Introduction
Synovial sarcoma (SS) is a malignant soft tissue
tumor that originates from synovial cells or mesenchymal cells that
differentiate into synovial cells (1). The symptoms of SS are pain and a
localized mass at an early stage, and joint damage can be found
when the tumor progresses. Many patients are diagnosed at an
advanced stage because of the atypical symptoms and the rapid
progression of the tumor (2).
Sustained proliferation of the tumor cells combined with
insufficient apoptosis plays an important role in tumor
progression. Therefore, it is of great importance to reveal the
underlying mechanism that regulates the proliferation and apoptosis
of tumor cells.
Autophagy is an evolutionarily conserved response at
the subcellular level. Autophagy degrades the cytoplasm and
organelles, releases amino acids and fatty acids, and plays an
important role in metabolism and organelle renewal in cells
(3). Normally, autophagy adapts
cells to the environment to help cells survive tough conditions.
Notably, autophagy may function as an alternative cell death
mechanism, similar to apoptosis under certain conditions, which is
described as type II programmed cell death (4). Autophagy is closely related to
carcinogenesis and tumor progression. Aberrant expression of
autophagic genes, changes in autophagic activity and conversion of
autophagic signaling pathways can affect the viability of tumor
cells (5).
The Beclin1 gene, also known as BECN1, is an
essential gene involved in the autophagy process in mammalian
cells. Liang et al (6) found
a new protein (molecular weight 60 ku) in rats with encephalitis
caused by the fatal Sinbis virus in 1998 and named the gene that
coded this protein Beclin1. Beclin1 is a homologue of yeast Atg6,
located on the human chromosome 17q21. Beclin1 codes a sequence
with 450 amino acid residues, which contains three special domains:
The conserved BH3 domain (residues 107–135), the coiled coil domain
(residues 140–268) and the evolutionarily conserved domain
(residues 244–337) (7). Some
studies have confirmed that Beclin1 can induce and regulate
autophagy by binding to Vps34p through the evolutionarily conserved
domain and UVRAG through the coiled coil domain (8). Moreover, the function of Beclin1 in
apoptosis has been investigated in many studies. A recent research
showed that Beclin1 regulated apoptosis by binding to the
anti-apoptotic members of the Bcl family such as Bcl-2, Bcl-xl and
Bcl-w through the BH3 domain (9).
The antitumor effect of Beclin1 has been confirmed
in many types of tumors such as breast (10,11),
colon (12,13), cervical (14,15)
ovarian cancer (16,17) and glioblastoma (18,19).
Some studies have reported that the expression level of Beclin1 is
significantly lower in ovarian cancer tissue than in normal ovarian
tissue (20,21); moreover, inhibited proliferation was
observed in breast cancer cells with high expression level of
Beclin1 (22,23). However, the underlying mechanism by
which Beclin1 promotes tumor cell death remains unclear. Some
studies have suggested that Beclin1 inhibits the viability of tumor
cells by inducing autophagic cell death (24,25);
some studies indicate that Beclin1 directly induces the apoptosis
of tumor cells in an autophagy-independent manner (26,27).
In the present study, we explored the function of Beclin1 in SW982
synovial sarcoma cells and investigated the mechanism by which
Beclin1 regulates cell proliferation, apoptosis and autophagy.
Materials and methods
Cell culture
The human synovial sarcoma cell line SW982 was
obtained from the Shanghai Institute of Cell Biology, Chinese
Academy of Sciences (Shanghai, China). The SW982 cells were
cultured in Dulbecco's modified Eagle's medium (DMEM)/F12 medium
(Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented
with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific,
Inc.), 100 U/ml penicillin and 100 µg/ml streptomycin in a humid
atmosphere containing 5% CO2 at 37°C.
Establishment of stable cell lines
overexpressing Beclin1
The lentiviruses expressing the Beclin1 sequence
(OE) and the negative control lentiviruses (NC) were constructed by
Hanbio Co. (Shanghai, China). The lentiviral vector contains a GFP
marker for indicating the transfection efficiency and a
puromycin-resistant marker for selecting the transfected cells. The
virus titer was raised to 108 transfection units
(TU)/ml. Cells were seeded in 6-well plates and infected with
viruses and polybrene on the following day. A total of 24 h later,
the medium containing the viruses was removed and replaced with
fresh medium. The infected cells were treated with puromycin for 7
days to obtain the positive clones. Positive clones were selected
and purified to establish the stable cell line. The expression
level of Beclin1 was determined by immunofluorescence staining,
RT-qPCR and western blot analysis.
Immunofluorescence staining
Cells were seeded in 24-well plates and maintained
for 48 h. After being washed 3 times with phosphate-buffered saline
(PBS), cells were fixed in a 4% paraformaldehyde solution for 15
min, permeabilized with 0.3% Triton X-100, blocked with 5% BSA
blocking reagent for 30 min and then incubated with the
anti-Beclin1 monoclonal primary antibody (dilution 1:50; cat. no.
BM5181; Wuhan Boster Biological Technology, Ltd., Wuhan, China)
overnight at 4°C. After another 3 washes with PBS, the cells were
incubated with tetraethyl rhodamine isothiocyanate
(TRITC)-conjugated secondary antibody (dilution 1:200; cat. no.
BA1090; Wuhan Boster Biological Technology, Ltd.) for 1 h at room
temperature. DAPI reagent was used to counterstain the nuclei. The
result of the staining was observed with an inverted fluorescence
microscope system (Nikon ECLIPSE Ti-S; Nikon, Tokyo, Japan).
Total RNA extraction and quantitative
real-time polymerase chain reaction (RT-qPCR) analysis
Total RNA was extracted from SW982 cells using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. Total RNA was
reverse-transcribed into cDNA using a PrimeScript RT reagent kit
(Takara Biotechnology, Co., Ltd., Dalian, China). RT-qPCR was
performed with the SYBR Premix Ex Taq (Takara Biotechnology, Co.,
Ltd.). PCR primers were as follows: Forward,
5′-GGTGTCTCTCGCAGATTCATC-3′ and reverse,
5′-TCAGTCTTCGGCTGAGGTTCT-3′. Glyceraldehyde-3-phosphate
dehydrogenase (GAPDH) was used as an internal control. The cycling
conditions were as follows: The initial denaturation at 95°C for 5
min, and followed by 40 cycles at 95°C for 15 sec, 55°C for 30 sec
and 72°C for 30 sec. The relative mRNA expression was calculated
using the 2−ΔΔCq method (28).
