Introduction
Hepatoblastoma is the most common malignant liver
tumor in children <5 years old (1,2). The
prognosis of children with hepatoblastoma is favorable if a
complete surgical resection of the tumor is possible; however, for
advanced and unresectable tumors, and for relapsed cases, the
prognosis is much worse (2,3), and surgery combined with chemotherapy
is required for long-term survival (1). The most commonly studied agents in the
treatment of hepatoblastoma include cisplatin (4) and doxorubicin (dox) (5). Dox is commonly used in the treatment
of a wide range of cancers, with the most serious adverse effect
being life-threatening heart damage. Since multidrug resistance is
a common problem encountered in response to chemotherapy for the
treatment of hepatoblastoma (6,7), the
development of novel therapeutic strategies is critical.
The orphan nuclear receptor liver receptor homolog-1
[LRH-1, also known as nuclear receptor subfamily 5 group A member 2
(NR5A2)] is a member of a subfamily of nuclear receptors that binds
to identical DNA consensus sequences (8). LRH-1 is primarily expressed in
secretory tissues or tissues with high rates of protein production,
such as the liver (9), pancreas
(10,11), breast (12) and muscle (13). LRH-1 has prominent roles in
development, metabolism (8), stem
cell pluripotency (14) and
tumorigenesis, including in breast cancer (12), pancreatic cancer (15) and endometrial cancers (16). In the liver, LRH-1 regulates
cholesterol metabolism and bile acid homeostasis (17). Transcriptional targets of LRH-1
include cyclin D1 (CCND1), cyclin E1 (CCNE1) and c-Myc, which are
known to control cell differentiation, growth and proliferation
(15). Inhibition of LRH-1
signaling has been successful in preclinical studies of some cancer
types (12,14,16);
however, the role of LRH-1 in hepatoblastoma remains unclear.
Development of small molecule agonists is a promising area of
research (17,18) and antagonists for LRH-1 may work as
potent anticancer agents (19,20).
The present study assessed the antitumorigenic efficacy of the
recently developed LRH-1 antagonist (LRA),
pyrazolylbiphenylethanone compound
1-(3′-(1-(2-(4-Morpholinyl)ethyl)-1H-pyrazol-3-yl)-3-biphenylyl)
ethanone, which can bind to the LRH-1 ligand binding domain and
block LRH-1 from forming an active conformation (20).
In the present study, the expression levels of LRH-1
were examined in a panel of hepatoblastoma cell lines in
vitro; the mRNA and protein expression levels were upregulated
in HepG2 and Huh6 cells. Specific inhibition of LRH-1 using LRA
inhibited proliferation of these cells through downregulation of
CCND1 and c-Myc, and via induction of cell cycle arrest at
G1 phase. LRA also increased the antitumor effects of
dox in these cells. Overall, the present study supports a role for
LRH-1 in liver cancer and raises the possibility that inhibition of
LRH-1 may be effective in the treatment of hepatoblastoma.
Materials and methods
Cell culture
The hepatoblastoma cell line HepG2 was grown in
Eagle's Minimum Essential Medium (Lonza, Salisbury, MD, USA), HepT1
cells were grown in RPMI 1640 (Lonza), and HuH6 and 293T cells were
grown in Dulbecco's modified Eagle's medium (DMEM; Lonza); all
media were supplemented with 10% heat-inactivated fetal bovine
serum (FBS, SAFC Biosciences, Inc., Lenexa, KS, USA), 2 mM
L-glutamine (Thermo Fisher Scientific, Inc., Waltham, MA, USA) and
100 U/ml penicillin G/streptomycin (Thermo Fisher Scientific,
Inc.). THLE-2 cells [American Type Culture Collection (ATCC),
Manassas, VA, USA] were grown in Bronchial Epithelial Cell Growth
Medium (Lonza) supplemented with 10% FBS, 1%
penicillin-streptomycin and 50 µg ml−1 gentamycin
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) in
PureCol/fibronectin-coated T-75 flasks. HepG2, THLE-2 and 293T
cells were purchased from ATCC; HuH6 cells were purchased from
Riken BioResource Center (Tsukaba, Japan). HepT1 cells were a
generous gift from Dr Stefano Cairo (Department of Morphology,
Surgery and Experimental Medicine, University of Ferrara, Ferrara,
Italy). All cells were incubated at 37°C in a humidified atmosphere
containing 5% CO2 and 95% O2.
Establishment of stable short hairpin
(sh)RNA-mediated LRH-1 knockdown hepatoblastoma cell lines
shRNA-induced knockdown of LRH-1 expression was
achieved using the lentiviral expression system from GE Healthcare
Dharmacon, Inc. (Lafayette, CO, USA). The shLRH-1/shNR5A2
constructs used in the present study were as follows: #1,
V2LHS_17029; #2, V2LHS_17033; #3, RHS4 430-98486912 (GE Healthcare
Dharmacon, Inc.). The shCCND1 construct used was RHS4531-EG595 (GE
Healthcare Dharmacon, Inc.). The shc-Myc construct used was
RHS4531-EG4609 GE Healthcare Dharmacon, Inc.). The control vector
used was RHS4346 (GE Healthcare Dharmacon, Inc.). Viral particles
were generated by co-transfecting 293T cells (ATCC) with the shRNAs
and the Lenti-vpak packaging kit, which contains packaging plasmids
and a transfection reagent (cat. no. TR30037; OriGene Technologies,
Inc., Rockville, MD, USA) according to the manufacturer's protocol.
