Introduction
Breast cancer is the second most common cancer
worldwide and, by far, the most frequent cancer among women
(1). Approximately 15–20% of all
patients with breast cancer are diagnosed with triple-negative
breast cancer (TNBC), named after its lack of estrogen receptor
alpha (ERα), progesterone receptor (PR) and epidermal growth factor
receptor 2 (HER2) expression (2).
While several treatment options are available to treat
ERa+ and HER2-driven breast cancers, options are
somewhat limited for patients with TNBC, who cannot take advantage
of these targeted therapies. There is therefore a pressing clinical
need to identify novel drug targets and therapeutic strategies for
women with TNBC. Recent years have witnessed major clinical
breakthroughs in cancer immunotherapies, which include cancer
peptide vaccines, dendritic cell vaccines, adoptive transfer of
cytotoxic T lymphocytes (CTL) and blockade of immune-suppressive
checkpoint molecules (3–8). However, despite success with
immunotherapies in treating melanoma and lung cancer, breast cancer
has proven to be particularly difficult to treat with checkpoint
blockade or other immunotherapies (9–13). The
current clinical trials for patients with TNBC utilizing
checkpoint-blockade immunotherapy have demonstrated only modest
efficacy (14).
Discoidin domain receptor tyrosine kinase (DDR) 1 is
a cell-surface tyrosine kinase, which can be activated by
collagens, and regulates cell growth, adhesion, migration and
matrix remodeling. DDR1 is predominantly expressed in normal
epithelial cells and its aberrant expression in a variety of human
cancers is associated with tumor progression, including breast,
lung, ovary, liver, gastric cancer and glioma (15–21).
Accumulating evidence has revealed DDR1 mutations in breast cancer,
schwannoma, endometrial cancer, lung cancer and acute leukemia
(22–26). Since DDR1 is being considered as a
potential novel therapeutic target in cancer, defining its precise
function during breast cancer progression is of critical importance
for the development of associated therapies alone and in
combination with immunotherapy.
In the present study, to further investigate the
role of DDR1 in breast cancer progression and to validate DDR1 as a
novel target for breast cancer therapy, two murine breast cancer
cell lines, 4T1 and EMT6, were used to mimic human TNBC. It was
demonstrated that DDR1 was frequently upregulated in patients with
breast cancer. Furthermore, it was revealed that tumor cell DDR1
promoted breast cancer growth in vivo by modulating CTLs.
Lastly, inhibition of DDR1 by using a specific extracellular domain
(ECD) neutralizing antibody decreased breast cancer growth and
increased CTL tumor infiltration in vivo.
Materials and methods
Patients and tissue samples
A total of 30 samples of breast cancer tissues and
adjacent tissues were surgically removed from patients (age range,
37–56 years) in Jiangxi Cancer Hospital between March 2013 and
December 2016. Ethical approval for the present study was provided
by the Ethics Committee of Jiangxi Cancer Hospital. Written
informed consent was obtained from all the study participants.
Bioinformatic analysis
For analysis of gene expression, The Cancer Genome
Atlas (TCGA) BRCA datasets were downloaded from TCGA (https://tcga-data.nci.nih.gov) (27). UALCAN, an easy to use, interactive
web-portal was used to analyze DDR1 mRNA gene expression in the
breast cancer datasets (28).
Mice, cell lines and chemicals
A total of 32 female BALB/c mice (age, 6–8 weeks;
weight, 22–28 g) were obtained from the Animal Center of the
Chinese Academy of Medical Sciences (Beijing, China). Mice were
provided with free access to food, water and bedding at all time,
and were housed in filter top cages (maximum 5 mice per cage) at
21–23°C with 45–55% humidity and a 12-h light/dark cycle. All
animal experiments were performed in accordance with the National
Institute of Health Guide for the Care and Use of Laboratory
Animals, with the approval of the Ethics Committee of Jiangxi
Cancer Hospital. Mouse mammary cancer cell lines 4T1 and EMT6,
derived from the BALB/c mouse strain, were obtained from the
American Type Culture Collection and maintained in RPMI-1640 medium
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal
bovine serum (FBS; Hyclone Laboratories; GE Healthcare Life
Sciences), 1% penicillin/streptomycin (Invitrogen; Thermo Fisher
Scientific, Inc.) at 37°C in 5% CO2.
