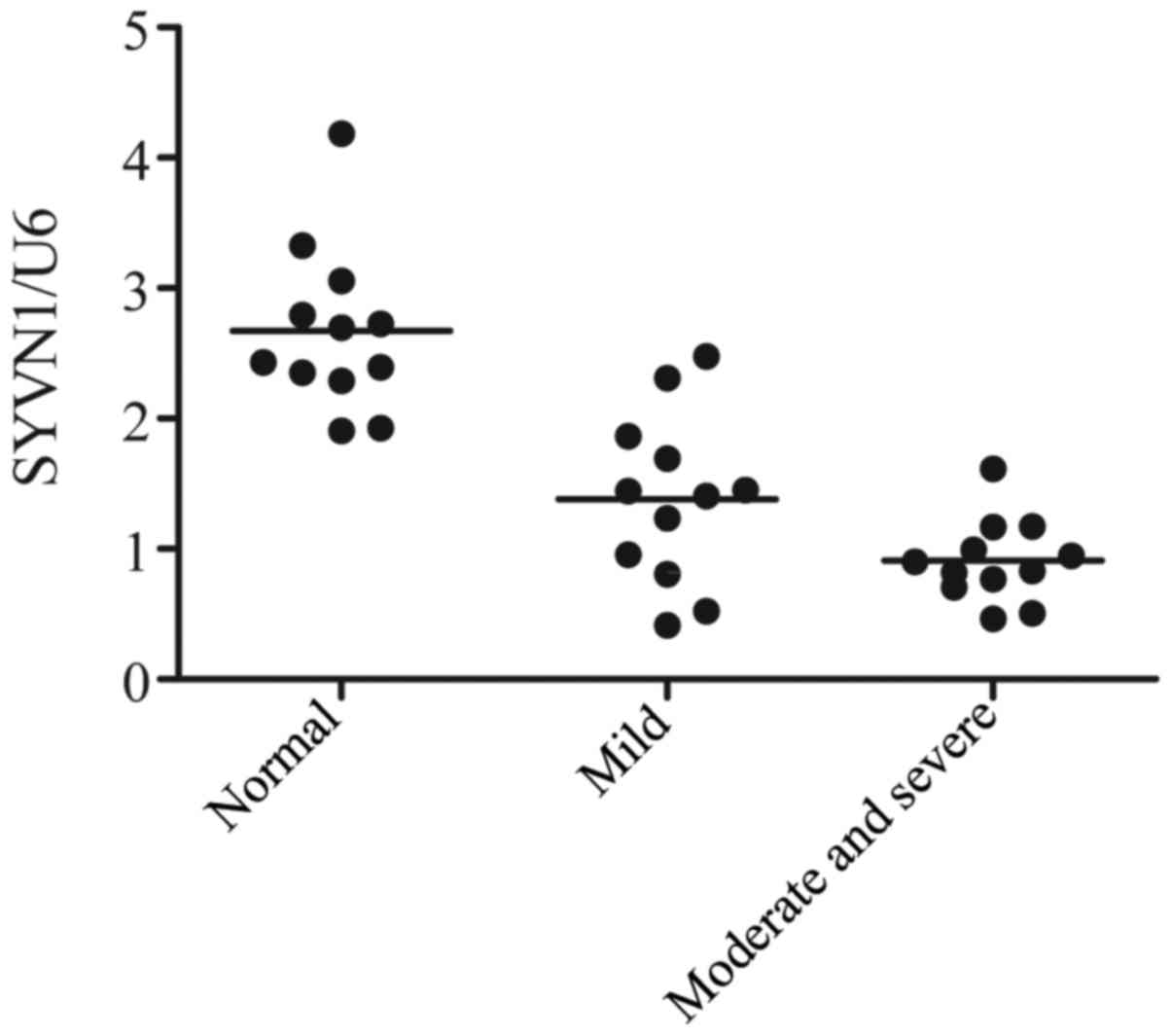
The level of SYVN1 was detected with the use of
quantitative PCR (qPCR) technique. As shown in Fig. 2, the gene level of SYVN1 in severe
OA group was the lowest in the three groups, while the gene level
of SYVN1 in normal group was the highest in the three groups. The
data thus indicated that the downregulation of SYVN1 induce the
development of OA.