Subsequently, the shRNA viral particles were transduced into HepG2
and HuH6 cells with 8 µg/ml hexadimethrine bromide (Polybrene; cat.
no. H9268; Sigma-Aldrich; Merck KGaA), and stable cell lines were
established after 10 days of puromycin (2 µg/ml) selection.
Knockdown was confirmed using quantitative polymerase chain
reaction (qPCR) or immunoblotting. The selected cell lines were
routinely cultured in puromycin-containing media until 2 days prior
to experimentation.
RNA extraction and reverse
transcription (RT)-qPCR
The Direct-zol RNA miniprep kit (Zymo Research
Corp., Irvine, CA, USA) was used to extract total RNA, according to
the manufacturer's protocol. RT was conducted according to
manufacturer's protocol, briefly, using random hexamer primers and
the Transcriptor First Strand cDNA Synthesis kit (Roche
Diagnostics, Indianapolis, IN, USA), and the resultant cDNA was
subjected to qPCR analysis using TaqMan Universal PCR Master Mix
(Applied Biosystems; Thermo Fisher Scientific, Inc.). Thermal
cycling conditions were performed as follows: Pre-denaturation at
95°C for 5 min, followed by 40 cycles of denaturation at 95°C for
10 sec, annealing at 60°C for 30 sec and extension at 72°C for 30
sec after which, a melting curve analysis was conducted. TaqMan
assay mixtures targeting LRH-1/NR5A2 (Hs00187067_m1), GAPDH
(Hs02758991), CCNE1 (Hs01026536_m1), CCND1 (Hs00765553_m1) and MYC
(Hs00905030_m1) (Applied Biosystems; Thermo Fisher Scientific,
Inc.) were used for the detection of mRNA expression. Amplification
and quantification were performed using the PRISM 7000 Real-Time
PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.).
The mRNA expression levels of LRH-1, CCND1, CCNE1 and MYC were
normalized to the mRNA levels of GAPDH, which was used as an
internal control. The 2−ΔΔCq method (21) was used to quantify the mRNA
expression levels. Data were analyzed by one-way analysis of
variance and a Dunnett's multiple comparison post hoc test, or
Student's t-test.
Antibodies and reagents
Antibodies against CCND1 (cat. no. sc-8396), CCNE1
(cat. no. sc-247), c-Myc (cat. no. sc-764) and LRH-1 (cat. no.
sc-25389) were obtained from Santa Cruz Biotechnology, Inc.
(Dallas, TX, USA). Anti-mouse (cat. no. 7076S) and anti-rabbit
(cat. no. 7074S) immunoglobulin G secondary antibodies were
obtained from Cell Signaling Technology, Inc. (Danvers, MA, USA).
Anti-LRH-1/NR5A2 (cat. no. PA5-28347) was obtained from Thermo
Fisher Scientific, Inc. Dox (cat. no. D1515) and anti-β-actin (cat.
no. A2228) antibody were obtained from Sigma-Aldrich; Merck KGaA.
LRA (cat. no. 505601) was purchased from Calbiochem; EMD Millipore
(Billerica, MA, USA).
Protein isolation and western
blotting
After each treatment, cells were harvested in
ice-cold PBS (pH 7.4) and spun down. The pellets were dissolved in
lysis buffer [50 mM Tris-HCl, pH 7.4; 150 mM NaCl; 1 mM EDTA; 1%
IGEPAL; 0.25% Na-deoycholate; 1 mM phenylmethyl-sulfonyl fluoride;
1 mM dithiothreitol; 10 mg/ml aprotinin; 10 mg/ml leupeptin; 1 mM
benzamidine; 20 mM disodium p-nitrophenylphosphate; 0.1 mM sodium
orthovanadate; 10 mM sodium fluoride; phosphatase inhibitor
cocktail A and B (Sigma-Aldrich; Merck KGaA)]. Proteins (50–100 µg)
were separated by 4–12% gradient SDS-PAGE (Invitrogen; Thermo
Fisher Scientific, Inc.), transferred to nitrocellulose membranes
using iBlot™ 2 Transfer Stacks (Invitrogen; Thermo Fisher
Scientific, Inc.) and were blocked overnight at 4°C in 5%
milk/TBS-0.1% Tween. The membranes were then probed with antibodies
against proteins of interest. Membranes were incubated with primary
antibodies at 1:1,000 dilution (vol/vol) overnight at 4°C, and with
secondary antibodies at 1:5,000 dilution at room temperature for 1
h. To verify protein loading, each sample was re-probed with
β-actin. Western blots were visualized using the ECL-Plus Western
blotting system (GE Healthcare Biosciences, Pittsburgh, PA, USA) or
IRDye® Infrared Dye exposed on the Odyssey image system
(Li-COR Biosciences, Lincoln, NE, USA). Blots were semi-quantified
using ImageJ (Version 1.43; National Institutes of Health,
Bethesda, MD, USA).
Cell viability assay
HepG2, HepT1 and HuH6 cells were plated in 96-well
plates at a density of 2×104 cells/well. After they were
allowed to settle for 24 h, the cells were treated with increasing
concentrations of LRA (0.1–100 µM) for 72 h. Cell viability was
measured for consecutive days using the Cell Counting Kit-8 (CCK-8)
assay (Dojindo Molecular Technologies Inc., Rockville, MD, USA),
according to the manufacturer's protocol, by replacing the medium
in each well with 10% CCK-8 solution (10 µl/100 µl)/media (v/v).
After 2 h incubation at 37°C, absorbance was measured at 450 nm
using a standard plate reader (Beckman Coulter, Inc., Brea, CA,
USA).