Western blot analysis
Total protein was isolated using RIPA lysis buffer
(Beyotime Institute of Biotechnology, Shanghai, China) supplemented
with protease inhibitor cocktail (Beyotime Institute of
Biotechnology) on ice, and the concentration of the protein was
determined using a BCA protein assay kit (Beyotime Institute of
Biotechnology) according to the manufacturer's instructions. Equal
amounts of protein lysates (40–60 µg) were loaded per lane and
separated by 8–12% sodium dodecyl sulfate-polyacrylamide gel
electrophoresis (SDS-PAGE), and then electrotransferred onto
nitrocellulose membranes (Millipore, Bedford, MA, USA). The
membranes were blocked with 5% skimmed milk for 2 h at room
temperature and then incubated with the following primary
antibodies overnight at 4°C: Anti-PARP monoclonal antibody (1:1,000
dilution; cat. no. ab32561; Abcam, Cambridge, UK), anti-p62
monoclonal antibody (1:1,000 dilution; cat. no. ab207305; Abcam),
anti-PCNA monoclonal antibody (1:500 dilution; cat. no. 13110; Cell
Signaling Technology, Inc., Inc., Danvers MA, USA), anti-Atg5
monoclonal antibody (1:1,000 dilution; cat. no. 9980; Cell
Signaling Technology, Inc., Inc.), anti-Bcl-2 monoclonal antibody
(1:1,000 dilution; cat. no. 4223; Cell Signaling Technology, Inc.),
anti-Bax monoclonal antibody (1:1,000 dilution; cat. no. 5023; Cell
Signaling Technology, Inc.), anti-Beclin1 monoclonal antibody
(1:1,000 dilution; cat. no. ab210498; Abcam), anti-β-actin
monoclonal antibody (1:1,000 dilution; cat. no. 4970; Cell
Signaling Technology, Inc.), anti-caspase-3 monoclonal antibody
(1:1,000 dilution; cat. no. 9665; Cell Signaling Technology, Inc.),
anti-cleaved-caspase-3 monoclonal antibody (1:500 dilution; cat.
no. 9664; Cell Signaling Technology, Inc.), anti-caspase-9
monoclonal antibody (1:500 dilution; cat. no. ab32539; Abcam),
anti-LC3 monoclonal antibody (1:1,000 dilution; cat. no. 12741;
Cell Signaling Technology, Inc.), anti-Akt monoclonal antibody
(1:1,000 dilution; cat. no. ab179463; Abcam) and anti-phospho-Akt
(Ser-473) monoclonal antibody (1:1,000 dilution; cat. no. ab81283;
Abcam). After being washed with TBST buffer (Tris-buffered saline
supplemented with Tween-20) 3 times (10 min for each time), the
membranes were incubated with goat anti-rabbit horseradish
peroxidase-conjugated secondary antibody (1:5,000 dilution; cat.
no. BA1055; Wuhan Boster Biological Technology, Ltd.) for 1 h at
room temperature. After another 3 washes, the proteins that had
bound with the antibodies were visualized with an enhanced
electrochemiluminescence (ECL) system (GeneTools software version
4.03.05.0; Synoptics Ltd., Cambridge, UK).
Cell viability assay
The viability of the cells was detected using the
Cell Counting Kit-8 (CCK-8) assay. Cells were seeded in 96-well
plates (104 cells/well) and allowed to adhere overnight
before the assay. At 24, 48 and 72 h, 10 µl of CCK-8 reagent
(Nanjing KeyGen Biotech, Co., Ltd., Nanjing, China) was added to
each well and incubated for 4 h. The optical density (OD) values
were measured at a wavelength of 450 nm with a microplate reader
(Thermo Fisher Scientific, Inc.).
EdU cell proliferation assay
The proliferation of cells was detected using EdU
cell proliferation assay according to the manufacturer's
instructions. Cells were seeded in 24-well plates and maintained
for 48 h before the assay. A total of 500 µl EdU (10 µM) reagent
(Biotime, Shanghai, China) was added to each well and incubated for
4 h to label the cells. After 3 times washing with PBS, cells were
fixed in a 4% paraformaldehyde solution for 15 min, permeabilized
with 0.3% Triton X-100 for another 15 min, and then incubated with
the click-reaction reagent for 30 min at room temperature in the
dark. DAPI reagent was used to counterstain the nucleus. The result
of staining was observed with a fluorescence inversion microscope
system (Nikon ECLIPSE Ti-S; Nikon, Tokyo, Japan).
Cell apoptosis assay
An Annexin V-PE/7-AAD apoptosis detection kit
(Nanjing KeyGen Biotech) was used according to the manufacturer's
instructions to measure the apoptotic rate of cells. Cells
(106 cells/well) were seeded into 6-well plates and
allowed to adhere overnight. After being cultured for 48 h, cells
(including the floating ones) were harvested and washed twice with
PBS, and then resuspended with 400 µl Annexin V binding buffer at a
density of 4×105 cells/ml. Subsequently, the cells were
incubated with phycoerythrin (PE) labelled Annexin V for 15 min at
room temperature in the dark, and then with 7-amino-actinomycin D
(7-AAD) for another 5 min on ice in the dark. Flow cytometry (Guava
easyCyte HT; EMD Millipore, Temecula, CA, USA) was applied to
distinguish early apoptotic, late apoptotic, necrotic and viable
cells.
Plate clone formation assay
Cells were seeded and maintained in 6-well plates at
a density of 200 cells/well. A total of 10 ml of medium was added
into each well to supply sufficient nutrition for clone formation.
After 3 weeks, cells were fixed in a 4% paraformaldehyde solution
for 30 min, and then stained with Giemsa dye. The quantity and
diameter of the clones were considered as the evaluating indicators
of the proliferation ability of the cells (Nikon d610 camera;
Nikon).
Transmission electron microscopy
Cells were fixed with ice-cold 2% glutaraldehyde and
then 1% osmium tetroxide. After 2 washes with PBS and dehydration
with gradient ethanol (30–100%), the cells were embedded in
propylene oxide/embedding resin (1:1). The resin blocks were cut
into ultra-thin (60 nm) sections with a LKB-V ultramicrotome (LKB,
Broma, Sweden). The sections were fixed on 200 mesh copper standard
grids and stained with uranyl acetate and lead citrate. The cell
ultrastructure was observed through an H-7650 transmission electron
microscope (Hitachi, Ibaraki, Japan).
RNAi experiment
Small interfering RNA (siRNA) targeting Atg5 and
control siRNA were synthesized by Shangha GenePharma Co., Ltd.
(Shanghai, China). The siRNA sequences were as follows: 1#siRNA,
5′-GACGUUGGUAACUGACAAATT-3′; 2#siRNA, 5′-GUCCAUCUAAGGAUGCAAUTT-3′;
3#siRNA, 5′-GACCUUUCAUUCAGAAGCUTT-3′; and control siRNA,
5′-TTCTCCGAACGTGTCACGTTT-3′.
SW982 cells were seeded in 6-well plates at a
density of 5×105 cells/well and allowed to adhere for 24
h. Then, the cells were transfected with Atg5 siRNA or control
siRNA. X-tremeGENE siRNA Transfection reagent (Roche Diagnostics,
Mannheim, Germany) was used to improve the transfection efficiency,
according to the manufacturer's protocol. The suppressing
efficiency on Atg5 was determined by RT-qPCR and western blot
analysis.
Statistical analysis
Statistical analysis was performed using the SPSS
software version 18.0 (SPSS, Inc., Chicago, IL, USA). Data are
presented as the means ± standard deviation (SD). All data were
analyzed with one-way ANOVA tests followed by Tukey's correction
for multiple comparisons. All statistical tests were two-sided, and
P<0.05 was considered to indicate a statistically significant
result.