miR-125b-5p targets SYVN1 in synovial
cells
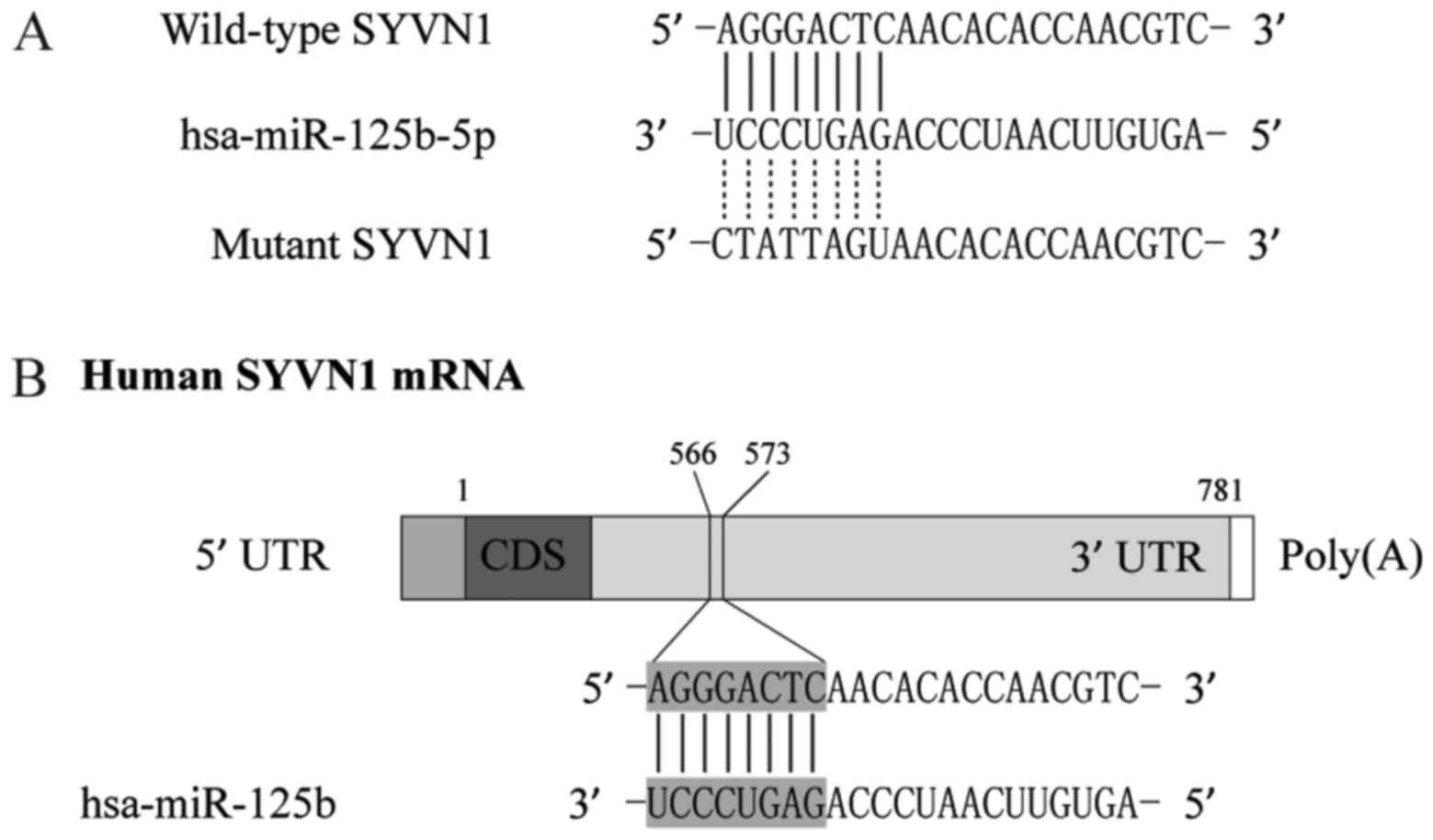
By using computational analysis, such as
Diana-MICROT, miRDB, TargetMiner, and TargetScan, were chosen to
determine the possible target genes of miR-125b-5p, we identified
that SYVN1 is a virtual target of miR-125b-5p with binding sites in
the 3′UTR of the gene, as shown in Fig.
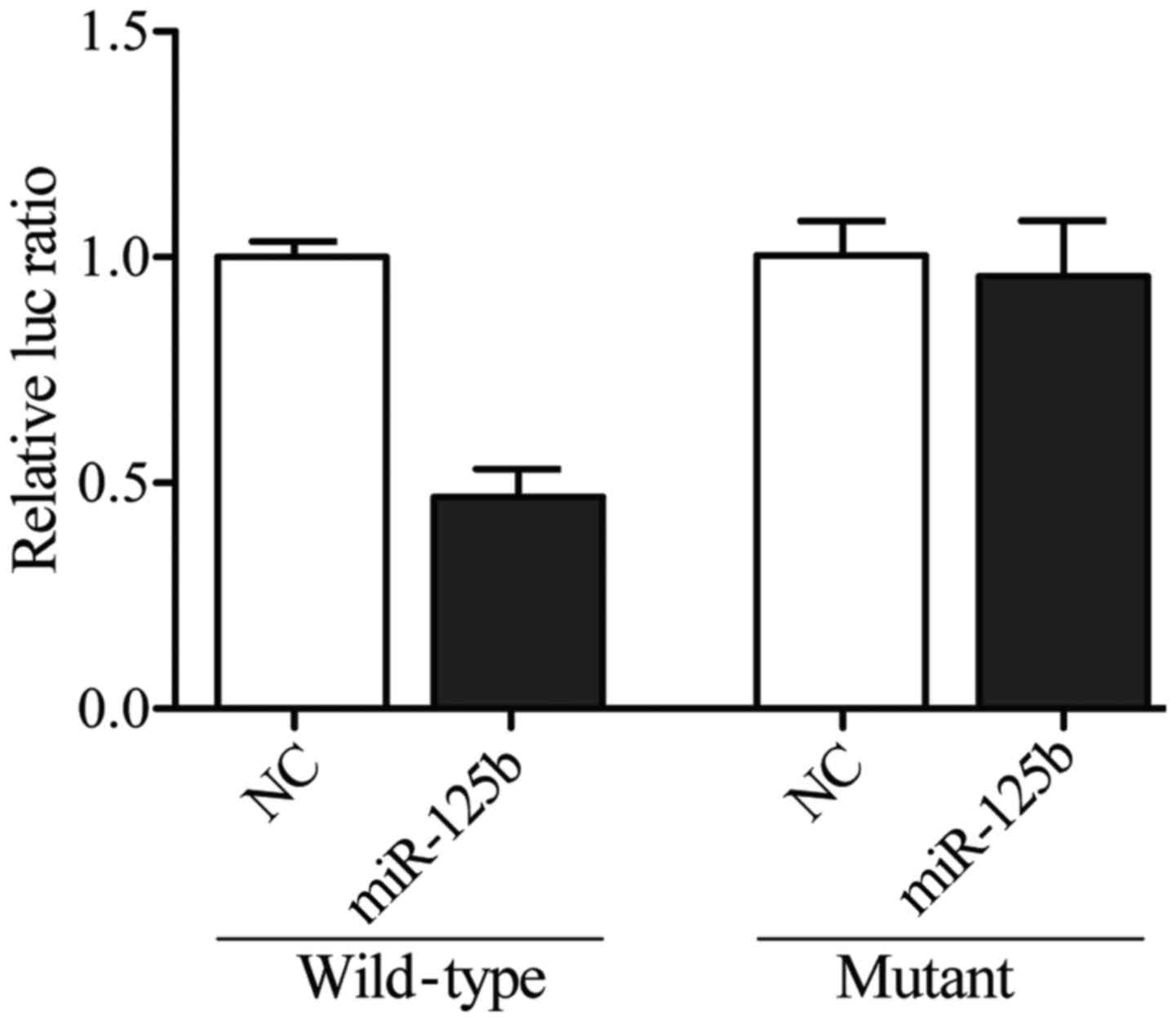
3. To further confirm the interaction between SYVN1 3′TUR and
miR-125b-5p, we sub-cloned the full length SYVN1 3′UTR into a
vector and co-transfected it with miR-125b-5p mimics or inhibitors
prior to dual-luciferase analysis, and we found that the luciferase