In experiments analyzing the combinatory effects of
LRA with dox, the cells were seeded in 96-well plates and were
treated with 10 µM LRA combined with increasing concentrations of
dox (0–10 µM) for 48 h at 37°C, after which, cell viability was
measured using an MTT assay. Briefly, the medium in each well was
replaced with 9% MTT (5 mg/ml)/media (v/v). After 4 h incubation at
37°C, 85 µl MTT/media was aspirated and 50 µl dimethyl sulfoxide
(DMSO) was added. The plate was then read at 550 nm using a
multimode plate reader (Beckman Coulter, Inc.) within 10 min.
Cell proliferation assay
HepG2, HepT1 and HuH6 cells were plated in 96-well
plates at a density of 2×103 cells/well. After they were
allowed to settle for 24 h, the cells were treated with increasing
concentrations of LRA (1–10 µM) for various durations. The CCK-8
assay (Dojindo Molecular Technologies Inc.), which is a sensitive
colorimetric assay used for the determination of cell viability in
cell proliferation assays, was used for quantification of the
number of proliferating cells, according to the manufacturer's
protocol. Briefly, the medium in each well was replaced with 10%
CCK-8 solution (10 µl/100 µl)/media (v/v) for 2 h at 37°C, after
which, absorbance was measured at 450 nm using a standard plate
reader (Beckman Coulter, Inc.). For the cell proliferation assay,
wells that contained known numbers of viable cells were used to
create a calibration curve.
Colony formation assay
For soft agar assays, a base layer of 1% (w/v)
agarose (cat. no. 214220; Difco; BD Biosciences, Franklin Lakes,
NJ, USA) mixed with cell culture medium was plated into 6-well
plates and allowed to solidify. The 1.5 ml top agar layer, which
was added on the top of the base layer, was made of 0.3% agar and
media solution, and the HepG2, HepT1 and HuH6 cells cultured in
regular medium were washed, counted and added to the mixture at
1×104 cells/well with increasing concentrations of LRA
(1–10 µM) at 37°C for 21 days. Culture medium (500 µl) was added on
top of the agarose to prevent drying of the soft agar. Cells were
grown at 37°C. Cells were treated with 1 ml 5 mg/ml MTT (cat. no.
M5655; Sigma-Aldrich; Merck KGaA) per well for 2 h after 21 days of
growth. All experiments were conducted in triplicate, and the means
and standard deviation were determined. One-way ANOVA followed by
Dunnett's multiple comparison post hoc test was used to determine
statistical significance.
For clonogenic assays, the cells were plated at
5,000 cells/well in 6-well culture dishes and left to form colonies
over a period of 3 weeks. The colonies produced were fixed with
methanol and stained with 0.05% crystal violet dye (cat. no. C3886;
Sigma-Aldrich; Merck KGaA) for 10 min, and the number of colonies
was scored to determine the colony-forming ability of the cells.
Colonies containing >30 cells were counted. Images of the plates
were also captured.
Propidium iodide (PI) staining and
flow cytometric analysis of the cell cycle
HepG2 and HuH6 cells were cultured in 6-well plates
at 3×105 cells/well overnight and were then treated for
24 h with LRA at a final concentration of 0, 1 or 10 µM.
Subsequently, the cells were harvested, washed with PBS and fixed
with 70% ethanol. The cells were then resuspended in PBS containing
100 µg/ml RNase A (Sigma-Aldrich; Merck KGaA) and 10 µg/ml PI for
30 min at room temperature. DNA content was determined by
fluorescence-activated cell analysis of PI-stained cells on a
LSR-II flow cytometer (BD Biosciences). Subsequently, analysis was
performed using BD FACDiva software v. 6.0 (BD Biosciences) and
FlowJo_v10 (FlowJo LLC, Ashland, OR, USA).
Statistical analysis
Statistical analysis was performed using GraphPad
Prism 5 software (GraphPad Software, Inc., La Jolla, CA, USA). All
values were presented as the means ± standard deviation. One-way
ANOVA and Dunnett's multiple comparison post hoc test, or
Student'st-test (two-tailed) were used to analyze the differences
between the experimental and control groups. P<0.05 was
considered to indicate a statistically significant difference. Half
maximal inhibitory concentration (IC50) values were
calculated using nonlinear regression in GraphPad Prism.
Results
LRH-1 expression is increased in
hepatoblastoma cell lines
To explore the role of LRH-1 in hepatoblastoma, the
expression levels of LRH-1 were detected in three hepatoblastoma
cell lines, HepG2, HuH6 and HepT1, and one control cell line,
THLE-2. The THLE-2 cell line consists of epithelial cells from the
left liver lobe transformed with SV40 large T antigen. The RT-qPCR
analysis demonstrated that the mRNA expression levels of LRH-1 were
significantly increased in HepG2 and HuH6 cells compared with in
HepT1 and THLE-2 cells (Fig. 1A).
Western blot analysis also revealed a similar expression pattern of
LRH-1 protein in these cell lines (Fig.
1B).
Knockdown of LRH-1 decreases cell
proliferation via the suppression of CCND1 and c-Myc expression in
hepatoblastoma cells
To examine whether silencing LRH-1 inhibits
hepatoblastoma cell proliferation, stable LRH-1 knockdown cell
lines were generated using lentiviral-based shRNAs in HepG2 and
HuH6 cells. RT-qPCR and western blot analyses were conducted to
verify the effectiveness of the shRNA sequences; shRNA3 was
revealed to the most efficient, clearly depleting LRH-1 at the mRNA
and protein levels (Fig. 2A-C).
