Introduction
With an estimated 500,000 new cases and 300,000
deaths per year, cervical cancer is one of the most common female
malignancies worldwide (1).
Approximately 80% of cases occur in developing countries, where
extensive screening by cervical cytology is unavailable (2). Cervical cancer morbidity is low in
developed countries due to available cervical screening and ongoing
active health education programs (3). Currently, several therapeutic
strategies, including surgery, chemotherapy and radiotherapy, are
utilised to treat patients with cervical cancer (4,5).
Although tremendous advances have been made in conventional
treatments, the prognosis of cervical cancer remains poor due to
development of resistance to radiotherapy and chemotherapy
(6). The overall 5-year survival
rate is <40%, particularly for patients presenting with advanced
stage disease (7). Therefore,
identification of the mechanisms underlying the formation and
progression of cervical cancer may significantly promote early
diagnosis, prognosis and development of novel therapeutic methods
for patients with this malignancy.
Numerous studies have reported that abnormal
expression of microRNAs (miRNAs) is significantly involved in the
pathogenesis of human cancers, including cervical cancer (8–10).
miRNAs comprise a large group of small and endogenous RNAs
measuring 18–23 nucleotides in length and are unable to encode for
proteins (11). miRNAs can modulate
expression of their target genes by binding to target mRNAs at the
3′-untranslated region (3′-UTR), forming stable duplexes in a
partial complementary manner and inducing mRNA degradation or
interfering with translation (12).
Through these regulatory roles, miRNAs play key roles in many
cellular biological processes, such as cellular development,
growth, differentiation, epithelial-mesenchymal transition and
apoptosis (13). In recent years, a
wide variety of miRNAs were discovered to be abnormally expressed
in various types of human cancer, such as breast (14), lung (15), prostate (16), cervical (17) and ovarian cancer (18). Accumulated evidence also suggests
that miRNA dysregulation contributes to initiation and progression
of various human malignancies (19–21).
In human cancer, miRNAs may function as oncogenes by inhibiting
tumour-suppressor genes or as tumour suppressors by downregulating
oncogenes (22,23). Therefore, miRNA regulation may be a
potential therapeutic strategy for human cancer treatment.
miRNA-433 (miR-433) has been studied in several
types of human cancer (24–26). However, little information is
available concerning the expression pattern and biological roles of
miR-433 in cervical cancer. In the present study, we investigated
the miR-433 expression pattern in cervical cancer, the effects of
miR-433 on cervical cancer cells and the underlying molecular
mechanisms. Metadherin (MTDH) (also known as AEG-1 or
LYRIC) was predicted as a potential target of miR-433 and
was selected for further target identification; this gene is
upregulated in cervical cancer tissues and contributes to cervical
cancer occurrence and progression (27,28).
The present study may provide novel insights into cervical cancer
initiation and progression and strategies for cervical cancer
treatment.
Materials and methods
Ethics statement and tissue
samples
The present study was performed according to the
principles of the Declaration of Helsinki and approved by the
Ethics Committees of The Third Affiliated Hospital of Sun Yat-Sen
University. Written informed consent for research purposes was also
provided by each participant. Cervical cancer tissues and
corresponding adjacent normal tissues were collected from 65
patients who underwent surgical resection at The Third Affiliated
Hospital of Sun Yat-sen University between September 2014 and
January 2016. All the patients did not receive prior radiotherapy
or chemotherapy. All tissue samples were stored in liquid nitrogen
until use.
Cell lines
Cervical cancer cell lines (HeLa, C-33A, SiHa and
Ca-Ski) and a human normal cervical epithelial cell line
(Ect1/E6E7) were purchased from the American Type Culture
Collection (ATCC; Manassas, VA, USA). Cervical cancer cells were
grown in Dulbecco's modified Eagle's medium (DMEM) supplemented
with 10% fetal bovine serum (FBS), 100 IU/ml penicillin and 100
µg/ml streptomycin (Gibco, Grand Island, NY, USA). Ect1/E6E7 cells
were maintained in keratinocyte serum-free medium (Gibco)
containing 0.1 ng/ml human recombinant epithelial growth factor,
0.05 mg/ml bovine pituitary extract, 100 IU/ml penicillin, and 100
µg/ml streptomycin (Gibco) at 37°C in a humidified incubator with
5% CO2.
Cell transfection
The miR-433 mimics and negative control miRNA mimics
(miR-NC) were obtained from GeneCopoeia (Guangzhou, China).
MTDH-targeted small interfering RNA (si-MTDH) and the negative
control siRNA (si-NC) were chemically synthesized by GenePharma
Co., Ltd. (Shanghai, China). MTDH overexpressed vector
(pCDNA3.1-MTDH) and corresponding blank vector (pCDNA3.1) were
obtained from the Chinese Academy of Sciences (Changchun, China).
Cells were seeded into 6-well plates 18–24 h before transfection.
Following the protocols for the use of Lipofectamine 2000
(Invitrogen, Carlsbad, CA, USA), miR-433 mimics (60 nM), miR-NC (60
nM), si-MTDH (60 nM), si-NC (60 nM), pcDNA3.1-MTDH (2 mg/ml) or
pcDNA3.1 (2 mg/ml) was transfected into the cells. After incubation
for 6 h, the culture medium was replaced with fresh DMEM with 10%
FBS. Reverse transcription-quantitative polymerase chain reaction
(RT-qPCR) and western blotting were performed to determine the
transfection efficiency.