activity of the cells cotransfected with wild-type SYVN1 3′UTR was
significantly lower than that of the cells transfected with mutant
SYVN1 3′UTR and scramble controls, while the luciferase activity of
the cells transfected with mutant SYVN1 3′UTR was substantially
comparable with that of the cells transfected with scramble
controls (Fig. 4), indicating that
the SYVN1 was the target gene of miR-125b-5p, and the upregulation
of miR-125b-5p suppressed the expression of SYVN1.
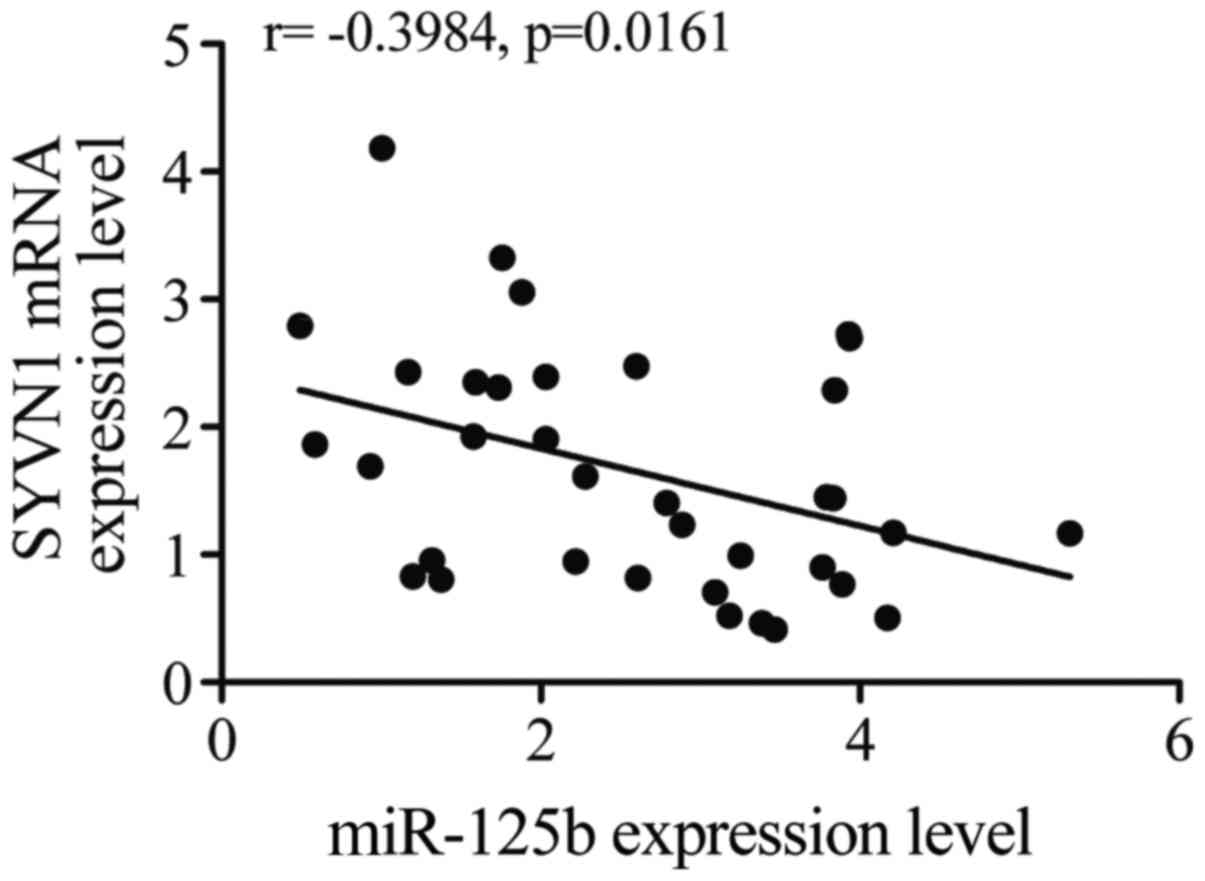
Negative regulatory relationship
between SYVN1 and miR-125b-5p