Therefore, shRNA3 was used for the subsequent experiments. Since
HepT1 had low baseline LRH-1 expression, the effects of shLRH-1
were only detected on HepG2 and HuH6 cells. RT-qPCR analyses
demonstrated that the mRNA expression levels of LRH-1 were reduced
by 70–80% in response to shLRH-1 in both HepG2 and HuH6 cells
compared with the vector control shRNA (shCTL) cells (Fig. 2A and D). Furthermore, western blot
analysis revealed that the protein expression levels of LRH-1 were
significantly reduced in shLRH-1 cells by ~80% compared with the
shCTL cells (Fig. 2C and E).
To determine whether knockdown of LRH-1 had an
inhibitory effect on hepatoblastoma cell growth, CCK-8 assays were
conducted with HepG2 and HuH6 cells infected with shLRH-1 compared
with cells infected with shCTL. After 5 days, both shLRH-1 cell
lines exhibited significant decreases in cell proliferation
(Fig. 2F and G). Colony formation
assays were performed to determine the long-term effects of LRH-1
knockdown on the proliferation of hepatoblastoma cells. After 3
weeks, there were significantly fewer colonies (P<0.05) of HepG2
and HuH6 shLRH-1 cells compared with the shCTL cells (Fig. 2H and I). Taken together, these
results indicated that LRH-1 may serve an important role in
hepatoblastoma cell proliferation, since specific knockdown of
LRH-1 with shRNA resulted in the inhibition of proliferation and
colony formation in vitro.
It has been demonstrated that LRH-1 regulates the
expression of the cell cycle proteins CCND1 (14,15),
CCNE1 (14,15) and c-Myc (15). To explore the molecular mechanisms
by which LRH-1 regulates hepatoblastoma cell proliferation, RT-qPCR
and western blot analyses were performed on HepG2 and HuH6 shLRH-1
and shCTL cells. The results demonstrated that the mRNA and protein
expression levels of CCND1 and c-Myc were significantly decreased
in response to LRH-1 knockdown, whereas no marked alterations were
observed in CCNE1 expression following LRH-1 knockdown (Fig 3A-D). These results suggested that
LRH-1 knockdown-induced decreases in cell proliferation may be
associated with the downregulation of CCND1 and c-Myc in
hepatoblastoma cells.
 | Figure 3.Silencing LRH-1 gene expression
affects cell cycle gene expression in hepatoblastoma cells. (A and
B) mRNA expression levels of CCND1, CCNE1 and c-Myc in shCTL and
shLRH-1 cells, as determined by reverse transcription-quantitative
polymerase chain reaction. *P<0.05, **P<0.001 compared with
the shCTL group 1. (C and D) CCND1, CCNE1 and c-Myc protein
expression levels were determined by western blotting in shLRH-1
and shCTL HepG2 and HuH6 cells. Relative protein expression levels
are shown in the lower panels. Data are presented as the means ±
standard deviation, n=3 independent experiments. *P<0.05
compared with the shCTL group. CCND1, cyclin D1; CCNE1, cyclin E1;
CTL, control; LRH-1, liver receptor homolog-1; sh, short hairpin
RNA. |
LRA inhibits cell viability,
proliferation and anchorage-independent growth
In order to determine the anticancer effects of
targeting LRH-1 in hepatoblastoma cells, the specific LRH-1
antagonist LRA was used (20). Cell
viability assays were performed to determine the IC50
values at which HepG2, HuH6 and HepT1 hepatoblastoma cells respond
to LRH-1 inhibition. The results revealed that the treatment
markedly reduced the cell viability of all three hepatoblastoma
cell lines in a dose-dependent manner, although HepT1 was
relatively resistant to LRA treatment, compared with cells treated
with vehicle (DMSO) only. Specifically, the IC50 values
of LRA in hepatoblastoma cell lines were 40.11 µM in HepG2, 29.66
µM in HuH6 and 53.23 µM in HepT1 cells (Fig. 4A and B).
 | Figure 4.LRA inhibits cell viability,
proliferation and soft agar colony formation of hepatoblastoma
cells. (A) HepG2, HepT1 and HuH6 cells were plated in 96-well
flat-bottomed plates and were treated with increasing
concentrations of LRA for 72 h. Cytotoxic activity was determined
with CCK-8 assays. Data are presented as the means ± standard
deviation, n=3–5 independent experiments. *P<0.05, **P<0.01
compared with vehicle-treated cells. (B) IC50 values of
LRA in hepatoblastoma cell lines. (C) HepG2, HepT1 and HuH6 cells
were treated with LRA at the indicated concentrations. Cell
proliferation was detected by CCK-8 assays. Data are presented as
the means ± standard deviation, n=3–5 independent experiments.
*P<0.05, **P<0.01, as analyzed by ANOVA multiple comparison
analysis testing. (D) HepG2, HepT1 and HuH6 cells were seeded in
6-well plates with LRA (0, 1, 10 and 50 µM), media and agar, and
were grown for 3 weeks. The colonies were stained with MTT and
images were captured. (E) Colonies were counted and are presented
as the means ± standard deviation, n=3 independent experiments.
**P<0.01, ***P<0.001; IC50, half maximal
inhibitory concentration; LRA, LRH-1 antagonist; LRH-1, liver
receptor homolog-1. |
The present study also aimed to determine if LRA
could inhibit cell proliferation of the three hepatoblastoma cell
lines at sub-IC50 concentrations. HepG2, HepT1 and HuH6
cells were exposed to lower doses of LRA and their growth was
observed for 7 days. LRA induced dose-dependent inhibition of HepG2
and HuH6 cell proliferation; these were the two cell lines in which
baseline LRH-1 expression levels were high. Specifically, in these
two cell lines, the proliferation rates were significantly
decreased after 5 days of exposure to 10 µM LRA and after 7 days of
exposure to 1 µM LRA (Fig. 4C).