RT-qPCR
Total RNA was isolated from tissue samples or cells
using TRIzol (Thermo Fisher Scientific, Waltham, MA, USA) according
to the manufacturer's instructions. For miR-433 expression, reverse
transcription was performed using TaqMan MicroRNA Reverse
Transcription kit (Applied Biosystems, Foster City, CA, USA).
Quantitative PCR was performed to detect the miR-433 expression
level using TaqMan MicroRNA PCR kit (Applied Biosystems). To
quantify MTDH mRNA expression, PrimeScript RT reagent kit
was used to synthesize cDNA, which was then amplified using SYBR
Premix Ex Taq™ kit (both from Takara Bio, Dalian, China). GAPDH and
U6 were used for normalization of MTDH mRNA and miR-433,
respectively. Primers used in the present study were purchased from
Guangzhou RiboBio Co., Ltd. (Guangzhou, China) and shown in
Table I. Each sample was performed
in triplicate, and relative expression changes were calculated
using the 2−ΔΔCt method (29).
 | Table I.RT-qPCR primers. |
Table I.
RT-qPCR primers.
| Gene |
| Sequences
(5→3) |
|---|
| MicroRNA-433 | F |
TGCGGTACGGTGAGCCTGTC |
|
| R |
CCAGTGCAGGGTCCGAGGT |
| U6 | F |
CTTCAAGTAATCCAGGATAGGC |
|
| R |
ATTGGAACGATACAGAGAAGATT |
| MTDH | F |
TGCCTCCTTCACAGACCAA |
|
| R |
TCGGCTGCAGATGAGATAG |
| GAPDH | F |
CATGAGAAGTATGACAACAGCCT |
|
| R |
AGTCCTTCCACGATACCAAAGT |
MTT assay
Cell proliferation was determined using the MTT
assay (Sigma, St. Louis, MO, USA). At 24 h post-transfection,
transfected cells were collected and seeded into 96-well plates at
a density of 3.0×103/well. The plates were incubated for
0, 24, 48 or 72 h after transfection. At each time point, cells
were treated with 20 µl MTT assay reagent (5 mg/ml) for additional
4 h. The supernatant was removed, and 150 µl of dimethyl sulfoxide
(Sigma) was added into each well. Cellular proliferation was
determined by detecting the optical density (OD) at a wavelength of
490 nm. Each assay was performed in triplicate and repeated 3
times.
Cell invasion assay
The Matrigel invasion chambers were utilized to
assess cell invasion ability (8 µm; Corning, Cambridge, MA, USA).
At 48 h post-transfection, cells were incubated with FBS-free
culture medium. On the following day, cells were harvested and
suspended in FBS-free culture medium. Cells (1×105) were
placed intothe upper chambers, and the lower chambers were filled
with DMEM containing 10% FBS. After incubation for 48 h, the cells
remaining on the top of the chambers were removed. Cells that
invaded to the bottom of the membranes were fixed, stained with
0.5% crystal violet and washed. The invasive cells in at least 5
randomly selected fields were photographed and counted under an
inverted microscope (Olympus Corp., Tokyo, Japan). The present
study was performed in triplicate and repeated 3 times.
Flow cytometric analysis
The cell apoptosis rate was determined using the
Dead Cell Apoptosis Kit with Annexin V Alexa Fluor™ 488 and
propidium iodide (PI) (catalog no. V13241; Thermo Fisher
Scientific, Waltham, MA, USA), according to the manufacturer's
instructions. Subsequent to a 72-h incubation, transfected cells
were harvested. After washing 3 times with ice-cold
phosphate-buffered saline (PBS), the transfected cells were fixed
in 80% ice-cold ethanol in PBS. Subsequently, the cells were
resuspended in 100 µl Annexin-binding buffer and incubated with 5
µl Annexin V-FITC and 3 µl PI (50 µg/ml). After incubation at room
temperature in the dark for 20 min, cell apoptosis was examined
using flow cytometry and analyzed using FACSCalibur and CellQuest
software (Beckman Coulter, Inc., Miami, FL, USA).
Bioinformatic predication
TargetScan (http://www.targetscan.org/index.html) and miRanda
(http://www. microrna.org/microrna/)
were adopted to analyze the potential targets of miR-433 (30). ‘Human’ was selected as the species,
and ‘miR-433’ was entered. Putative miRNA-mRNA interaction was
based on the total context score. The more negative the total
context score, the higher the probability of miRNA-mRNA binding.
Relevant targets predicted by all 2 databases were chosen for
laboratory experimentation.
Luciferase reporter assay
Luciferase reporter plasmids,
pMIR-Report-MTDH-3′-UTR-wild-type (Wt) and
pMIR-Report-MTDH-3′-UTR-mutant (Mut), were synthesized and
confirmed by GenePharma. Cervical cancer cells were co-transfected
with pMIR-Report-MTDH-3′-UTR-Wt or pMIR-Report-MTDH-3′-UTR-Mut and
miR-433 mimics or miR-NC using Lipofectamine 2000. After incubation
for 48 h, luciferase reporter assays were conducted using the
Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA)
following the manufacturer's instructions. Firefly luciferase
activity was used as an internal control for Renilla
luciferase activity. All experiments were carried out in triplicate
and repeated 3 times.