Plasmid transfection
The full-length cDNA of mouse DDR1 (NM_172962.1;
2,625 bp) cloned in the pCMV3 backbone vector (cat. no. MG50829-UT)
was obtained from Sino Biological, Inc. 4T1 cells (2×105
cells in 2 ml) were seeded into a 6-well plate, one day prior to
transfection. A total of 5 µg DDR1-expressing plasmid DNA or empty
vector, with 10 µl Lipofectamine® 2000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) in 200 µl
Opti-MEM® medium, was added into each well. After 6 h,
the transfection medium was replaced with fresh medium. After 72 h,
4T1-DDR1 cells and control 4T1-vector cells were selected with 500
µg/ml hygromycin for 1 week.
CRISPR/Cas9-mediated knockout (KO) of
mouse DDR1
DDR1-deficient cells were generated using the
CRISPR/Cas9 system, in order to investigate the role of DDR1 in the
interactions between immune cells and mammary tumor cells. The
target sequences for mouse DDR1 were: i, GCAGCAGCAGTAGAGATGAG; and
ii, GCAGTGATGGAGATGGGGCT. The sequences for DDR1 were selected
using the CRISPR MultiTargeter tool (http://www.multicrispr.net/) (29) and off-targets were excluded using
GT-Scan tools (30). Oligonucleotides
with BsmB1 restriction sites for guide RNAs were synthesized
by SBS Genetech Co., Ltd., then phosphorylated using T4 kinase
(31). The phosphorylated
oligonucleotides were cloned into LentiCRISPR v2 (Plasmid no.
52961; Addgene) and the sequences of the cloned plasmids that were
extracted from numerous selected colonies were confirmed by SBS
Genetech Co., Ltd. EMT6 cells were transfected with
pLentiCRISPR-single guide RNA (sgRNA) DDR1 using Lipofectamine™
2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. The cells were cultured with 1 µg/ml
puromycin for 2 weeks, starting at 3 days post-transfection. Single
cells were then sorted by fluorescence-activated cell sorting
(FACS) into 96-well plates. The depletion of DDR1 in the surviving
cells was validated via western blotting. KO1 and KO2 clones were
from two independent sgRNA sequences of mouse DDR1.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted using TRIzol®
(Thermo Fisher Scientific, Inc.). Reverse transcription reactions
were performed using M-MLV reverse transcriptase (Promega
Corporation). For mRNA detection, DDR1 and GAPDH mRNA expression
levels were analyzed using Luminaris Color HiGreen qPCR Master Mix
(Thermo Fisher Scientific, Inc.), according to the manufacturer's
protocol (Applied Biosystems; Thermo Fisher Scientific). The
following primers were used: GAPDH, forward
5′-GACTCATGACCACAGTCCATGC-3′ and reverse
5′-AGAGGCAGGGATGATGTTCTG-3′; DDR1, forward
5′-GATTTCCCCTTAATGTGCGT-3′ and reverse 5′-TGGCATCTGGCCGTAAGATC-3′.
Relative gene expression was calculated by the 2−ΔΔCq
method (32).
Western blotting
The cells were washed twice with phosphate buffered
saline (PBS) and lysed in ice-cold radio immunoprecipitation assay
buffer (RIPA; Beyotime Institute of Biotechnology) with freshly
added 0.01% protease inhibitor PMSF (Amresco) and incubated on ice
for 20 min. The total cell protein was collected by centrifugation
at 10,000 × g for 10 min at 4°C. Protein concentrations were
determined using a Bradford assay (Bio-Rad Laboratories, Inc.).