The regulatory relationship between SYVN1 and
miR-125b-5p was tested with the use of quantitative PCR, as shown
in Fig. 5, the negative regulatory
relationship between miR-125b-5p and SYVN1 was validated, and the
negative correlation coefficient was −0.40 (r= −0.40).
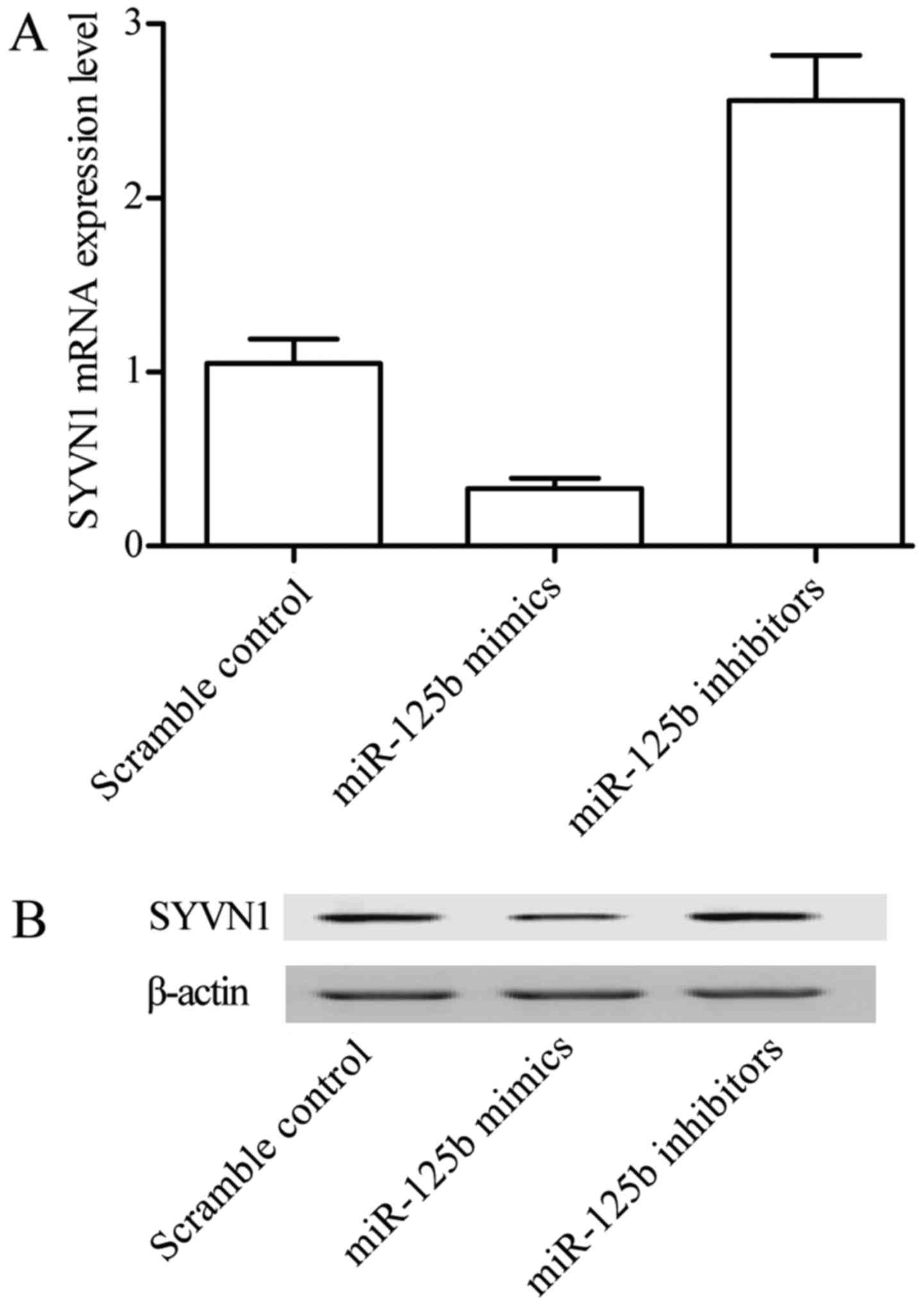
Overexpression of miR-125b-5p
decreases expression of SYVN1
To investigate whether miR-125b-5p has an inhibitory
effect on the expression of SYVN1, quantitative PCR and western
blot analysis were used. Cells were transfected with miR-125b-5p
mimic, miR-125b-5p inhibitor and scramble control in synovial
cells, respectively. As showed in Fig.