However, HepT1 cells, which had a lower baseline expression of
LRH-1, were less sensitive to LRA treatment; the proliferation rate
in these cells was significantly decreased only after 7 days of
exposure to 10 µM LRA (Fig. 4C).
These data indicated that LRA may inhibit hepatoblastoma cell
proliferation as both a cytotoxic and cytostatic agent.
Cancer cells have the unique ability to grow in soft
agar without being anchored to a surface. Therefore, the present
study evaluated the effects of LRA on this anchorage-independent
growth capability using soft agar growth assays. HepG2, HuH6 and
HepT1 hepatoblastoma cells growing on soft agar were treated with
increasing concentrations of LRA for 3 weeks, after which, their
ability to form colonies was assessed, as compared with the
vehicle-treated control cells. Following treatment with LRA, colony
formation was significantly decreased in all tested hepatoblastoma
cells in a dose-dependent manner (Fig.
4D and E), thus suggesting that LRA impaired
anchorage-independent growth of hepatoblastoma cells.
LRA inhibits cell proliferation
through downregulation of CCND1 and c-Myc in hepatoblastoma cell
lines
LRH-1 is known to function upstream of CCND1, CCNE1
and c-Myc. Therefore, it was hypothesized that the effects of LRA
on hepatoblastoma cell lines were induced through one or more of
these proteins. To test this hypothesis, the expression levels of
these proteins were detected in HepG2 and HuH6 hepatoblastoma cells
incubated with increasing doses of LRA for 24 h. Western blot
analyses of these proteins revealed that LRA dose dependently
inhibited CCND1 and c-Myc expression, whereas LRA did not seem to
markedly affect expression levels in HepT1 cells (Fig. 5A and B). To further determine
whether LRA affects the cell cycle, HepG2 and HuH6 cells exposed to
various concentrations of LRA for 24 h were stained with PI and
subjected to flow cytometry for cell cycle analysis. Following LRA
treatment, the percentage of cells in G1/G0
phase was increased in both cell lines, thus indicating that LRA
treatment may lead to cell cycle arrest in hepatoblastoma cells
(Fig. 5C and D). These results
suggested that LRA may inhibit hepatoblastoma cell proliferation by
inhibiting CCND1 and c-Myc expression, and by inducing cell cycle
arrest at G1 phase.
To further confirm that the LRH-1 downstream
targets, CCND1 and c-Myc, decreased proliferation of hepatoblastoma
cells, the present study examined whether silencing CCND1 and c-Myc
inhibited hepatoblastoma cell proliferation. Stable CCND1 and c-Myc
knockdown cell lines were generated using lentiviral-based shRNAs
in HepG2 and HuH6 cells. Western blot analyses were conducted to
verify the effectiveness of the shRNA sequences (Fig. 6A and B). To determine whether
knockdown of CCND1 and c-Myc exerted an inhibitory effect on HepG2
and HuH6 hepatoblastoma cell growth, CCK-8 assays were conducted.
After 7 days, both CCND1 and c-Myc knockdown cell lines exhibited
significant decreases in cell proliferation compared with cells
infected with shCTL (Fig. 6C and
D).
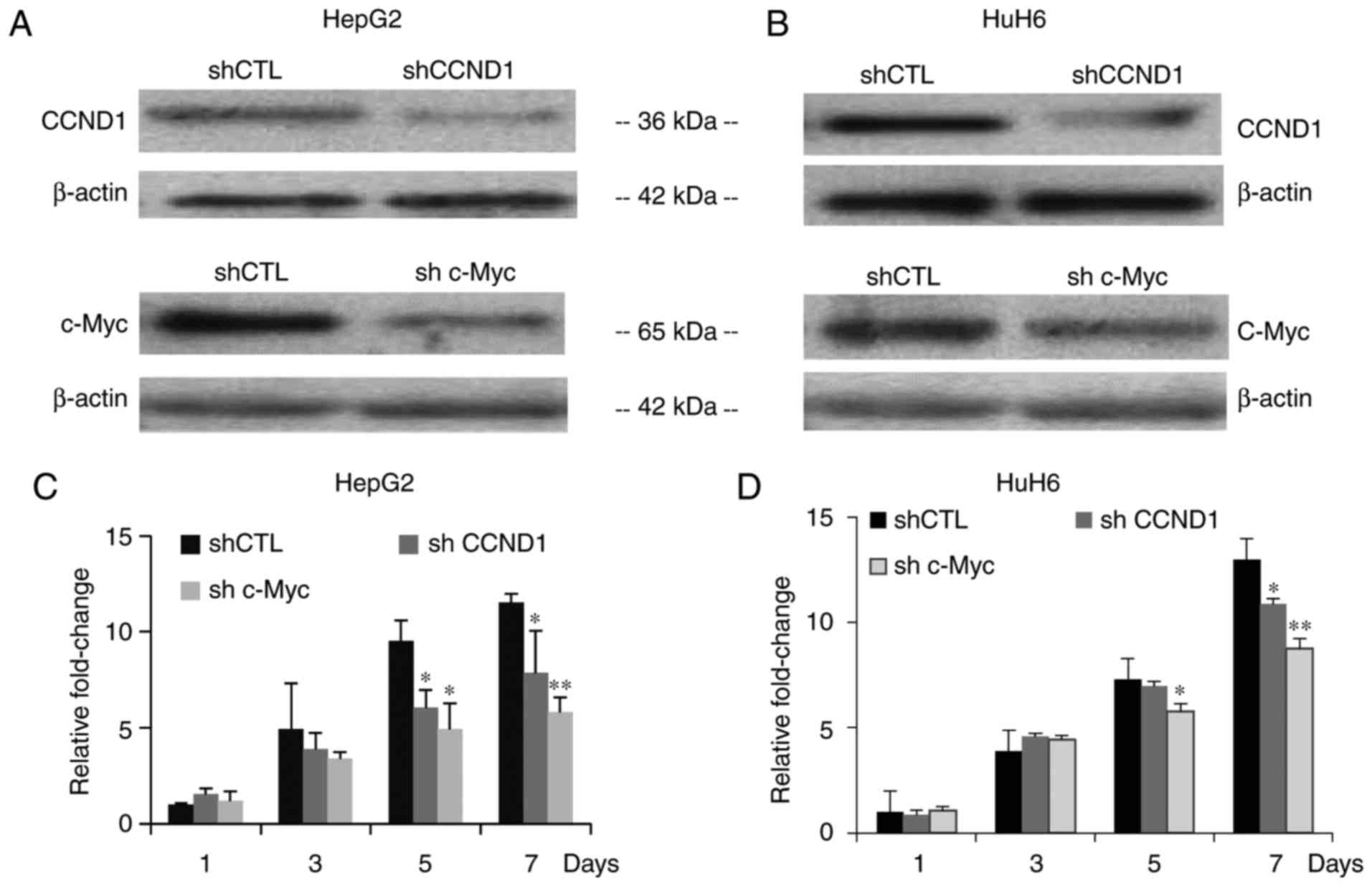 | Figure 6.Lentivirus-mediated shRNA suppresses
CCND1 and c-Myc expression, and inhibits proliferation. (A and B)
Western blotting was performed on HepG2 and HuH6 cells infected
with shCTL, shCCND1 or shc-Myc. β-actin was used as a control
protein to determine relative expression levels. (C and D)
Proliferation of HepG2 and HuH6 cells infected with shCTL, shCCND1
or shc-Myc, as assessed with Cell Counting Kit-8 assays. Data are
presented as the means ± standard deviation, n=3 independent
experiments. *P<0.05, **P<0.01. CCND1, cyclin D1; CTL,
control; LRH-1, liver receptor homolog-1; sh/shRNA, short hairpin