Results
A stable Beclin1-overexpressing cell
line is established
We established the Beclin1-overexpressing SW982
cells and selected the stable cells. We divided the SW982 cells
into 3 groups: Control, NC (transfected with the negative control
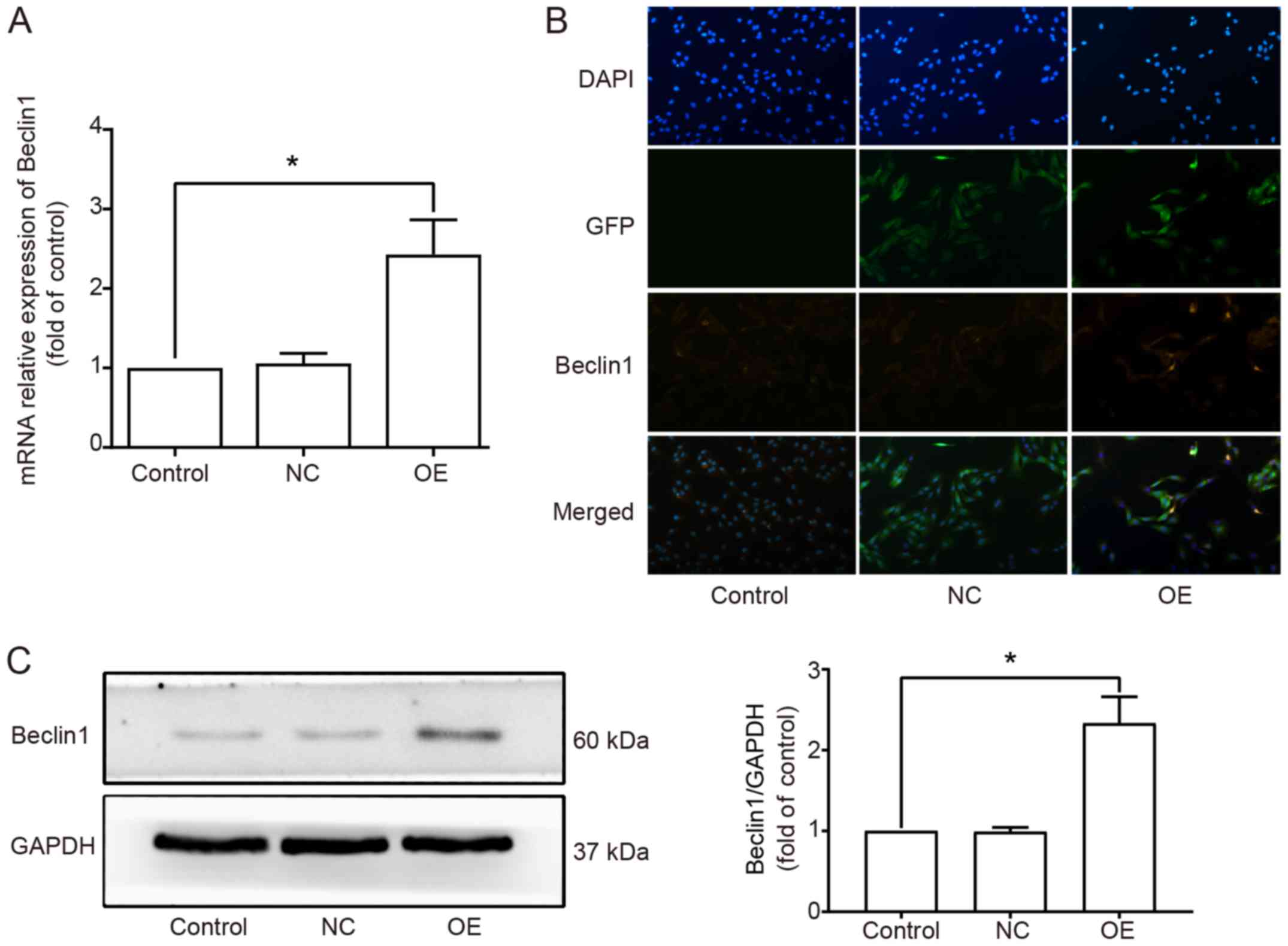
lentivirus) and OE (Beclin1-overexpressing) groups. The expression
level of Beclin1 in each group was detected by RT-qPCR (Fig. 1A), immunofluorescence staining
(Fig. 1B) and western blot analysis
(Fig. 1C). The results indicated
that the expression level of Beclin1 was significantly increased in
the OE group compared to that noted in the Control and NC groups.
No significant difference was observed between the Control and NC
groups.
Beclin1 overexpression inhibits cell
viability in SW982 cells
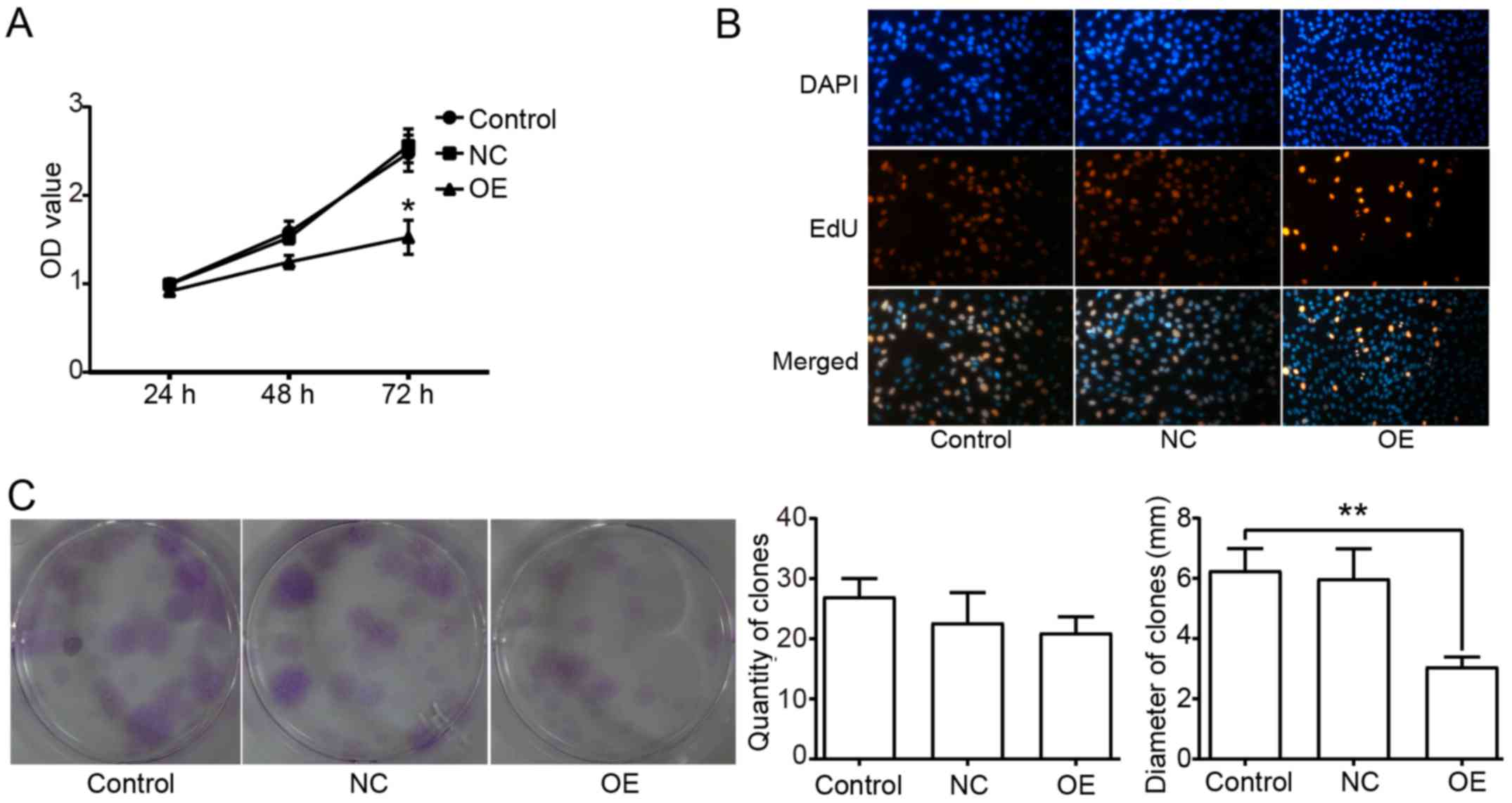
The antitumor effect of Beclin1 has been confirmed
in many tumor cells. To explore the function of Beclin1 in the
viability of SW982 cells, a CCK-8 assay was used to detect the cell
viability, an EdU cell proliferation assay was used to detect the
synthesis of DNA, and a plate clone formation assay was performed
to detect the proliferation ability. The result of the CCK-8 assay
showed that the viability of SW982 cells in the OE group was
significantly inhibited (Fig. 2A).