6, both mRNA (Fig. 6A) and
protein (Fig. 6B) SYVN1 expression
were significantly decreased in cells in which miR-125b-5p was
overexpressed (transfected with miR-125b-5p), in comparison with
similar cells transfected with scramble control. On the other hand,
downregulation of miR-125b-5p via transfection of the cells with
miR-125b-5p inhibitors significantly upregulated the mRNA (Fig. 6A) and protein (Fig. 6B) expression of SYVN1. These
findings further demonstrated that SYVN1 is a direct target of
miR-125b-5p in PTENCE cell lines.
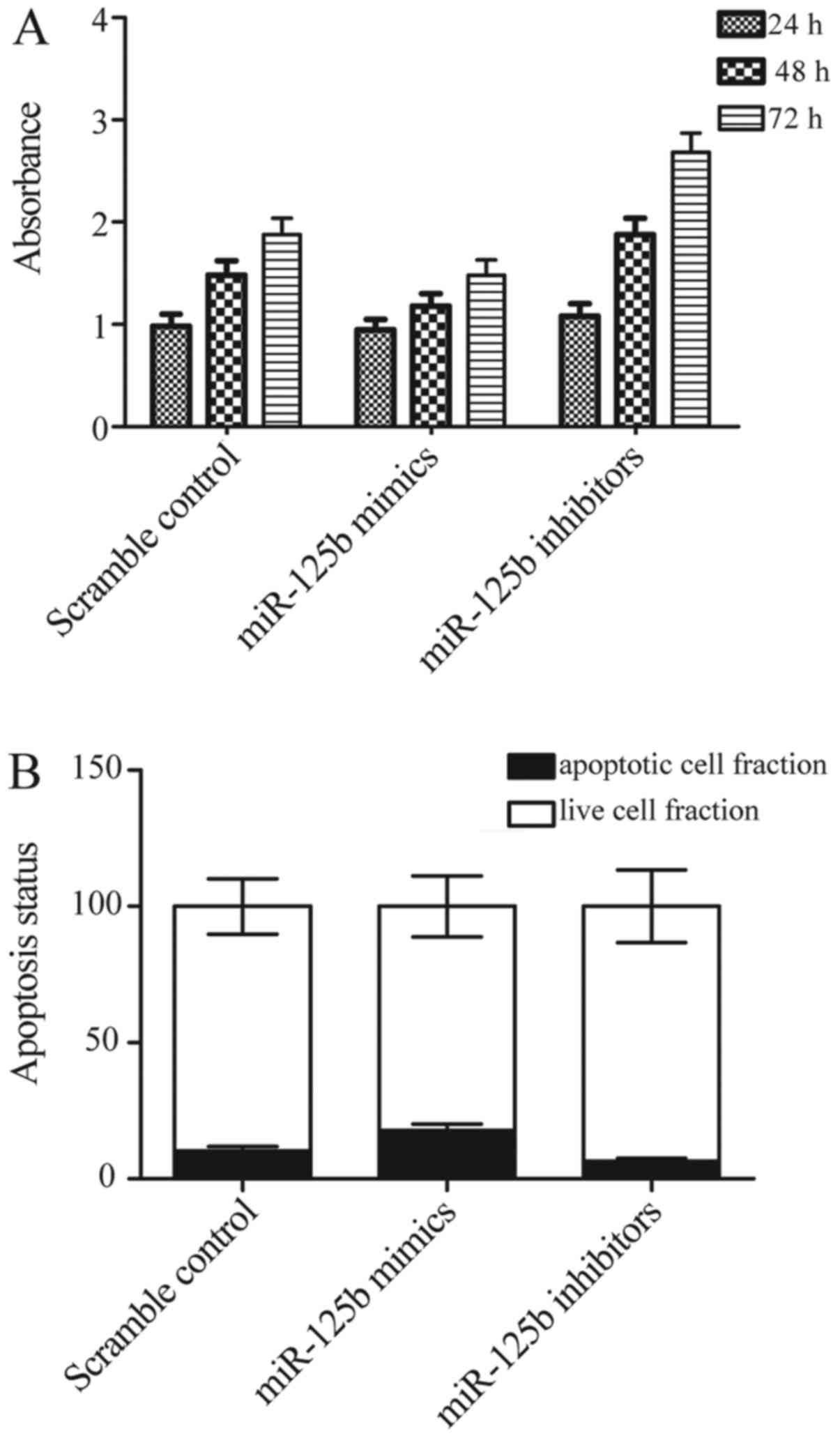
Effect of miR-125b-5p on proliferation
and apoptosis of synovial cells
As synovial cells apoptosis and proliferation are
crucial elements in OA pathogenesis, the effect of miR-125b-5p on
synovial cell apoptosis and proliferation were investigated using
CCK8 assay. The results of CCK8 assay showed that miR-125b-5p mimic
inhibited cell proliferation of OA synovial cells (Fig. 7A), In contrast, miR-125b-5p
inhibitor promoted cell proliferation of OA synovial cells at the
same time (Fig. 7A) compared with
scramble control. Moreover, to explore the underlying molecular
mechanisms, we evaluated the apoptosis status of the differently
treated synovial cells, and found that overexpression of
miR-125b-5p promotes apoptosis of synovial cells, while
downregulation of miR-125b-5p significantly inhibits apoptosis of
synovial cells. These data suggested that proliferation was
inhibited and apoptosis was promoted by miR-125b-5p.