Total protein (30 µg) from each sample were separated via SDS-PAGE
(10% gel) and electrophoretically transferred onto a polyvinylidene
difluoride membrane (EMD Millipore). The immunoblots were blocked
by incubation in 5% skimmed milk, 25 mM Tris (hydroxymethyl)
aminomethane-HCl (pH 8.0), 150 mM NaCl, and 0.1% Tween®
20 for 1 h at 25°C. Membranes were incubated with the following
primary antibodies: DDR1 (1:1,000; cat. no. sc-532; Santa Cruz
Biotechnology, Inc.), DDR1 (1:1,000; cat. no. AF2396; R&D
Systems, Inc.), and β-actin (1:10,000; cat. no. sc-47778; Santa
Cruz Biotechnology, Inc.), followed by the corresponding
horseradish peroxidase-conjugated secondary antibodies (1:10,000;
cat. nos. 115-035-003 and 111-035-003; Jackson ImmunoResearch
Laboratories, Inc.). Protein detection was performed using an
enhanced chemiluminescence kit (ECL; Thermo Fisher Scientific,
Inc.).
Cell Counting Kit-8 (CCK-8)
cytotoxicity assay
The proliferation of cells was measured using a
CCK-8 cytotoxicity assay (Dojindo Molecular Technologies, Inc.).
EMT6 or 4T1 cells were seeded in 96-well plates at a density of
2,000 cells/well and cultured with RPMI-1640 medium containing 10%
FBS. CCK-8 solution (20 µl) was added to each well, and the plates
were incubated for 2 h at 37°C at the indicated time points. The
optical density levels were measured using a microplate reader
scanning at 450 nm according to the manufacturer's protocol.
Detection of released DDR1
extracellular domain in the media
EMT6 cells were washed twice with warm PBS, and the
cell monolayers were incubated (37°C) in serum-free media for
another 24 h. The conditioned media were then collected and
clarified by a spin at 10,000 × g for 10 min at 4°C to remove cell
debris. The supernatants were supplemented with EDTA to a final
concentration of 2 mM, before a high-speed centrifugation (20,000 ×
g, 30 min, 4°C) to remove cell membranes and vesicles. A total of
200 µl of each media were then precipitated with trichloroacetic
acid (24), and the resultant pellets
were washed with acetone and resuspended in 2× reducing Laemmli
SDS-sample buffer. The immunoblots were probed with antibody AF2396
(as aforementioned), which is directed against the N-terminal
region of DDR1.
In vivo tumor studies
The tumor cells in log phase growth were centrifuged
at 500 × g for 5 min at room temperature, washed once with Hank's
balanced salt solution (HBSS), counted, and re-suspended in HBSS
with 50% Matrigel (BD Biosciences) at a concentration of
5×105 cells/ml (4T1) or 1×106 cells/ml
(EMT-6). Cell suspensions (100 µl) were injected subcutaneously
near the fourth mammary gland fat pad. The mice were acclimated to
the study conditions for at least 1 week prior to tumor cell
implantation. The animals were randomly distributed into treatment
groups (5 mice per group) and treated with mouse immunoglobulin
(Ig) G1 isotype control (cat. no. BE0093; Bio X Cell), or anti-DDR1
(mouse IgG1 clone 5D5; cat. no. MABT333; EMD Millipore; 200
µg/mouse intraperitoneally twice per week for 4 weeks). Tumors were
measured once per week using a caliper, and tumor volumes were
calculated using the modified ellipsoid formula 1/2 × (length ×
width2).
Preparation of single cell suspension
and antibody staining for flow cytometry
The tumors were collected, weighed and enzymatically
digested using a cocktail of dispase (Thermo Fisher Scientific,
Inc.), collagenase P (Roche Diagnostics) and DNaseI (Roche
Diagnostics) (33) for 45 min at
37°C, to obtain a single cell suspension. Cells were counted using
a Vi-CELL XR (Beckman Coulter, Inc.). For T-cell staining, cells
were first incubated with mouse BD Fc block (1:100; clone 2.4G2;
cat. no. 553141; BD Biosciences) and LIVE/DEAD® Fixable
Dead Cell Stain (Invitrogen; Thermo Fisher Scientific, Inc.) for 30
min on ice. The cells were then stained for 30 min on ice with the
following antibodies: Anti-CD45-BV605 (cat. no. 563053; clone
30-F11; 1:200; BD Biosciences), anti-CD3e-FITC (cat. no. 553061;
clone 145-2C11; 1:100; BD Biosciences), anti-CD8-PerCP (cat. no.