The EdU-positive rate was much lower in the OE group compared to
the other groups (Fig. 2B).
Meanwhile, the clones in the OE group had a significantly smaller
mean diameter than those in the other two groups. However, there
was no significant difference in the quantity of clones among the 3
groups (Fig. 2C).
Beclin1 overexpression induces
apoptosis in the SW982 cells
To determine the apoptotic rate, flow cytometry
(FCM) was performed, and the result showed that the apoptotic rate
in the OE group was much higher than the rate in the other two
groups (Fig. 3A). For further
confirmation, western blot assays were performed to detect the
expression levels of proteins such as PCNA, cleaved-PARP,
cleaved-caspase-3, Bcl-2 and Bax. The result showed that the
expression levels of PCNA and Bcl-2 were decreased, while the
cleaved-PARP and cleaved-caspase-3 were increased in the OE group.
However, there was no difference in the expression level of the
pro-apoptotic protein Bax among the 3 groups (Fig. 3B).
Beclin1 overexpression increases the
autophagic activity in SW982 cells
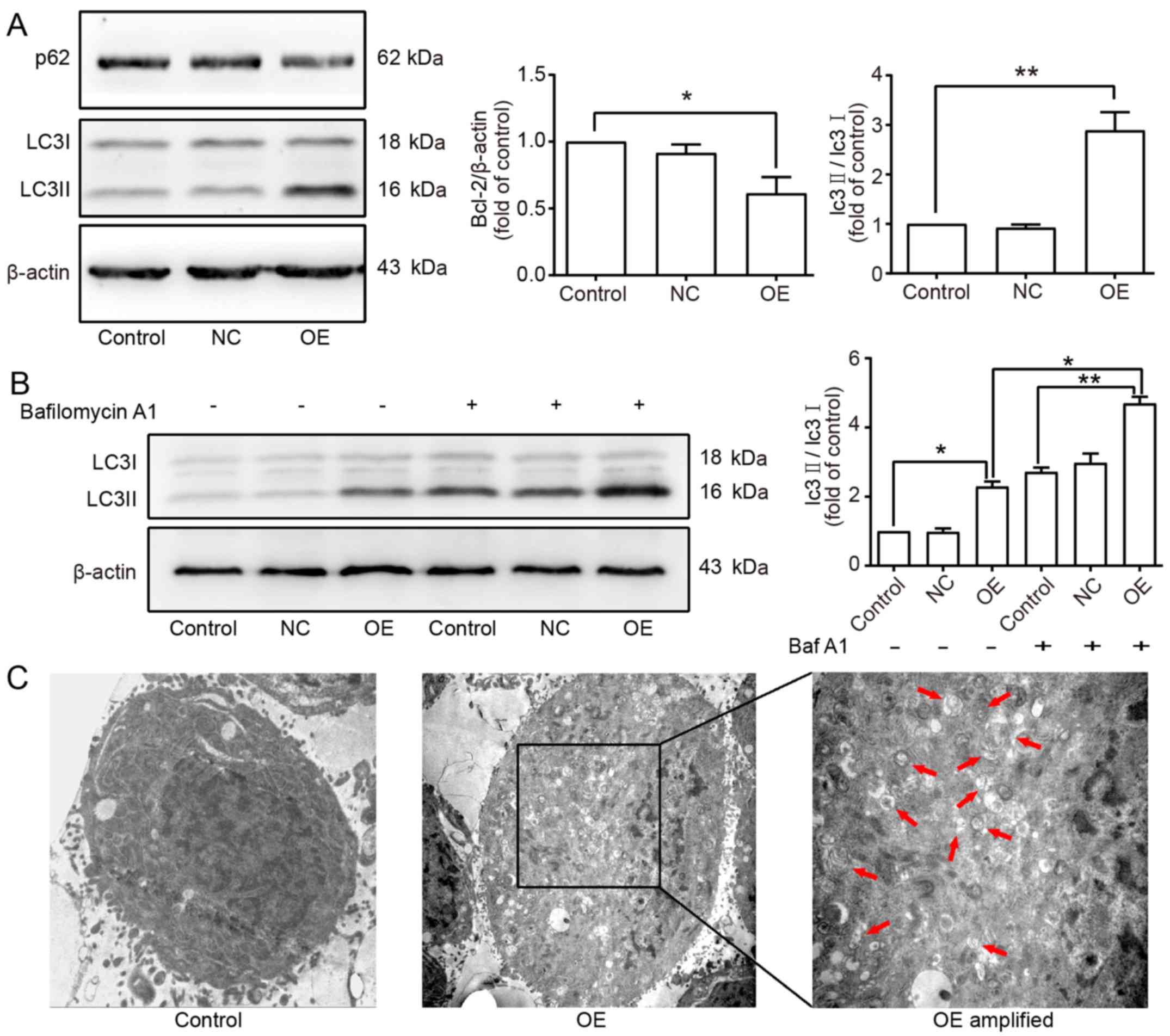
It is commonly known that Beclin1 is a vital gene
involved in the autophagy process. To explore the effect of Beclin1
overexpression on the autophagy in SW982 cells, the
autophagy-related markers LC3 and p62 were detected by western blot
assay. The results showed that the expression level of LC3II was
increased and that of p62 was decreased in the OE group (Fig. 4A). To confirm that Beclin1
overexpression could increase the autophagic flux, Bafilomycin A1
(Baf A1) which is a specific autophagic inhibitor was applied to
inhibit the degradation of autophagy-lysosomes. The results showed
that the expression level of LC3II was further increased in the
presence of Baf A1, which indicated that Beclin1 overexpression
enhanced the autophagic activity in SW982 cells (Fig. 4B). Moreover, the formation of
autophagosomes was observed by transmission electron microscopy,
and more double membrane-enclosed autophagic vesicles containing
engulfed organelles were found in the SW982 cells of the OE group
compared to the Control group (Fig.
4C).