Western blot analysis
Total cell lysates were prepared by incubating cells
in RIPA buffer (Beyotime, Shanghai, China) on ice for 1 h. The
concentration of protein was determined using the BCA protein assay
kit according to the manufacturer's protocol (Pierce Biotechnology,
Inc., Rockford, IL, USA). Equal amounts of proteins were separated
on 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis
gels, transferred onto polyvinylidene difluoride membranes
(Millipore, Billerica, MA, USA), blocked with 5% skim milk in
Tris-buffered saline containing 0.05% Tween-20 (TBST), and
incubated with primary antibodies overnight at 4°C as follows:
mouse anti-human monoclonal MTDH (sc-517220; 1:1,000 dilution),
mouse anti-human monoclonal p-AKT (sc-271966; 1:1,000 dilution),
mouse anti-human monoclonal AKT (sc-56878; 1:1,000 dilution),
rabbit anti-human monoclonal p-β-catenin (ab75777; 1:1,000
dilution) (all from Santa Cruz Biotechnology, Santa Cruz, CA, USA),
mouse anti-human monoclonal β-catenin antibody (ab22656; Abcam,
Cambridge, UK, USA), and mouse anti-human monoclonal GAPDH
(sc-32233; 1:1,000 dilution; Santa Cruz Biotechnology). In the
following steps, membranes were washed with TBST and probed with
corresponding horseradish peroxidase (HRP)-conjugated secondary
antibody (1:5,000 dilution; Santa Cruz Biotechnology) for 2 h at
room temperature. The protein bands were detected using enhanced
chemiluminescence (ECL) solution (Pierce Biotechnology, Inc.).
GAPDH was used as an internal control.
Statistical analysis
All data are expressed as mean ± SD, and were
compared with two-tailed Student's t-test or one way ANOVA using
SPSS 19.0 statistical software (SPSS, Inc., Chicago, IL, USA).
Student-Newman-Keuls (SNK) was used to compare differences between
2 groups in the multiple group study. P<0.05 was considered to
indicate a statistically significant result.
Results
miR-433 is frequently downregulated in
cervical tissues and cell lines
In the present study, we first examined miR-433
expression in cervical cancer and corresponding adjacent normal
tissues by RT-qPCR. Our results showed much higher miR-433
expression in the adjacent normal tissues than that observed in the
cervical cancer tissues (Fig. 1A;
P<0.05). We then investigated the association between miR-433
and clinicopathological features in cervical cancer. The median
miR-433 expression level (median=0.8902) was regarded as a cut-off
to divide all cervical cancer patients into either the miR-433
low-expression group (n=33) or miR-433 high-expression group
(n=32). As shown in Table II, low
miR-433 expression level was significantly correlated with tumour
size (P=0.018), FIGO stage (P=0.031), lymph node (P=0.001) and
distant metastases (P=0.032). However, no significant correlations
were noted between miR-433 expression and age (P=0.685), histology
(P=0.526), HPV infection (P=0.247) and family history of cancer
(P=0.163).
 | Table II.Correlations between miR-433
expression and the clinicopathologic features of the cervical
cancer cases. |
Table II.
Correlations between miR-433
expression and the clinicopathologic features of the cervical
cancer cases.
|
|
| miR-433
expression |
|
|---|
|
|
|
|
|
|---|
| Features | No. of cases | Low | High | P-value |
|---|
| Age (years) |
|
|
| 0.685 |
|
<50 | 26 | 14 | 12 |
|
|
≥50 | 39 | 19 | 20 |
|
| Histology |
|
|
| 0.526 |
|
SCC | 55 | 27 | 28 |
|
|
Adenocarcinoma | 10 | 6 | 4 |
|
| HPV infection |
|
|
| 0.247 |
|
Positive | 47 | 25 | 20 |
|
|
Negative | 18 | 8 | 12 |
|
| Tumour size
(cm) |
|
|
| 0.018 |
|
<4 | 29 | 10 | 19 |
|
| ≥4 | 36 | 23 | 13 |
|
| Family history of
cancer |
|
|
| 0.163 |
| No | 37 | 16 | 21 |
|
|
Yes | 28 | 17 | 11 |
|
| FIGO stage |
|
|
| 0.031 |
|
I–II | 24 | 8 | 16 |
|
|
III–IV | 41 | 25 | 16 |
|
| Lymph node
metastasis |
|
|
| 0.001 |
|
Yes | 30 | 22 | 8 |
|
| No | 35 | 11 | 24 |
|
| Distant
metastasis |
|
|
| 0.032 |
|
Yes | 18 | 13 | 5 |
|
| No | 47 | 20 | 27 |
|
We further measured expression levels of miR-433 in
cervical cancer cell lines (HeLa, C-33A, SiHa and Ca-Ski) and human
normal cervical epithelial cell line (Ect1/E6E7). As shown in
Fig. 1B, the expression level of
miR-433 was downregulated in cervical cancer cell lines compared
with that noted in the Ect1/E6E7 cells (P<0.05).
Upregulation of miR-433 inhibits cell
proliferation and invasion and promotes apoptosis in cervical
cancer
Downregulation of miR-433 in cervical cancer
prompted us to explore whether miR-433 acts as a tumour suppressor
in cervical cancer. Therefore, we examined the effects of miR-433
overexpression on cervical cancer cells. HeLa and SiHa cells, which
expressed relatively low miR-433 expression among the 4 examined
cervical cancer cell lines, were transfected with miR-433 mimics or
miR-NC. Expression levels determined through RT-qPCR confirmed
marked upregulation of miR-433 in the HeLa and SiHa cells
transfected with miR-433 mimics (Fig.