RNA. |
LRA significantly enhances the
cytotoxic effects of dox on hepatoblastoma cells
It is well known that monotherapies are less
effective in the treatment of high-risk cancer, due to the
acquisition of chemoresistance to drugs after prolonged exposure.
Therefore, the present study evaluated the effects of LRA in
combination with the established chemotherapeutic drug dox
(5). Cell viability assays were
conducted on HepG2 and HuH6 cells treated with 10 µM LRA and
increasing doses of dox, in order to determine if LRH-1 inhibition
affected the responsiveness of cells to chemotherapy. Notably, the
viability of these hepatoblastoma cell lines was much lower
following combination treatment compared with dox treatment alone
(Fig. 7). These data indicated that
LRA enhanced the cytotoxicity of dox in hepatoblastoma cells.
Discussion
Numerous studies have suggested that inhibition of
LRH-1 signaling is effective in the treatment of breast, pancreatic
and endometrial cancer (12,14,16).
The present results demonstrated that LRH-1 was upregulated in
HepG2 and HuH6 human hepatoblastoma cells compared with the control
THLE-2 cells; this finding is similar to the results of a previous
report studying pancreatic cancer cell lines and human tumor
samples (14). In addition,
silencing LRH-1 by shRNA inhibited cell proliferation and colony
formation of HepG2 and HuH6 cells, and, to a lesser extent, HepT1
cells, which have lower baseline levels of LRH-1. Subsequently,
LRH-1 was inhibited with the antagonist LRA, and the results
indicated that LRA may work as a cytostatic drug, blocking cell
proliferation through the suppression of CCND1 and c-Myc. Notably,
HepT1, the tested cell line with the lowest endogenous levels of
LRH-1, exhibited less sensitivity to LRA. Taken together, these
findings indicated that cells with higher expression levels of
LRH-1 may be more susceptible to LRH-1 inhibition.
Transcriptional targets of LRH-1 include c-Myc, and
the cell cycle regulators CCND1 and CCNE1, and these three genes
are known to control cell differentiation, growth and
proliferation. Both CCND1 and CCNE1 are frequently overexpressed in
gastrointestinal tumors and contribute to oncogenesis in animal
models (15,23,24).
c-Myc is a potent oncogene that promotes tumorigenesis in various
tissues, and its overexpression predicts poor clinical outcomes
(25). Other downstream targets of
LRH-1 include calpain 1, which upregulates the expression of CCNE1
truncated T1/T2 isoforms (14) and
Nanog, which serves a critical role in the reprograming of murine
somatic cells to pluripotent cells (26). In addition, scavenger receptor class
B member 1, low-density lipoprotein receptor and steroidogenic
acute regulatory protein are downstream targets of LRH-1 in the
progesterone synthesis pathway in ovulation (27).
In hepatoblastoma, downregulation of LRH-1 by
lentiviral shRNAs or inhibition of LRH-1 by LRA resulted in a
decrease in the mRNA and protein expression levels of CCND1 and
c-Myc, which may lead to the inhibition of cell proliferation and
colony formation. However, significant alterations were not
detected in CCNE1; these findings differ from the results observed
in pancreatic and colon cancer (28). In different cell types, diverse
effects can be seen on gene and protein expression. For example,
varying effects on CCNE1 expression may be due to different
regulation of the downstream LRH-1 signaling pathways. CCND1
inhibition-mediated cell cycle arrest at the G1 phase is
likely one of the major underlying mechanisms of action of LRA. In
addition, suppression of c-Myc by LRA may contribute to the
observed suppression of hepatoblastoma cell proliferation.