Autophagy contributes to the reduction
in cell viability in SW982 cells
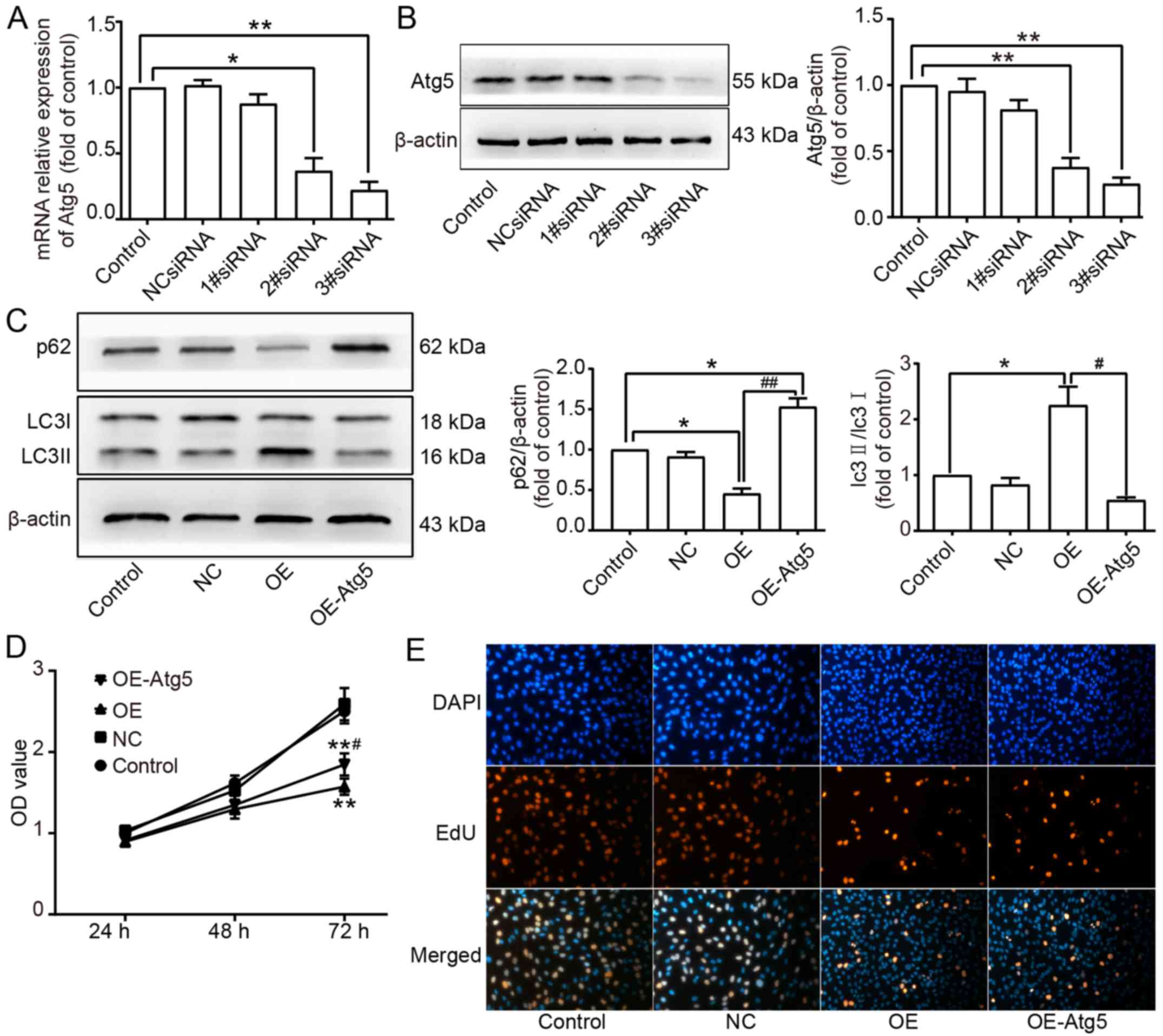
To investigate the relationship between apoptosis
and autophagy, both induced by Beclin1-overexpression, small
interfering RNA (siRNA) targeting Atg5 was used to inhibit the
expression of Atg5 to suppress the autophagic activity. The
efficiency of inhibition was determined by RT-qPCR and western blot
assay. The results showed that 2#siRNA and 3#siRNA effectively
suppressed the expression level of Atg5 (Fig. 5A and B). Therefore, 3#siRNA which
had the highest inhibition efficiency was used to knockdown Atg5 in
SW982 cells of the OE group, and the Atg5-knockdown cells in the OE
group were isolated as the OE-Atg5 group. Western blot assay showed
that the expression level of LC3II was decreased, but the
expression level of p62 was increased in the OE-Atg5 group
(Fig. 5C). A CCK-8 assay was
performed to detect the cell viability in each group, and the
result demonstrated that the viability in the OE-Atg5 group was
significantly increased compared to the OE group but lower than the
viability of the Control and NC groups (Fig. 5D). EdU assay showed that there was
no difference in the EdU-positive rate between the OE and OE-Atg5
groups (Fig. 5E).
Autophagy promotes the apoptosis
induced by Beclin1 overexpression through the Akt/Bcl-2/caspase-9
signaling pathway
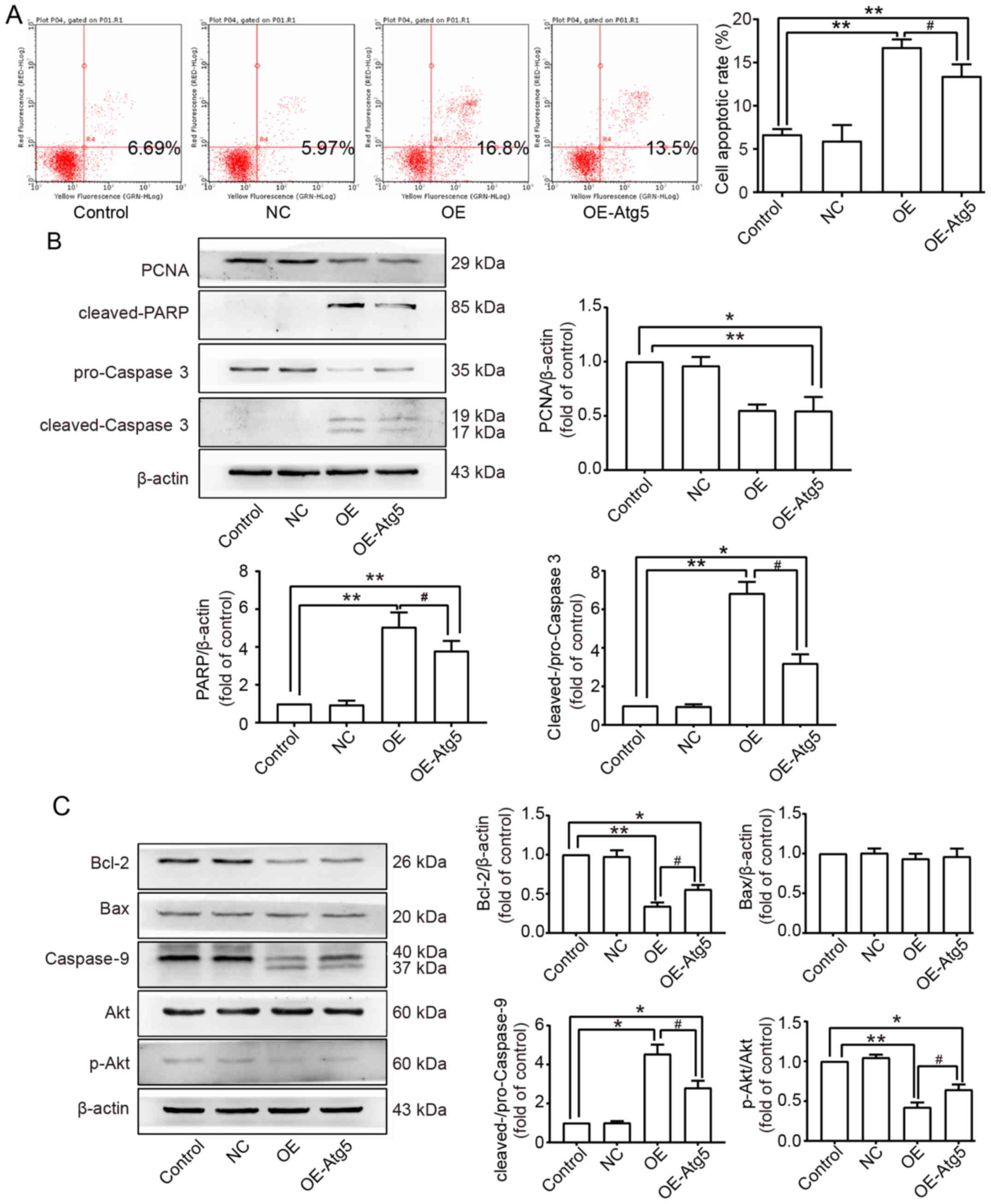
Since the relationship between apoptosis and
autophagy remains unclear, FCM was performed to determine the
change in the apoptotic rate when autophagy was inhibited. The
result showed that inhibition of autophagy suppressed apoptosis
induced by Beclin1 overexpression to some extent, but the effect
was not absolutely reversed (Fig.