2A and B; P<0.05). Firstly, we measured cellular
proliferation using MTT assay after transfection of HeLa and SiHa
cells with miR-433 mimics or miR-NC. The results showed that
restoration of the expression of miR-433 significantly decreased
proliferation of the HeLa and SiHa cells compared with that noted
in the miR-NC group (Fig. 2C and D;
P<0.05). HeLa and SiHa cells were transfected with miR-433
mimics or miR-NC; and cell invasion assay was performed to evaluate
the effects of miR-433 restoration on the invasion of these cells.
Invasion capabilities of HeLa and SiHa cells were significantly
suppressed when cells were transfected with the miR-433 mimics
(Fig. 2E and F; P<0.05). Flow
cytometric analysis was used to determine the apoptosis rates of
the HeLa and SiHa cells transfected with the miR-433 mimics or
miR-NC. The results revealed that at 72 h after transfection, the
apoptosis rate was significantly increased in the HeLa and SiHa
cells transfected with miR-433 mimics compared with that noted in
the miR-NC controls (Fig. 2G and H;
P<0.05). These results indicate that miR-433 acts as a tumour
suppressor in cervical cancer.
MTDH is a direct target gene of
miR-433 in cervical cancer
To further elucidate the underlying mechanisms
involved in the tumour-suppressive roles of miR-433 in cervical
cancer cells, bioinformatic analysis was performed to search for
potential downstream targets of miR-433. Hundreds of candidate
targets were predicted, such as PAK4, PAX6,
MACC1, MTDH, CREB1 and Notch1. Among
these candidate targets, MTDH was selected for further
target identification (Fig. 3A);
this gene was found to be upregulated in cervical cancer tissues
and to contribute to cervical cancer occurrence and progression
(27,28). We employed a luciferase reporter
assay to ascertain whether MTDH is a direct target gene of
miR-433. HeLa and SiHa cells were cotransfected with
pMIR-Report-MTDH-3′-UTR-wild-type (Wt) or
pMIR-Report-MTDH-3′-UTR-mutant-type (Mut) and miR-433 mimics or
miR-NC. The results showed that miR-433 introduction decreased
luciferase activity of pMIR-Report-MTDH-3′-UTR-Wt, but not
pMIR-Report-MTDH-3′-UTR-Mut (Fig.
3B; P<0.05).
We further investigated whether miR-433
overexpression can regulate MTDH expression in cervical cancer
cells. Results of RT-qPCR and western blot analysis showed that
ectopic expression of miR-433 in HeLa and SiHa cells significantly
reduced MTDH expression at both mRNA and protein levels (Fig. 3C and D; P<0.05). Collectively,
these results suggest that MTDH is a direct downstream
target of miR-433 in cervical cancer.
MTDH expression is upregulated in
cervical cancer tissues, and its expression is negatively
correlated with miR-433
To further explore the association between miR-433
and MTDH, MTDH expression was detected in cervical cancer and
corresponding adjacent normal tissues. Data from RT-qPCR and
western blot analysis demonstrated significantly increased
expression of MTDH in cervical cancer tissues compared with that in
corresponding adjacent normal tissues (Fig. 4A and B; P<0.05). We also
evaluated the correlation between MTDH mRNA and miR-433 expression
level in cervical cancer tissues and Spearman's correlation
analysis indicated a significantly negative correlation between
miR-433 and MTDH mRNA expression among the cervical cancer
tissues (Fig. 4C; r=−0.7038;
P<0.0001).
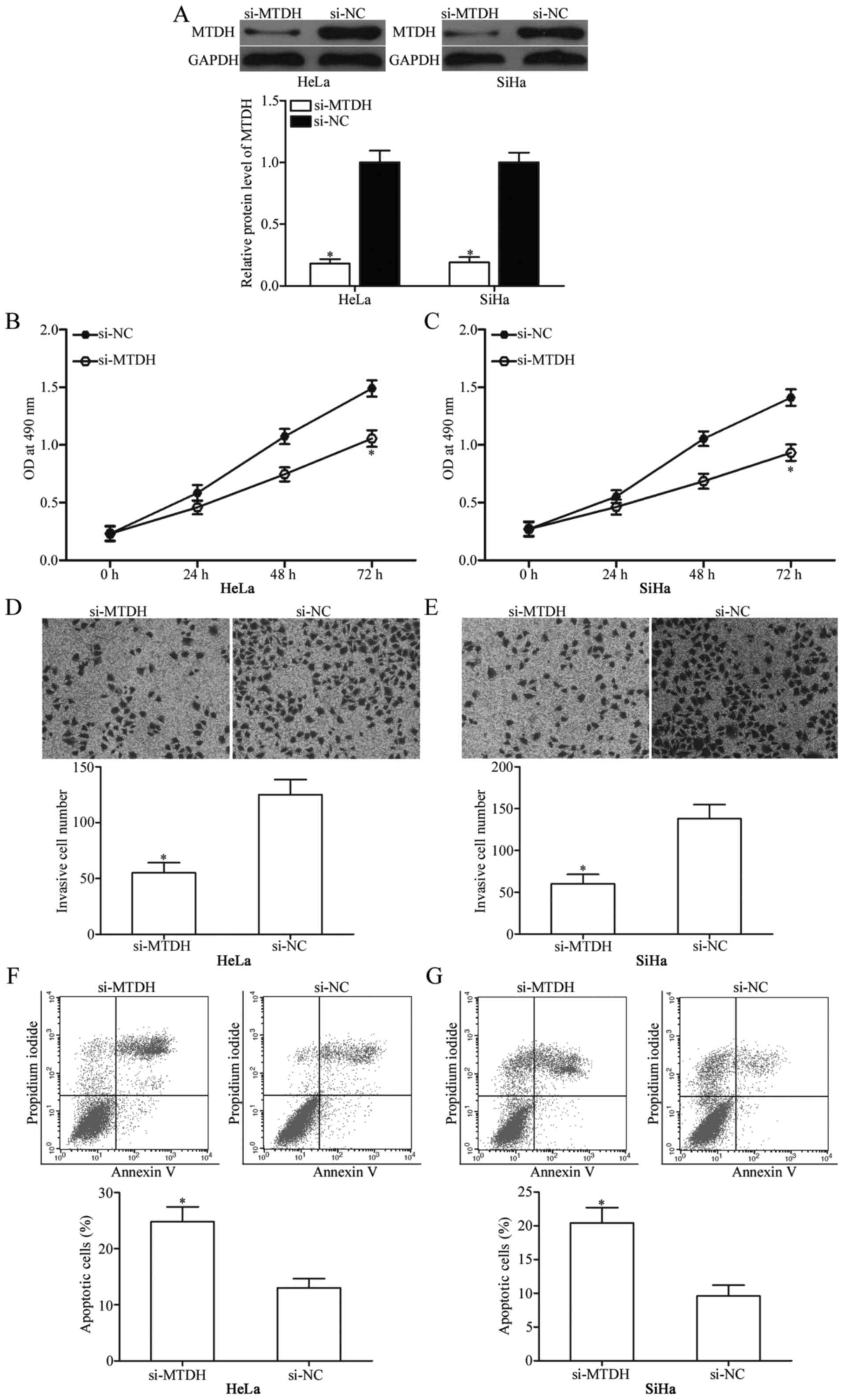
MTDH knockdown suppresses cervical
cancer cell proliferation and invasion, and induces apoptosis in
vitro
To investigate whether downregulation of MTDH