Discussion
Previously, miR-125b-5p was demonstrated to be
involved in atherosclerosis, at least partially, for the first time
by signaling for adhesion molecules and inflammatory cytokines
which participated in vascular atherosclerosis pathological
process. miR-125b-5p was also reported to be reduced during
vascular neointima formation and in calcified vessels in
atherosclerotic mice by Goettsch et al (26), and Tili et al (27) reported that the expression level of
MCP-1 and IL-6 was decreased by miR-125b-5p overexpression, which
agreed with their previous research results showing that expression
level of miR-125b-5p reduced in response to tumor necrosis factor-α
and lipopolysaccharide. Those findings suggested that miR-125b-5p
might serve as a small molecular target during atherosclerosis.
Moreover, the data indicated that exertion of miR-125b-5p function
might be related to inflammation (28,29).
It has been demonstrated previously that miR-125b-5p
served as a leukemogenesis oncomiR (30). The chief carcinogenic event is
miR-125b activation by the chromosomal translocation
t(2;11)(p21;q23) in acute myeloid leukemia and miR-125b locus
translocation into immunoglobulin heavy chain enhancer in
lymphoblastic leukemia [t(11;14)(q24;q32)] (31,32)
Targeting to multiple genes such as STAT3, PUMA, BCL3 and BAK1,
miR-125b-5p was involved in apoptosis regulation, however,
miR-125b-5p could lead to either tumor suppression or oncogenesis
depending on cell types (30).
miR-125b-5p overexpression leads to apoptosis in the majority of
myeloma cells, resulting in survival benefit in T-ALL (33–35).
In this study, we collected 36 participants
consisting of 12 normal control, 12 mild OA and 12 severe OA
samples based on the Kellgren/Lawrence Criterion prior to our
research, then we performed quantitative PCR on the expression
levels of miR-125b-5p and synoviolin 1 (SYVN1) among the three
groups, and found that the gene level of miR-125b-5p in severe OA
group was the highest in the three groups, while the gene level of
miR-125b-5p in normal group was the lowest. While the gene level of
SYVN1 in severe OA group was the lowest, while the gene level of
SYVN1 in normal group was the highest in the three groups.
Moreover, we then performed computational analysis and luciferase
analysis to confirm that SYVN1 is a virtual target of miR-125b-5p
with binding sites in the 3′UTR of the gene, and confirmed by the
luciferase activity of the cells co-transfected with wild-type
SYVN1 3′UTR were significantly lower than that of the cells
transfected with mutant SYVN1 3′UTR and scramble controls, while
the luciferase activity of the cells transfected with mutant SYVN1
3′UTR substantially comparable with that of the cells transfected
with scramble controls.
Synoviolin (SYVN1), the thologue or mammalian of
yeast Hrd1 (degradation of 3-hydroxy-3-methylglutaryl reductase) is
a multispanning membrane protein, and RING-H2 finger domain in the
carboxy-terminal located in the cytoplasm (36). Substantial research has indicated
that SYVN1 served as an E3 ubiquitin ligase for endoplasmic
reticulum (ER)-associated degradation (ERAD), a process
participating in quality control of cells and cellular adaptation
to ERAD misfolded proteins (37,38).
ERAD is involved in numerous human diseases, for example diabetes
and neurodegenerative diseases (such as Alzheimer disease, cerebral
ischaemia/hypoxia and Parkinson's disease) (39). In the ERAD pathway, SYVN1 is
involved in the harmful protein disposing, ubiquitination and
recognition, making sure that these proteins are delivered by ER
and degraded to reduce damage of ER. SYVN1 also suppress ER
stress-induced apoptosis (9,40).