6A). This conclusion was confirmed by western blot assay; the
expression levels of cleaved-PARP and cleaved-caspase-3 were both
decreased in the OE-Atg5 group compared to the OE group, but no
difference was found in the expression of PCNA between the OE and
OE-Atg5 groups (Fig. 6B). Akt is an
important gene involved in both apoptosis and autophagy through
various signaling pathways; we further studied whether this pathway
participated in the pathophysiological process mentioned above in
SW982 cells. Western blot assays were performed, and related
proteins were detected; the expression level of Bcl-2 was increased
in the OE-Atg5 group compared to the OE group; no difference was
found in the expression level of Bax among all groups. Activation
of caspase-9 was enhanced in the OE group, and Atg5-knockdown
inhibited this activation to a certain degree. Phosphorylation of
Akt was inhibited in the OE group, and Atg5 knockdown reversed this
inhibition (Fig. 6C).
Discussion
The antitumor effect of Beclin1 has been confirmed
in many previous studies, and Beclin1 gene deletion has been found
in ovarian (29–31), breast (32,33),
prostate (34,35) and other tumors (36–39).
However, the underlying mechanism of this phenomenon remains
unclear, which has caused a lot of controversy.
In the present study, we demonstrated that Beclin1
overexpression inhibited cell viability and induced apoptosis.
Beclin1 has a BH3 domain that binds to the anti-apoptotic protein
Bcl-2 to undermine the anti-apoptotic ability of tumor cells. Bcl-2
is mainly expressed in proliferating and differentiating cells.
However, high expression of Bcl-2 inhibits the apoptosis of tumor
cells, which is closely associated with tumorigenesis and tumor
progress (40). PCNA is closely
related to cellular DNA synthesis. The expression level of PCNA can
be regarded as an indicator of cell proliferation status. FCM assay
demonstrated that Beclin1 overexpression induced apoptosis in SW982
cells. Western blot assay showed that Beclin1 overexpression
decreased the expression levels of Bcl-2 and PCNA, and promoted the
activation of caspase-3 and PARP, but the expression level of Bax
did not significantly change. We assumed that the reason for the
decreased viability was the imbalance of Bcl-2/Bax, mainly because
of the downregulation of Bcl-2. In addition, similar findings have
been reported in previous studies (41–44).
The result of the plate clone formation assay showed that Beclin1
overexpression inhibited cell proliferation. Notably, the OE group
had a smaller mean diameter of clones due to the inhibition of
proliferation, but there was no significant difference in the
quantity of clones among the different groups. This result implied
that Beclin1 did not simply kill the tumor cells, but functioned
subtly to regulate cell proliferation and death within an
appropriate range.
Under normal conditions, autophagy protects cells
against pro-apoptotic factors (45,46).
However, autophagy can also play a role similar to apoptosis,
leading to autophagic cell death in some cases. This biological
process was named type II programmed cell death to distinguish it
from apoptosis (47). In recent
years, more and more cases of autophagic death have been reported,
especially in tumors (24). Yu
et al (48) found that Zinc
oxide nanoparticles induced autophagic cell death and mitochondrial
damage via the generation of reactive oxygen species. Li et
al (49) found that plumbagin
induced apoptotic and autophagic cell death through inhibition of
the PI3K/Akt/mTOR pathway in human non-small cell lung cancer
cells. Beclin1, as an important gene of autophagy, is related to
both apoptosis and autophagy. Mcl-1-dependent activation of Beclin1
mediated autophagic cell death induced by sorafenib and SC-59 in
hepatocellular carcinoma cells (50). Zoledronate induced autophagic cell
death in human umbilical vein endothelial cells via Beclin1
dependent pathway activation (25).
In this study, Beclin1 overexpression enhanced
autophagic activity, and when the autophagic activity was inhibited
by Atg5-knockdown, the cell viability was significantly increased.
We assumed that Beclin1 overexpression induced autophagic death in
SW982 cells. However, because the inhibition of autophagic activity
did not absolutely reverse the cell death induced by Beclin1
overexpression, there may be an autophagy-independent pathway that
led to cell death. The inhibition of autophagy did not change the
expression level of the PCNA, which implied that Beclin1 could
suppress the proliferation of SW982 in an autophagy-independent
way. In addition, EdU assays confirmed this result mentioned above.
Additionally, FCM assay and western blot analysis of the
apoptosis-related proteins caspase-3 and PARP showed that the
inhibition of autophagy also suppressed the apoptotic activity. It
was reported that autophagy could result in cell death alone
(51,52), and autophagy could also induce cell
death via activating apoptosis (53,54).
Moreover, we also found that Akt/Bcl-2/caspase-9 pathway was
involved in the cell death induced by Beclin1 overexpression, and
it is known that Akt affects both apoptosis and autophagy (55).
Autophagy is a double-edged sword for cell fate, and
its definite mechanism remains unclear. The role of Beclin1 in the
crosstalk between autophagy and apoptosis needs further
exploration. Altogether, the present study revealed the critical
antitumor effect of Beclin1 in the SW982 synovial sarcoma cells,
suggesting that Beclin1 might be an important target for synovial
sarcoma therapy. SW982 is one kind of synovial sarcoma cell lines,
which is most commonly used in studies because of its stable
biological characteristics. All of the results in this study were
observed in SW982 synovial sarcoma cells. Different cell lines of
synovial sarcoma vary in the biological characteristics, it would
be better to use more than 2 different cell lines to make the
conclusion. However, SW982 is the only cell line that can be
obtained from the ATCC bioresource center, and it is also the only
synovial sarcoma cell line exists in China over the course of this
study. We will use other synovial sarcoma cell lines or primary
synovial sarcoma cells to verified our present result in further
studies.
Acknowledgements
We wish to thank Dr Liang Bai for the technical
advice.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81271948, 81601877
and 81171742).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
JZ, PX, SL and JC conceived, designed and performed
the experiments. YC and KX analyzed the data. XR and JS performed
the electron microscopy analysis. JZ wrote the manuscript. All
authors read and approved the manuscript and agree to be
accountable for all aspects of the research in ensuring that the
accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Yaser S, Salah S, Al-Shatti M, Abu-Sheikha
A, Shehadeh A, Sultan I, Salem A, Sughayer M, Al-Loh S and Al-Mousa
A: Prognostic factors that govern localized synovial sarcoma: A
single institution retrospective study on 51 patients. Med Oncol.