expression exhibits tumour-suppressive functions similar to those
of miR-433 overexpression in cervical cancer, HeLa and SiHa cells
were transfected with si-MTDH to genetically knock down endogenous
MTDH expression (Fig. 5A;
P<0.05). Next, MTT assay, cell invasion assay and flow
cytometric analysis were conducted in HeLa and SiHa cells
transfected with si-MTDH or si-NC. The results showed that MTDH
knockdown exhibited tumour-suppressive roles similar to those of
miR-433 overexpression in cervical cancer cell proliferation
(Fig. 5B and C; P<0,05),
invasion (Fig. 5D and E; P<0,05)
and apoptosis (Fig. 5F and G;
P<0,05) and further suggest that MTDH is a functional downstream
target of miR-433 in cervical cancer.
Overexpression of MTDH reverses the
tumour-suppressive effects of miR-433 in cervical cancer cells
To further evaluate whether MTDH mediates the
effects of miR-433, which affects cervical cancer cell
proliferation, invasion and apoptosis, rescue experiments were
performed, and miR-433 mimics with or without pcDNA3.1-MTDH were
transfected into HeLa and SiHa cells. Western blot results
confirmed that MTDH expression was recovered in the miR-433
mimic-transfected cells after being transfected with pcDNA3.1-MTDH
(Fig. 6A; P<0.05). Rescue
experiments revealed that MTDH overexpression markedly reversed the
effects of miR-433 overexpression in regards to proliferation
(Fig. 6B and C; P<0,05),
invasion (Fig. 6D and E; P<0,05)
and apoptosis (Fig. 6F and G;
P<0,05) of HeLa and SiHa cells. These results indicate that
miR-433 exert its tumour-suppressing roles in cervical cancer
cells, at least in part, by suppressing MTDH.
miR-433 inactivates the AKT and
β-catenin signalling pathways in cervical cancer
MTDH was previously reported to play essential roles
inthe regulation of the AKT and β-catenin pathways (31,32).
Thus, we detected expression levels of p-AKT, AKT, p-β-catenin and
β-catenin in HeLa and SiHa cells after transfection with miR-433
mimics or miR-NC. As shown in Fig.
7, restoration of expression of miR-433 decreased p-AKT and
p-β-catenin expressions in the HeLa and SiHa cells. However, this
restored expression did not affect total AKT and β-catenin
expression. We also noted recovered expression levels of p-AKT and
p-β-catenin in the miR-433 mimic-transfected HeLa and SiHa cells
cotransfected with pcDNA3.1-MTDH. These results indicate that
miR-433 exerts tumour-suppressing roles in cervical cancer cells by
directly targeting MTDH and affecting downstream AKT and β-catenin
pathways.
Discussion
Emerging data have shown that miRNAs play
significant roles in various human cancers, including cervical
cancer (33,34). Aberrantly expressed miRNAs in
cervical cancer contribute to tumour occurrence and development as
either tumour suppressors or promoters (35). Therefore, identification of specific
miRNAs and their targets in cervical cancer may provide novel and
efficient therapeutic methods for patients with this malignancy. In
the present study, we observed downregulation of miR-433 in
cervical cancer tissues and cell lines compared with that in
respective controls. Low miR-433 expression was significantly
correlated with tumour size, FIGO stage, lymph node and distant
metastases of patients with cervical cancer. miR-433 overexpression
inhibited cell proliferation, and invasion and promoted apoptosis
of cervical cancer. MTDH was validated as a direct target of
miR-433 in cervical cancer. Therefore, our data indicate that
miR-433 may be associated with progression of cervical cancer
malignancy.