Development of novel targeted drugs is integral to
overcoming chemoresistance and improving the survival of patients
with hepatoblastoma. In order to identify a novel therapy, the
present study investigated the effects of LRA on hepatoblastoma
growth. LRA was able to decrease hepatoblastoma cell proliferation
in vitro, when used as a single agent. It was therefore
hypothesized that LRA may be combined with other chemotherapeutic
agents that regulate different signaling pathways to sensitize
tumors to various forms of therapy, impair tumor cell escape
mechanisms and increase efficacy for patients with hepatoblastoma.
The present study evaluated the effects of combination therapy of
LRA with dox, which is an anticancer drug that intercalates into
DNA and inhibits macromolecular biosynthesis. LRA enhanced the
cytotoxic effects of dox on hepatoblastoma cells. Based on these
findings, it is possible that LRA treatment may sensitize patients
to chemotherapy and thus decrease drug toxicity by allowing lower
concentrations of drug to be used.
Notably, HepT1 cells possessed lower levels of
LRH-1, which may be due to the involvement of other upstream
regulators. In PCR-based microsatellite analysis of chromosome arm
11p, a loss of heterozygosity at all informative loci, including
the Wilms tumor 1 homolog and insulin-like growth factor 2 genes,
was detected (29), which may
explain why HepT1 cells behave differently to other cells. It has
previously been reported that HepG2 and HuH6 hepatoblastoma cell
lines accurately resemble primary human hepatoblastoma samples to
varying degrees, and the HepG2 cell line most accurately mimics
human hepatoblastoma (22).
In conclusion, the present results suggested that
LRH-1 may contribute to cell proliferation in hepatoblastoma.
Hepatoblastoma cells with higher LRH-1 expression levels were more
susceptible to LRH-1 inhibition. In addition, it was revealed that
LRA may behave as a cytostatic compound at low concentrations, and
it may inhibit hepatoblastoma cell proliferation through the
suppression of CCND1 and c-Myc expression. LRA also exhibited
cytotoxic effects at higher concentrations. Furthermore, LRA
induced cell cycle arrest at G1 phase, and inhibited
colony forming ability and tumor growth. Analysis of a panel of
hepatoblastoma cell lines provided compelling evidence to suggest
that LRH-1 may serve an important role in the progression of
hepatoblastoma and implicated LRA as a novel potential agent for
innovative therapeutic strategies against hepatoblastoma. LRA might
serve as an effective drug in potential clinical trials for
patients with recurrent or refractory hepatoblastoma, particularly
in patients resistant to dox. Therefore, LRA may be considered a
promising novel candidate for innovative therapeutic strategies
against hepatoblastoma.
Acknowledgements
The authors would like to thank Dr. Amos Gaikward
and Dr. Tatiana Goltsova (Baylor College of Medicine, Texas
Children's Hospital, Houston, TX, USA) for their assistance with
flow cytometry.
Funding
The present study was supported by the Texas
Children's Department of Surgery Seed Grant (S.V.) and the Macy
Easom Cancer Research Foundation Grant (S.V.).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JiJ and SAV designed and performed the experiments
regarding the activity of LRA. JuJ, SEW, RHP, YS, NGJ, BL, WS, XC
and YY designed and performed the experiments regarding the LRH-1
expression and activity of LRA. JiJ and SAV generated the ideas,
supervised the study, and wrote the paper. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
LRH-1
|
liver receptor homolog-1
|
|
dox
|
doxorubicin
|
|
CCK-8
|
Cell Counting Kit-8 assay
|
|
IC50
|
half maximal inhibitory
concentration
|
|
PI
|
propidium iodide
|
References
|
1
|
Buendia MA: Unravelling the genetics of
hepatoblastoma: Few mutations, what else? J Hepatol. 6:1202–4.
2014. View Article : Google Scholar
|
|
2
|
von Schweinitz D: Hepatoblastoma: Recent
developments in research and treatment. Semin Pediatr Surg.
21:21–30. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
von Schweinitz D: Management of liver
tumors in childhood. Semin Pediatr Surg. 15:17–24. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Schmid I, Haberle B, Albert MH,
Corbacioglu S, Fröhlich B, Graf N, Kammer B, Kontny U, Leuschner I,
Scheel-Walter HG, et al: Sorafenib and cisplatin/doxorubicin
(PLADO) in pediatric hepatocellular carcinoma. Pediatr Blood
Cancer. 58:539–544. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Malogolowkin MH, Katzenstein HM, Krailo M,
Chen Z, Quinn JJ, Reynolds M and Ortega JA: Redefining the role of
doxorubicin for the treatment of children with hepatoblastoma. J
Clin Oncol. 26:2379–2383. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Warmann SW, Armeanu S, Frank H, Buck H,
Graepler F, Lemken ML, Heitmann H, Seitz G, Lauer UM, Bitzer M and
Fuchs J: In vitro gene targeting in human hepatoblastoma. Pediatr
Surg Int. 22:16–23. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Warmann S, Hunger M, Teichmann B, Flemming
P, Gratz KF and Fuchs J: The role of the MDR1 gene in the
development of multidrug resistance in human hepatoblastoma:
Clinical course and in vivo model. Cancer. 95:1795–1801. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fayard E, Auwerx J and Schoonjans K: LR
H-1 An orphan nuclear receptor involved in development, metabolism
and steroidogenesis. Trends Cell Biol. 14:250–260. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fayard E, Schoonjans K, Annicotte JS and
Auwerx J: Liver receptor homolog 1 controls the expression of
carboxyl ester lipase. J Biol Chem. 278:35725–35731. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Petersen GM, Amundadottir L, Fuchs CS,
Kraft P, Stolzenberg-Solomon RZ, Jacobs KB, Arslan AA,
Bueno-de-Mesquita HB, Gallinger S, Gross M, et al: A genome-wide
association study identifies pancreatic cancer susceptibility loci
on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat Genet. 42:224–228.