31:9582014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
El Beaino M, Araujo DM, Gopalakrishnan V,
Lazar AJ and Lin PP: Prognosis of T1 synovial sarcoma depends upon
surgery by oncologic surgeons. J Surg Oncol. 114:490–494. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Levine B and Klionsky DJ: Development by
self-digestion: Molecular mechanisms and biological functions of
autophagy. Dev Cell. 6:463–477. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wen YF, Zand B, Ozpolat B, Szczepanski MJ,
Lu CH, Yuca E, Carroll AR, Alpay N, Bartholomeusz C, Tekedereli I,
et al: Antagonism of tumoral prolactin receptor promotes
autophagy-related cell death. Cell Rep. 7:488–500. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Galluzzi L, Pietrocola F, Pedro Bravo-San
JM, Amaravadi RK, Baehrecke EH, Cecconi F, Codogno P, Debnath J,
Gewirtz DA, Karantza V, et al: Autophagy in malignant
transformation and cancer progression. EMBO J. 34:856–880. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liang XH, Kleeman LK, Jiang HH, Gordon G,
Goldman JE, Berry G, Herman B and Levine B: Protection against
fatal Sindbis virus encephalitis by Beclin, a novel
Bcl-2-interacting protein. J Virol. 72:8586–8596. 1998.PubMed/NCBI
|
|
7
|
Oberstein A, Jeffrey PD and Shi YG:
Crystal structure of the Bcl-XL-Beclin 1 peptide complex: Beclin 1
is a novel BH3-only protein. J Biol Chem. 282:13123–13132. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Park JM, Tougeron D, Huang S, Okamoto K
and Sinicrope FA: Beclin 1 and UVRAG confer protection from
radiation-induced DNA damage and maintain centrosome stability in
colorectal cancer cells. PLoS One. 9:e1008192014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang W, Fan H, Li X, Wu G, Zhao W, Zhang
G, Zhao G and Li L: Beclin 1 promotes apoptosis and decreases
invasion by upregulating the expression of ECRG4 in A549 human lung
adenocarcinoma cells. Mol Med Report. 14:355–360. 2016.
|
|
10
|
De Amicis F, Aquila S, Morelli C, Guido C,
Santoro M, Perrotta I, Mauro L, Giordano F, Nigro A, Andò S, et al:
Bergapten drives autophagy through the up-regulation of PTEN
expression in breast cancer cells. Mol Cancer. 14:1302015.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jung YY, Lee YK and Koo JS: The potential
of Beclin 1 as a therapeutic target for the treatment of breast
cancer. Expert Opin Ther Targets. 20:167–178. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wu S, Sun C, Tian D, Li Y, Gao X, He S and
Li T: Expression and clinical significances of Beclin1, LC3 and
mTOR in colorectal cancer. Int J Clin Exp Pathol. 8:3882–3891.
2015.PubMed/NCBI
|
|
13
|
Chen Z, Li Y, Zhang C, Yi H, Wu C, Wang J,
Liu Y, Tan J and Wen J: Downregulation of Beclin 1 and impairment
of autophagy in a small population of colorectal cancer. Dig Dis
Sci. 58:2887–2894. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hu YF, Lei X, Zhang HY, Ma JW, Yang WW,
Chen ML, Cui J and Zhao H: Expressions and clinical significance of
autophagy-related markers Beclin1, LC3, and EGFR in human cervical
squamous cell carcinoma. Onco Targets Ther. 8:2243–2249.
2015.PubMed/NCBI
|
|
15
|
Chen Z, Wang B, Yu F, Chen Q, Tian Y, Ma S
and Liu X: The roles of mitochondria in radiation-induced
autophagic cell death in cervical cancer cells. Tumour Biol.
37:4083–4091. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ju LL, Zhao CY, Ye KF, Yang H and Zhang J:
Expression and clinical implication of Beclin1, HMGB1, p62,
survivin, BRCA1 and ERCC1 in epithelial ovarian tumor tissues. Eur
Rev Med Pharmacol Sci. 20:1993–2003. 2016.PubMed/NCBI
|
|
17
|
Cai M, Hu Z, Liu J, Gao J, Liu C, Liu D,
Tan M, Zhang D and Lin B: Beclin 1 expression in ovarian tissues
and its effects on ovarian cancer prognosis. Int J Mol Sci.
15:5292–5303. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xue S, Xiao-Hong S, Lin S, Jie B, Li-Li W,
Jia-Yao G, Shun S, Pei-Nan L, Mo-Li W, Qian W, et al: Lumbar
puncture-administered resveratrol inhibits STAT3 activation,
enhancing autophagy and apoptosis in orthotopic rat glioblastomas.
Oncotarget. 7:75790–75799. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Huang X, Qi Q, Hua X, Li X, Zhang W, Sun
H, Li S, Wang X and Li B: Beclin 1, an autophagy-related gene,
augments apoptosis in U87 glioblastoma cells. Oncol Rep.
31:1761–1767. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bai H, Li H, Li W, Gui T, Yang J, Cao D
and Shen K: The PI3K/AKT/mTOR pathway is a potential predictor of
distinct invasive and migratory capacities in human ovarian cancer
cell lines. Oncotarget. 6:25520–25532. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Katagiri H, Nakayama K, Razia S, Nakamura
K, Sato E, Ishibashi T, Ishikawa M, Iida K, Ishikawa N, Otsuki Y,
et al: Loss of autophagy-related protein Beclin 1 may define poor
prognosis in ovarian clear cell carcinomas. Int J Oncol.
47:2037–2044. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Garbar C, Mascaux C, Giustiniani J,
Salesse S, Debelle L, Antonicelli F, Merrouche Y and Bensussan A:
Autophagy is decreased in triple-negative breast carcinoma
involving likely the MUC1-EGFR-NEU1 signalling pathway. Int J Clin
Exp Pathol. 8:4344–4355. 2015.PubMed/NCBI
|
|
23
|
Rohatgi RA, Janusis J, Leonard D, Bellvé
KD, Fogarty KE, Baehrecke EH, Corvera S and Shaw LM: Beclin 1
regulates growth factor receptor signaling in breast cancer.
Oncogene. 34:5352–5362. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shimizu S, Yoshida T, Tsujioka M and
Arakawa S: Autophagic cell death and cancer. Int J Mol Sci.
15:3145–3153. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lu Y, Wang Z, Han W and Li H: Zoledronate
induces autophagic cell death in human umbilical vein endothelial
cells via Beclin-1 dependent pathway activation. Mol Med Report.
14:4747–4754. 2016. View Article : Google Scholar
|
|
26
|
Elgendy M, Ciro M, Abdel-Aziz AK, Belmonte
G, Dal Zuffo R, Mercurio C, Miracco C, Lanfrancone L, Foiani M and
Minucci S: Beclin 1 restrains tumorigenesis through Mcl-1
destabilization in an autophagy-independent reciprocal manner. Nat
Commun. 5:56372014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Rohatgi RA and Shaw LM: An
autophagy-independent function of Beclin 1 in cancer. Mol Cell
Oncol. 3:pii: e1030539. 2016.PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Correa RJ, Valdes YR, Shepherd TG and
DiMattia GE: Beclin-1 expression is retained in high-grade serous
ovarian cancer yet is not essential for autophagy induction in
vitro. J Ovarian Res. 8:522015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang YF, Xu YL, Tang ZH, Li T, Zhang LL,
Chen X, Lu JH, Leung CH, Ma DL, Qiang WA, et al: Baicalein induces
Beclin 1- and extracellular signal-regulated kinase-dependent
autophagy in ovarian cancer cells. Am J Chin Med. 45:123–136. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ying H, Qu D, Liu C, Ying T, Lv J, Jin S
and Xu H: Chemoresistance is associated with Beclin-1 and PTEN
expression in epithelial ovarian cancers. Oncol Lett. 9:1759–1763.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li WL, Xiong LX, Shi XY, Xiao L, Qi GY and
Meng C: IKKβ/NFκBp65 activated by interleukin-13 targets the
autophagy-related genes LC3B and beclin 1 in fibroblasts
co-cultured with breast cancer cells. Exp Ther Med. 11:1259–1264.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
De Amicis F, Guido C, Santoro M, Giordano
F, Donà A, Rizza P, Pellegrino M, Perrotta I, Bonofiglio D, Sisci
D, et al: Ligand activated progesterone receptor B drives
autophagy-senescence transition through a Beclin-1/Bcl-2 dependent
mechanism in human breast cancer cells. Oncotarget. 7:57955–57969.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Baspinar S, Bircan S, Orhan H, Kapucuoglu
N and Bozkurt KK: The relation of beclin 1 and bcl-2 expressions in
high grade prostatic intraepithelial neoplasia and prostate
adenocarcinoma: A tissue microarray study. Pathol Res Pract.