Numerous studies have reported abnormal expression
of miR-433 in certain types of human cancer. For example, miR-433
was found to be significantly downregulated in gastric cancer
tissues. Aberrant expression of miR-433 was correlated with pM and
pTNM stage in clinical gastric cancer patients (36). In colorectal cancer, miR-433
expression was lower in tumour tissues and cell lines compared with
that in corresponding adjacent tissues and normal human colon
mucosal epithelial cell line. Low expression level of miR-433 was
associated with tumour size in patients with colorectal cancer
(37). Downregulation of miR-433
was also observed in glioma (24),
retinoblastoma (25), ovarian
cancer (26), hepatocellular
(38) and oral squamous cell
carcinoma (39). These findings
suggest that miR-433 may be a diagnostic and prognostic biomarker
for a number of cancer types.
miR-433 was reported to play important roles in the
formation and progression of various types of human cancer. For
example, Guo et al discovered that upregulation of miR-433
suppressed cell proliferation, migration, invasion and cell cycle
progression of gastric cancer (36). In colorectal cancer, overexpression
of miR-433 decreased cell viability and increased apoptosis
(37). Sun et al reported
that miR-433 overexpression suppressed glioma cell proliferation
and metastasis, induced apoptosis in vitro and reduced
tumour growth in vivo (24).
A previous functional study demonstrated that restoration of
miR-433 expression attenuated retinoblastoma cell proliferation and
motility and promoted cell cycle arrest and apoptosis (25). In ovarian cancer, ectopic expression
of miR-433 suppressed cell migration and invasion (26). In hepatocellular carcinoma,
restoration of miR-433 expression reduced cell proliferation and
migration (38,40). Wang et al revealed that
restoration of miR-433 expression inhibited cell proliferation and
motility in oral squamous cell carcinoma (39). These findings also suggest that
miR-433 may perform important functions in these types of cancer
and may be investigated as a potential therapeutic target for the
treatment of various types of cancer.
Identification of cancer-specific miRNAs and their
target genes is important for elucidating miRNA functions in
tumourigenesis and tumour development and may provide promising
therapeutic targets (41). Several
miR-433 targets were identified; such targets include KRAS in
gastric cancer (36), MACC1 in
colorectal cancer (37), cAMP
responsive element binding protein (CREB) in glioma (24), Notch1 and PAX6 in retinoblastoma
(25), Notch1 in ovarian cancer
(26), PAK4 and CREB1 in
hepatocellular carcinoma (38,40),
and histone deacetylase 6 in oral squamous cell carcinoma (39). In the present study, MTDH was
predicted as a potential target of miR-433 using bioinformatic
analysis. Subsequently, luciferase reporter assays indicated direct
binding of miR-433 to MTDH 3′-UTR. RT-qPCR and western blot
analysis revealed that miR-433 negatively regulates MTDH expression
at both the mRNA and protein levels in cervical cancer cells. Our
experimental data further revealed a significant increase in MTDH
in cervical cancer tissues, and this result was negatively
correlated with miR-433 expression patterns. MTDH knockdown showed
tumour-suppressive roles similar to those as miR-433 overexpression
in cervical cancer. Finally, rescue experiments revealed that MTDH
upregulation markedly reversed the effects of miR-433
overexpression on cervical cancer cells. These results strongly
demonstrated that MTDH is a direct target of miR-433 in cervical
cancer.
MTDH gene is located at chromosome 8q22, and
it was first discovered in human foetal astrocytes by Su et
al in 2002 (42). MTDH
encodes for a 582-amino acid protein and is ubiquitously expressed
in all organs and distributed in the cell cytoplasm, membrane,
nucleus and endoplasmic reticulum (43). Previous studies have reported that
MTDH is overexpressed in a variety of human cancers, such as
non-small-cell lung (44), gastric
(45), breast (46), ovarian (47) and bladder cancer (48). MTDH plays critical roles in multiple
biological processes in tumourigenesis and tumour development
through integration of oncogenic pathways, including PI3K/AKT,
nuclear factor-κB, mitogen-activated protein kinase and
Wnt/β-catenin signalling pathways (49–51).
In cervical cancer, MTDH is upregulated and is significantly
correlated with tumour size, lymph node metastasis, TNM stage and
tumour differentiation (27,28).
Univariate and multivariate analyses indicated a shortened survival
period of cervical cancer patients with high expression levels of
MTDH (28). Therefore, MTDH is
considered a valuable prognostic marker and therapeutic target for
several types of cancer.
In conclusion, the present study demonstrated that
miR-433 is significantly downregulated in cervical cancer, and low
expression level of this miRNA is associated with tumour size, FIGO
stage, lymph node and distant metastases. In vitro studies
demonstrated that miR-433 suppressed cellular proliferation and
invasion and increased apoptosis in cervical cancer cells.
Mechanistically, MTDH was validated as a direct target gene
of miR-433 in cervical cancer. miR-433 may be a novel target for
future cervical cancer therapy. In subsequent research, we may
analyze the connection of miR-433 with AKT and β-catenin in patient
samples, and the regulatory roles of miR-433 on other signalling
pathways.
Acknowledgements
The study was supported by grants from Guangdong
Province Science and Technology Pan Projects (nos. 2013B021800137,
2013B022000044 and 2016A020215074) and Guangdong Province Medical
Research Foundation (no. A2016060).