In this study, we investigated quantitative PCR to
confirm the negative regulatory relationship between SYVN1 and
miR-125b-5p based on the negative correlation coefficient (r=
−0.40). We then conducted quantitative PCR and western blot
analysis to detect the expression level of SYVN1 of cells induced
with miR-125b-5p mimics or inhibitors, we found overexpression of
miR-125b-5p suppressed the level of SYVN1, while downregulation of
miR-125b-5p upregulated the level of SYVN1. Finally, we performed
MTT assay to detect the effect of miR-125b-5p on apoptosis and
proliferation of synovial cells by use of CCK8 kit, and found that
miR-125b-5p mimic inhibited cell proliferation, and miR-125b-5p
inhibitor promoted cell proliferation of OA synovial cells. In
contrast, overexpression of miR-125b-5p promoted apoptosis of
synovial cells, while downregulation of miR-125b-5p significantly
inhibited the apoptosis of synovial cells. SYVN1 was confirmed as a
rheumatoid regulator and its expression was significantly related
to rheumatoid arthritis development. Spontaneous arthropathy was
observed in transgenic SYVN1 mice with SYVN1 overexpression,
however, mice with reduced SYVN1 (SYVN1+/− mice) were at lower risk
of developing collagen-induced arthritis (CIA) (9,41).
In conclusion, these findings demonstrated that
miR-125b-5p could promote apoptosis of synovial cells through
targeting the SYVN1 gene, and the excessive apoptosis of synovial
cells could contribute to the development of OA.
References
|
1
|
Schaible HG, Richter F, Ebersberger A,
Boettger MK, Vanegas H, Natura G, Vazquez E and von Segond Banchet
G: Joint pain. Exp Brain Res. 196:153–162. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lascelles BD, McFarland JM and Swann H:
Guidelines for safe and effective use of NSAIDs in dogs. Vet Ther.
6:237–251. 2005.PubMed/NCBI
|
|
3
|
Blanco FJ, Ochs RL, Schwarz H and Lotz M:
Chondrocyte apoptosis induced by nitric oxide. Am J Pathol.
146:75–85. 1995.PubMed/NCBI
|
|
4
|
Albina JE, Cui S, Mateo RB and Reichner
JS: Nitric oxide-mediated apoptosis in murine peritoneal
macrophages. J Immunol. 150:5080–5085. 1993.PubMed/NCBI
|
|
5
|
Fehsel K, Kröncke KD, Meyer KL, Huber H,
Wahn V and Kolb-Bachofen V: Nitric oxide induces apoptosis in mouse
thymocytes. J Immunol. 155:2858–2865. 1995.PubMed/NCBI
|
|
6
|
Farrell AJ, Blake DR, Palmer RM and
Moncada S: Increased concentrations of nitrite in synovial fluid
and serum samples suggest increased nitric oxide synthesis in
rheumatic diseases. Ann Rheum Dis. 51:1219–1222. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hayashi T, Abe E, Yamate T, Taguchi Y and
Jasin HE: Nitric oxide production by superficial and deep articular
chondrocytes. Arthritis Rheum. 40:261–269. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Firestein GS, Yeo M and Zvaifler NJ:
Apoptosis in rheumatoid arthritis synovium. J Clin Invest.
96:1631–1638. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Amano T, Yamasaki S, Yagishita N,
Tsuchimochi K, Shin H, Kawahara K, Aratani S, Fujita H, Zhang L,
Ikeda R, et al: Synoviolin/Hrd1, an E3 ubiquitin ligase, as a novel
pathogenic factor for arthropathy. Genes Dev. 17:2436–2449. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Murchison EP and Hannon GJ: miRNAs on the
move: miRNA biogenesis and the RNAi machinery. Curr Opin Cell Biol.
16:223–229. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J,
Lee J, Provost P, Rådmark O, Kim S, et al: The nuclear RNase III
Drosha initiates microRNA processing. Nature. 425:415–419. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Denli AM, Tops BB, Plasterk RH, Ketting RF
and Hannon GJ: Processing of primary microRNAs by the
microprocessor complex. Nature. 432:231–235. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nam JW, Shin KR, Han J, Lee Y, Kim VN and
Zhang BT: Human microRNA prediction through a probabilistic
co-learning model of sequence and structure. Nucleic Acids Res.
33:3570–3581. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Swingler TE, Wheeler G, Carmont V, Elliott
HR, Barter MJ, Abu-Elmagd M, Donell ST, Boot-Handford RP,
Hajihosseini MK, Münsterberg A, et al: The expression and function
of microRNAs in chondrogenesis and osteoarthritis. Arthritis Rheum.
64:1909–1919. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hong E and Reddi AH: MicroRNAs in
chondrogenesis, articular cartilage, and osteoarthritis:
Implications for tissue engineering. Tissue Eng Part B Rev.
18:445–453. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dong S, Yang B, Guo H and Kang F:
MicroRNAs regulate osteogenesis and chondrogenesis. Biochem Biophys
Res Commun. 418:587–591. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dunn W, DuRaine G and Reddi AH: Profiling
microRNA expression in bovine articular cartilage and implications
for mechanotransduction. Arthritis Rheum. 60:2333–2339. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Iliopoulos D, Malizos KN, Oikonomou P and
Tsezou A: Integrative microRNA and proteomic approaches identify
novel osteoarthritis genes and their collaborative metabolic and
inflammatory networks. PLoS One. 3:e37402008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yu C, Chen WP and Wang XH: MicroRNA in
osteoarthritis. J Int Med Res. 39:1–9. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Goldring MB and Marcu KB: Epigenomic and
microRNA-mediated regulation in cartilage development, homeostasis,
and osteoarthritis. Trends Mol Med. 18:109–118. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Crowe N, Swingler TE, Le LT, Barter MJ,
Wheeler G, Pais H, Donell ST, Young DA, Dalmay T1 and Clark IM:
Detecting new microRNAs in human osteoarthritic chondrocytes
identifies miR-3085 as a human, chondrocyte-selective, microRNA.