210:412–418. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lian J, Wu X, He F, Karnak D, Tang W, Meng
Y, Xiang D, Ji M, Lawrence TS and Xu L: A natural BH3 mimetic
induces autophagy in apoptosis-resistant prostate cancer via
modulating Bcl-2-Beclin1 interaction at endoplasmic reticulum. Cell
Death Differ. 18:60–71. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jung G, Roh J, Lee H, Gil M, Yoon DH, Suh
C, Jang S, Park CJ, Huh J and Park CS: Autophagic markers BECLIN 1
and LC3 are associated with prognosis of multiple myeloma. Acta
Haematol. 134:17–24. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Fukui M, Yamabe N, Choi HJ, Polireddy K,
Chen Q and Zhu BT: Mechanism of ascorbate-induced cell death in
human pancreatic cancer cells: Role of Bcl-2, Beclin 1 and
autophagy. Planta Med. 81:838–846. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Al-Shenawy HA: Expression of Beclin-1, an
autophagy-related marker, in chronic hepatitis and hepatocellular
carcinoma and its relation with apoptotic markers. APMIS.
124:229–237. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ma K, Zhang C, Huang MY, Guo YX and Hu GQ:
Crosstalk between Beclin-1-dependent autophagy and
caspase-dependent apoptosis induced by tanshinone IIA in human
osteosarcoma MG-63 cells. Oncol Rep. 36:1807–1818. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sun C, Liu Z, Li S, Yang C, Xue R, Xi Y,
Wang L, Wang S, He Q, Huang J, et al: Down-regulation of c-Met and
Bcl2 by microRNA-206, activates apoptosis, and inhibits tumor cell
proliferation, migration and colony formation. Oncotarget.
6:25533–25574. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu LS, Bai XQ, Gao Y, Wu Q, Ren Z, Li Q,
Pan LH, He NY, Peng J and Tang ZH: PCSK9 promotes oxLDL-induced
PC12 cell apoptosis through the Bcl-2/Bax-Caspase 9/3 signaling
pathway. J Alzheimers Dis. 57:723–734. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lončarević-Vasiljković N, Milanović D,
Pešić V, Tešić V, Brkić M, Lazić D, Avramović V and Kanazir S:
Dietary restriction suppresses apoptotic cell death, promotes Bcl-2
and Bcl-xl mRNA expression and increases the Bcl-2/Bax protein
ratio in the rat cortex after cortical injury. Neurochem Int.
96:69–76. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Song S, Jacobson KN, McDermott KM, Reddy
SP, Cress AE, Tang HY, Dudek SM, Black SM, Garcia JG, Makino A, et
al: ATP promotes cell survival via regulation of cytosolic [Ca2+]
and Bcl-2/Bax ratio in lung cancer cells. Am J Physiol Cell
Physiol. 310:C99–C114. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Jiang N, Li Y and Ruan DY: Sa1688
aberrantly regulated dysadherin and Bcl-2/Bax2 enhances
tumorigenesis and DNA targeting drug resistance of liver cancer
stem cells. Gastroenterology. 148:S1012. 2015. View Article : Google Scholar
|
|
45
|
Cai Y, Xu P, Yang L, Xu K, Zhu J, Wu X,
Jiang C, Yuan Q, Wang B, Li Y, et al: HMGB1-mediated autophagy
decreases sensitivity to oxymatrine in SW982 human synovial sarcoma
cells. Sci Rep. 6:378452016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Xu K, Cai YS, Lu SM, Li XL, Liu L, Li Z,
Liu H and Xu P: Autophagy induction contributes to the resistance
to methotrexate treatment in rheumatoid arthritis fibroblast-like
synovial cells through high mobility group box chromosomal protein
1. Arthritis Res Ther. 17:3742015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Liu Y and Levine B: Autosis and autophagic
cell death: The dark side of autophagy. Cell Death Differ.
22:367–376. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Yu KN, Yoon TJ, Minai-Tehrani A, Kim JE,
Park SJ, Jeong MS, Ha SW, Lee JK, Kim JS and Cho MH: Zinc oxide
nanoparticle induced autophagic cell death and mitochondrial damage
via reactive oxygen species generation. Toxicol In Vitro.
27:1187–1195. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Li YC, He SM, He ZX, Li M, Yang Y, Pang
JX, Zhang X, Chow K, Zhou Q, Duan W, et al: Plumbagin induces
apoptotic and autophagic cell death through inhibition of the
PI3K/Akt/mTOR pathway in human non-small cell lung cancer cells.
Cancer Lett. 344:239–259. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Tai WT, Shiau CW, Chen HL, Liu CY, Lin CS,
Cheng AL, Chen PJ and Chen KF: Mcl-1-dependent activation of Beclin
1 mediates autophagic cell death induced by sorafenib and SC-59 in
hepatocellular carcinoma cells. Cell Death Dis. 4:e4852013.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wong VK, Li T, Law BY, Ma ED, Yip NC,
Michelangeli F, Law CK, Zhang MM, Lam KY, Chan PL, et al:
Saikosaponin-d, a novel SERCA inhibitor, induces autophagic cell
death in apoptosis-defective cells. Cell Death Dis. 4:e7202013.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Liu C, Yan X, Wang HQ, Gao YY, Liu J, Hu
Z, Liu D, Gao J and Lin B: Autophagy-independent enhancing effects
of Beclin 1 on cytotoxicity of ovarian cancer cells mediated by
proteasome inhibitors. BMC Cancer. 12:6222012. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Lamy L, Ngo VN, Emre NC, Shaffer AL III,
Yang Y, Tian E, Nair V, Kruhlak M, Zingone A, Landgren O, et al:
Control of autophagic cell death by caspase-10 in multiple myeloma.
Cancer Cell. 23:435–449. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Yu FS, Yu CS, Chen JC, Yang JL, Lu HF,
Chang SJ, Lin MW and Chung JG: Tetrandrine induces apoptosis Via
caspase-8, −9, and −3 and poly (ADP ribose) polymerase dependent
pathways and autophagy through beclin-1/LC3-I, II signaling
pathways in human oral cancer HSC-3 cells. Environ Toxicol.
31:395–406. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zhao ZQ, Yu ZY, Li J and Ouyang XN:
Gefitinib induces lung cancer cell autophagy and apoptosis via
blockade of the PI3K/AKT/mTOR pathway. Oncol Lett. 12:63–68. 2016.
View Article : Google Scholar : PubMed/NCBI
|