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sankaranarayanan R: Overview of cervical
cancer in the developing world. FIGO 26th Annual Report on the
Results of Treatment in Gynecological Cancer. Int J Gynaecol
Obstet. 95 Suppl 1:S205–S210. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cuzick J, Bergeron C, von Knebel Doeberitz
M, Gravitt P, Jeronimo J, Lorincz AT, Meijer J L M C,
Sankaranarayanan R, Snijders J F P and Szarewski A: New
technologies and procedures for cervical cancer screening. Vaccine.
30 Suppl 5:F107–F116. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Crafton SM and Salani R: Beyond
chemotherapy: An overview and review of targeted therapy in
cervical cancer. Clin Ther. 38:449–458. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kokka F, Bryant A, Brockbank E, Powell M
and Oram D: Hysterectomy with radiotherapy or chemotherapy or both
for women with locally advanced cervical cancer. Cochrane Database
Syst Rev. 4:CD0102602015.
|
|
6
|
de Freitas AC, Gomes Leitão MC and Coimbra
EC: Prospects of molecularly-targeted therapies for cervical cancer
treatment. Curr Drug Targets. 16:77–91. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dai S, Lu Y, Long Y, Lai Y, Du P, Ding N
and Yao D: Prognostic value of microRNAs in cervical carcinoma: A
systematic review and meta-analysis. Oncotarget. 7:35369–35378.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fan JY, Fan YJ, Wang XL, Gao HJ, Zhang Y,
Liu M and Tang H: miR-429 is involved in regulation of NF-κB
activity by targeting IKKβ and suppresses oncogenic activity in
cervical cancer cells. FEBS Lett. 591:118–128. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yan S, Li X, Jin Q and Yuan J:
MicroRNA-145 sensitizes cervical cancer cells to low-dose
irradiation by downregulating OCT4 expression. Exp Ther Med.
12:3130–3136. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sun P, Shen Y, Gong JM, Zhou LL, Sheng JH
and Duan FJ: A new microRNA expression signature for cervical
cancer. Int J Gynecol Cancer. 27:339–343. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Brodersen P and Voinnet O: Revisiting the
principles of microRNA target recognition and mode of action. Nat
Rev Mol Cell Biol. 10:141–148. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Harries LW: Long non-coding RNAs and human
disease. Biochem Soc Trans. 40:902–906. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hong Y, Liang H, Uzair-Ur-Rehman, Wang Y,
Zhang W, Zhou Y, Chen S, Yu M, Cui S, Liu M, et al: miR-96 promotes
cell proliferation, migration and invasion by targeting PTPN9 in
breast cancer. Sci Rep. 6:374212016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu Y, Hu X, Xia D and Zhang S:
MicroRNA-181b is downregulated in non-small cell lung cancer and
inhibits cell motility by directly targeting HMGB1. Oncol Lett.
12:4181–4186. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bucay N, Sekhon K, Yang T, Majid S,
Shahryari V, Hsieh C, Mitsui Y, Deng G, Tabatabai ZL, Yamamura S,
et al: MicroRNA-383 located in frequently deleted chromosomal locus
8p22 regulates CD44 in prostate cancer. Oncogene. 36:2667–2679.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lai XJ, Cheng XY and Hu LD: microRNA 421
induces apoptosis of c-33a cervical cancer cells via
down-regulation of Bcl-xL. Genet Mol Res. 15:152016. View Article : Google Scholar
|
|
18
|
Li X, Chen W, Zeng W, Wan C, Duan S and
Jiang S: microRNA-137 promotes apoptosis in ovarian cancer cells
via the regulation of XIAP. Br J Cancer. 116:66–76. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang J, Liu H, Tian L, Wang F, Han L,
Zhang W and Bai YA: miR-15b inhibits the progression of
glioblastoma cells through targeting insulin-like growth factor
receptor 1. Horm Cancer. 8:49–57. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li Y, Jiang W, Hu Y, Da Z, Zeng C, Tu M,
Deng Z and Xiao W: MicroRNA-199a-5p inhibits cisplatin-induced drug
resistance via inhibition of autophagy in osteosarcoma cells. Oncol
Lett. 12:4203–4208. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang P, Deng Y and Fu X: MiR-509-5p
suppresses the proliferation, migration, and invasion of non-small
cell lung cancer by targeting YWHAG. Biochem Biophys Res Commun.
2016.
|
|
22
|
Hwang HW and Mendell JT: MicroRNAs in cell
proliferation, cell death, and tumorigenesis. Br J Cancer. 96
Suppl:R40–R44. 2007.PubMed/NCBI
|
|
23
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sun S, Wang X, Xu X, Di H, Du J, Xu B,
Wang Q and Wang J: MiR-433-3p suppresses cell growth and enhances
chemosensitivity by targeting CREB in human glioma. Oncotarget.
8:5057–5068. 2017.PubMed/NCBI
|
|
25
|
Li X, Yang L, Shuai T, Piao T and Wang R:
MiR-433 inhibits retinoblastoma malignancy by suppressing Notch1
and PAX6 expression. Biomed Pharmacother. 82:247–255. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liang T, Guo Q, Li L, Cheng Y, Ren C and
Zhang G: MicroRNA-433 inhibits migration and invasion of ovarian
cancer cells via targeting Notch1. Neoplasma. 63:696–704. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Long M, Dong K, Gao P, Wang X, Liu L, Yang
S, Lin F, Wei J and Zhang H: Overexpression of astrocyte-elevated
gene-1 is associated with cervical carcinoma progression and
angiogenesis. Oncol Rep. 30:1414–1422. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Huang K, Li LA, Meng Y, You Y, Fu X and
Song L: High expression of astrocyte elevated gene-1 (AEG-1) is
associated with progression of cervical intraepithelial neoplasia
and unfavorable prognosis in cervical cancer. World J Surg Oncol.