2010. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Annicotte JS, Fayard E, Swift GH, Selander
L, Edlund H, Tanaka T, Kodama T, Schoonjans K and Auwerx J:
Pancreatic-duodenal homeobox 1 regulates expression of liver
receptor homolog 1 during pancreas development. Mol Cell Biol.
23:6713–6724. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Thiruchelvam PT, Lai CF, Hua H, Thomas RS,
Hurtado A, Hudson W, Bayly AR, Kyle FJ, Periyasamy M and Photiou A:
The liver receptor homolog-1 regulates estrogen receptor expression
in breast cancer cells. Breast Cancer Res Treat. 127:385–396. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bolado-Carrancio A, Riancho JA, Sainz J
and Rodriguez-Rey JC: Activation of nuclear receptor NR5A2
increases Glut4 expression and glucose metabolism in muscle cells.
Biochem Biophys Res Commun. 446:614–619. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lin Q, Aihara A, Chung W, Li Y, Huang Z,
Chen X, Weng S, Carlson RI, Wands JR and Dong X: LRH1 as a driving
factor in pancreatic cancer growth. Cancer Lett. 345:85–90. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Botrugno OA, Fayard E, Annicotte JS, Haby
C, Brennan T, Wendling O, Tanaka T, Kodama T, Thomas W, Auwerx J
and Schoonjans K: Synergy between LRH-1 and beta-catenin induces G1
cyclin-mediated cell proliferation. Mol Cell. 15:499–509. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dube C, Bergeron F, Vaillant MJ, Robert
NM, Brousseau C and Tremblay JJ: The nuclear receptors SF1 and LRH1
are expressed in endometrial cancer cells and regulate
steroidogenic gene transcription by cooperating with AP-1 factors.
Cancer Lett. 275:127–138. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Whitby RJ, Stec J, Blind RD, Dixon S,
Leesnitzer LM, Orband-Miller LA, Williams SP, Willson TM, Xu R,
Zuercher WJ, et al: Small molecule agonists of the orphan nuclear
receptors steroidogenic factor-1 (SF-1, NR5A1) and liver receptor
homologue-1 (LRH-1, NR5A2). J Med Chem. 54:2266–2281. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Whitby RJ, Dixon S, Maloney PR, Delerive
P, Goodwin BJ, Parks DJ and Willson TM: Identification of small
molecule agonists of the orphan nuclear receptors liver receptor
homolog-1 and steroidogenic factor-1. J Med Chem. 49:6652–6655.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Corzo CA, Mari Y, Chang MR, Khan T,
Kuruvilla D, Nuhant P, Kumar N, West GM, Duckett DR, Roush WR and
Griffin PR: Antiproliferation activity of a small molecule
repressor of liver receptor homolog 1. Mol Pharmacol. 87:296–304.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Benod C, Carlsson J, Uthayaruban R, Hwang
P, Irwin JJ, Doak AK, Shoichet BK, Sablin EP and Fletterick RJ:
Structure-based discovery of antagonists of nuclear receptor LRH-1.
J Biol Chem. 288:19830–19844. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2-ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Woodfield SE, Shi Y, Patel RH, Jin J,
Major A, Sarabia SF, Starosolski Z, Zorman B, Gupta SS, Chen Z, et
al: A novel cell line based orthotopic xenograft mouse model that
recapitulates human hepatoblastoma. Sci Rep. 7:177512017.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Heng JC, Feng B, Han J, Jiang J, Kraus P,
Ng JH, Orlov YL, Huss M, Yang L, Lufkin T, et al: The nuclear
receptor Nr5a2 can replace Oct4 in the reprogramming of murine
somatic cells to pluripotent cells. Cell Stem Cell. 6:167–174.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wagner RT, Xu X, Yi F, Merrill BJ and
Cooney AJ: Canonical Wnt/beta-catenin regulation of liver receptor
homolog-1 mediates pluripotency gene expression. Stem Cells.
28:1794–1804. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pelengaris S, Khan M and Evan G: c-MYC:
More than just a matter of life and death. Nat Rev Cancer.
2:764–776. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Annicotte JS, Chavey C, Servant N,
Teyssier J, Bardin A, Licznar A, Badia E, Pujol P, Vignon F,
Maudelonde T, et al: The nuclear receptor liver receptor homolog-1
is an estrogen receptor target gene. Oncogene. 24:8167–8175. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bertolin K, Gossen J, Schoonjans K and
Murphy BD: The orphan nuclear receptor Nr5a2 is essential for
luteinization in the female mouse ovary. Endocrinology.
155:1931–1943. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bayrer JR, Mukkamala S, Sablin EP, Webb P
and Fletterick RJ: Silencing LRH-1 in colon cancer cell lines
impairs proliferation and alters gene expression programs. Proc
Natl Acad Sci USA. 112:2467–2472. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pietsch T, Fonatsch C, Albrecht S, Maschek
H, Wolf HK and von Schweinitz D: Characterization of the continuous
cell line HepT1 derived from a human hepatoblastoma. Lab Invest.
74:809–818. 1996.PubMed/NCBI
|





