Osteoarthritis Cartilage. 24:534–543. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yamasaki S, Yagishita N, Tsuchimochi K,
Kato Y, Sasaki T, Amano T, Beppu M, Aoki H, Nakamura H, Nishioka K,
et al: Resistance to endoplasmic reticulum stress is an acquired
cellular characteristic of rheumatoid synovial cells. Int J Mol
Med. 18:113–117. 2006.PubMed/NCBI
|
|
25
|
Yagishita N and Nakajima T: Synoviolin as
a causative factor of arthropathy. Tanpakushitsu Kakusan Koso.
51:(Suppl 10). 1298–1303. 2006.(In Japanese). PubMed/NCBI
|
|
26
|
Goettsch C, Rauner M, Pacyna N, Hempel U,
Bornstein SR and Hofbauer LC: miR-125b regulates calcification of
vascular smooth muscle cells. Am J Pathol. 179:1594–1600. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tili E, Michaille JJ, Cimino A, Costinean
S, Dumitru CD, Adair B, Fabbri M, Alder H, Liu CG, Calin GA, et al:
Modulation of miR-155 and miR-125b levels following
lipopolysaccharide/TNF-alpha stimulation and their possible roles
in regulating the response to endotoxin shock. J Immunol.
179:5082–5089. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li X, Yao N, Zhang J and Liu Z:
MicroRNA-125b is involved in atherosclerosis obliterans in
vitro by targeting podocalyxin. Mol Med Rep. 12:561–568.
2015.PubMed/NCBI
|
|
29
|
Luo X, Ranade K, Talker R, Jallal B, Shen
N and Yao Y: microRNA-mediated regulation of innate immune response
in rheumatic diseases. Arthritis Res Ther. 15:2102013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Shaham L, Binder V, Gefen N, Borkhardt A
and Izraeli S: MiR-125 in normal and malignant hematopoiesis.
Leukemia. 26:2011–2018. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bousquet M, Quelen C, Rosati R, Mansat-De
Mas V, La Starza R, Bastard C, Lippert E, Talmant P,
Lafage-Pochitaloff M, Leroux D, et al: Myeloid cell differentiation
arrest by miR-125b-1 in myelodysplastic syndrome and acute myeloid
leukemia with the t(2;11)(p21;q23) translocation. J Exp Med.
205:2499–2506. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chapiro E, Russell LJ, Struski S, Cavé H,
Radford-Weiss I, Valle VD, Lachenaud J, Brousset P, Bernard OA,
Harrison CJ, et al: A new recurrent translocation t(11;14)(q24;q32)
involving IGH@ and miR-125b-1 in B-cell progenitor acute
lymphoblastic leukemia. Leukemia. 24:1362–1364. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang GQ, Kai M, Nakano M and Ohkura Y:
Pre-column fluorescence derivatization high-performance liquid
chromatography of opioid peptides in rat brain and its use for
enzymatic peptide characterization. Chem Pharm Bull (Tokyo).
39:126–129. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Schotte D, De Menezes RX, Akbari Moqadam
F, Khankahdani LM, Lange-Turenhout E, Chen C, Pieters R and Den
Boer ML: MicroRNA characterize genetic diversity and drug
resistance in pediatric acute lymphoblastic leukemia.
Haematologica. 96:703–711. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ooi AG, Sahoo D, Adorno M, Wang Y,
Weissman IL and Park CY: MicroRNA-125b expands hematopoietic stem
cells and enriches for the lymphoid-balanced and lymphoid-biased
subsets. Proc Natl Acad Sci USA. 107:21505–21510. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hampton RY, Gardner RG and Rine J: Role of
26S proteasome and HRD genes in the degradation of
3-hydroxy-3-methylglutaryl-CoA reductase, an integral endoplasmic
reticulum membrane protein. Mol Biol Cell. 7:2029–2044. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gardner RG, Swarbrick GM, Bays NW, Cronin
SR, Wilhovsky S, Seelig L, Kim C and Hampton RY: Endoplasmic
reticulum degradation requires lumen to cytosol signaling.
Transmembrane control of Hrd1p by Hrd3p. J Cell Biol. 151:69–82.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Friedlander R, Jarosch E, Urban J,
Volkwein C and Sommer T: A regulatory link between ER-associated
protein degradation and the unfolded-protein response. Nat Cell
Biol. 2:379–384. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yoshida H: ER stress and diseases. FEBS J.
274:630–658. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kaneko M, Ishiguro M, Niinuma Y, Uesugi M
and Nomura Y: Human HRD1 protects against ER stress-induced
apoptosis through ER-associated degradation. FEBS Lett.
532:147–152. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ritchlin C: Fibroblast biology. Effector
signals released by the synovial fibroblast in arthritis. Arthritis
Res. 2:356–360. 2000. View
Article : Google Scholar : PubMed/NCBI
|