11:2972013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li WF, Dai H, Ou Q, Zuo GQ and Liu CA:
Overexpression of microRNA-30a-5p inhibits liver cancer cell
proliferation and induces apoptosis by targeting MTDH/PTEN/AKT
pathway. Tumour Biol. 37:5885–5895. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shen X, Si Y, Yang Z, Wang Q, Yuan J and
Zhang X: MicroRNA-542-3p suppresses cell growth of gastric cancer
cells via targeting oncogene astrocyte-elevated gene-1. Med Oncol.
32:3612015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hu X, Schwarz JK, Lewis JS Jr, Huettner
PC, Rader JS, Deasy JO, Grigsby PW and Wang X: A microRNA
expression signature for cervical cancer prognosis. Cancer Res.
70:1441–1448. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lee JW, Choi CH, Choi JJ, Park YA, Kim SJ,
Hwang SY, Kim WY, Kim TJ, Lee JH, Kim BG, et al: Altered microRNA
expression in cervical carcinomas. Clin Cancer Res. 14:2535–2542.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Guo LH, Li H, Wang F, Yu J and He JS: The
tumor suppressor roles of miR-433 and miR-127 in gastric cancer.
Int J Mol Sci. 14:14171–14184. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li J, Mao X, Wang X, Miao G and Li J:
miR-433 reduces cell viability and promotes cell apoptosis by
regulating MACC1 in colorectal cancer. Oncol Lett. 13:81–88. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xue J, Chen LZ, Li ZZ, Hu YY, Yan SP and
Liu LY: MicroRNA-433 inhibits cell proliferation in hepatocellular
carcinoma by targeting p21 activated kinase (PAK4). Mol Cell
Biochem. 399:77–86. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang XC, Ma Y, Meng PS, Han JL, Yu HY and
Bi LJ: miR-433 inhibits oral squamous cell carcinoma (OSCC) cell
growth and metastasis by targeting HDAC6. Oral Oncol. 51:674–682.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yang Z, Tsuchiya H, Zhang Y, Hartnett ME
and Wang L: MicroRNA-433 inhibits liver cancer cell migration by
repressing the protein expression and function of cAMP response
element-binding protein. J Biol Chem. 288:28893–28899. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chen X, Bo L, Lu W, Zhou G and Chen Q:
MicroRNA-148b targets Rho-associated protein kinase 1 to inhibit
cell proliferation, migration and invasion in hepatocellular
carcinoma. Mol Med Rep. 13:477–482. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Su ZZ, Kang DC, Chen Y, Pekarskaya O, Chao
W, Volsky DJ and Fisher PB: Identification and cloning of human
astrocyte genes displaying elevated expression after infection with
HIV-1 or exposure to HIV-1 envelope glycoprotein by rapid
subtraction hybridization, RaSH. Oncogene. 21:3592–3602. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Lee SG, Kang DC, DeSalle R, Sarkar D and
Fisher PB: AEG-1/MTDH/LYRIC, the beginning: Initial cloning,
structure, expression profile, and regulation of expression. Adv
Cancer Res. 120:1–38. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ke ZF, Mao X, Zeng C, He S, Li S and Wang
LT: AEG-1 expression characteristics in human non-small cell lung
cancer and its relationship with apoptosis. Med Oncol. 30:3832013.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Dong L, Qin S, Li Y, Zhao L, Dong S, Wang
Y, Zhang C and Han S: High expression of astrocyte elevated gene-1
is associated with clinical staging, metastasis, and unfavorable
prognosis in gastric carcinoma. Tumour Biol. 36:2169–2178. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Tokunaga E, Nakashima Y, Yamashita N,
Hisamatsu Y, Okada S, Akiyoshi S, Aishima S, Kitao H, Morita M and
Maehara Y: Overexpression of metadherin/MTDH is associated with an
aggressive phenotype and a poor prognosis in invasive breast
cancer. Breast Cancer. 21:341–349. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhou B, Yang J, Shu B, Liu K, Xue L, Su N,
Liu J and Xi T: Overexpression of astrocyte-elevated gene-1 is
associated with ovarian cancer development and progression. Mol Med
Rep. 11:2981–2990. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhou J, Li J, Wang Z, Yin C and Zhang W:
Metadherin is a novel prognostic marker for bladder cancer
progression and overall patient survival. Asia Pac J Clin Oncol.
8:e42–e48. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Ge X, Lv X, Feng L, Liu X, Gao J, Chen N
and Wang X: Metadherin contributes to the pathogenesis of diffuse
large B-cell lymphoma. PLoS One. 7:e394492012. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhang J, Zhang Y, Liu S, Zhang Q, Wang Y,
Tong L, Chen X, Ji Y, Shang Q, Xu B, et al: Metadherin confers
chemoresistance of cervical cancer cells by inducing autophagy and
activating ERK/NF-κB pathway. Tumour Biol. 34:2433–2440. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Hu G, Wei Y and Kang Y: The multifaceted
role of MTDH/AEG-1 in cancer progression. Clin Cancer Res.
15:5615–5620. 2009. View Article : Google Scholar : PubMed/NCBI
|