100732; clone 53–6.7; 1:50; BioLegend, Inc.), anti-CD4-BV711 (cat.
no. 100557; clone RM4-5; 1:200; BioLegend, Inc.) and anti-CD69-APC
(cat. no. 104514; clone H1.2F3; 1:25; BioLegend, Inc.). For
intracellular staining, the cells were washed with 1 ml PBS, and
incubated for 30 min at 4°C with the surface marker antibodies.
After washing, cells were fixed and permeabilized with fixation
concentrate and permeabilization diluent (eBioscience) and stained
in 10X permeabilization buffer (eBioscience) for the intracellular
proteins, according to the manufacturer's protocol. The following
antibodies were used for 30 min at room temperature in the dark:
Anti-Ki67-PE antibody (cat. no. 652404; clone 16A8; 1:50;
BioLegend, Inc.) and anti-T-box transcription factor 21 (also known
as T-bet)-PE-Cy7 (cat. no. 644824; clone 4B10; 1:100; eBioscience).
Flow cytometry data were collected with a BD LSRFortessa cell
analyzer and analyzed using FlowJo Software (version 10.2; FlowJo
LLC).
Statistical analysis
Data were analyzed using GraphPad Prism software
(version 6; GraphPad Software, Inc.) and are presented as the mean
± standard deviation. Two-tailed Student's t-tests were used to
compare two groups, and one-way ANOVA followed by Dunett's test was
used to compare multiple groups. P<0.05 was considered to
indicate a statistically significant difference.
Results
DDR1 is frequently upregulated in
breast cancer
RT-qPCR analyses were performed to determine the
expression levels of DDR1 in 20 primary breast cancer samples and
their matched adjacent normal tissues. The results demonstrated
that the mRNA expression levels of DDR1 were significantly higher
in tumor tissues compared with the matched adjacent tissues
(Fig. 1A). Further analysis of DDR1
expression levels using the TCGA breast cancer database revealed
that DDR1 was expressed at higher levels in breast cancer samples
compared with normal samples (Fig.
1B). In addition, DDR1 mRNA expression levels were higher in
different subtypes (luminal, HER2+ and triple negative),
compared with those in the normal breast tissues (Fig. 1C). Furthermore, the protein expression
levels of DDR1 were examined by western blotting in another 10
primary breast cancer samples. The results demonstrated that the
protein expression of DDR1 was also markedly increased in the
breast cancer tissues collected in the present study compared with
their matched adjacent normal tissues (Fig. 1D). Overall, these data demonstrated
that DDR1 was upregulated in breast cancer.
Ectopic expression of DDR1 promotes
tumor growth in vivo by suppressing antitumor immunity
Consistent with previously published results
(34), mouse mammary tumor cells
4T1/Vector (4T1 cells stably transfected with empty vector)
expressed undetectable levels of endogenous DDR1 protein. In order
to determine whether DDR1 could affect tumor cell growth, mouse
DDR1 was transfected into 4T1 cells and stable cells overexpressing
DDR1 were obtained (Fig. 2A). The
effects of DDR1 overexpression on the cell proliferation were then
investigated using a CCK-8 assay. It was revealed that
overexpression of DDR1 did not affect cancer cell growth in
vitro (Fig. 2B). To further
investigate the effects of DDR1 on tumor cell growth in
vivo, a tumorigenesis assay was performed in BALB/c female mice
using 4T1 cells with or without stable DDR1 overexpression. The
tumors in the 4T1/DDR1 group grew faster compared with the
4T1/Vector group (P<0.01; Fig. 2C and
D). These results suggested that high DDR1 expression in breast
cancer may be associated with a more aggressive state. When
investigating models for immuno-oncology purposes, it is also
important to know the composition of infiltrated immune cells into
a tumor. To this end, it was demonstrated that there was a lower
percentage of CD4+ and CD8+ T cells in the
tumors of the 4T1/DDR1 group compared with those in the 4T1/Vector
group (P=0.014 and P=0.037, respectively; Fig. 2E and F).
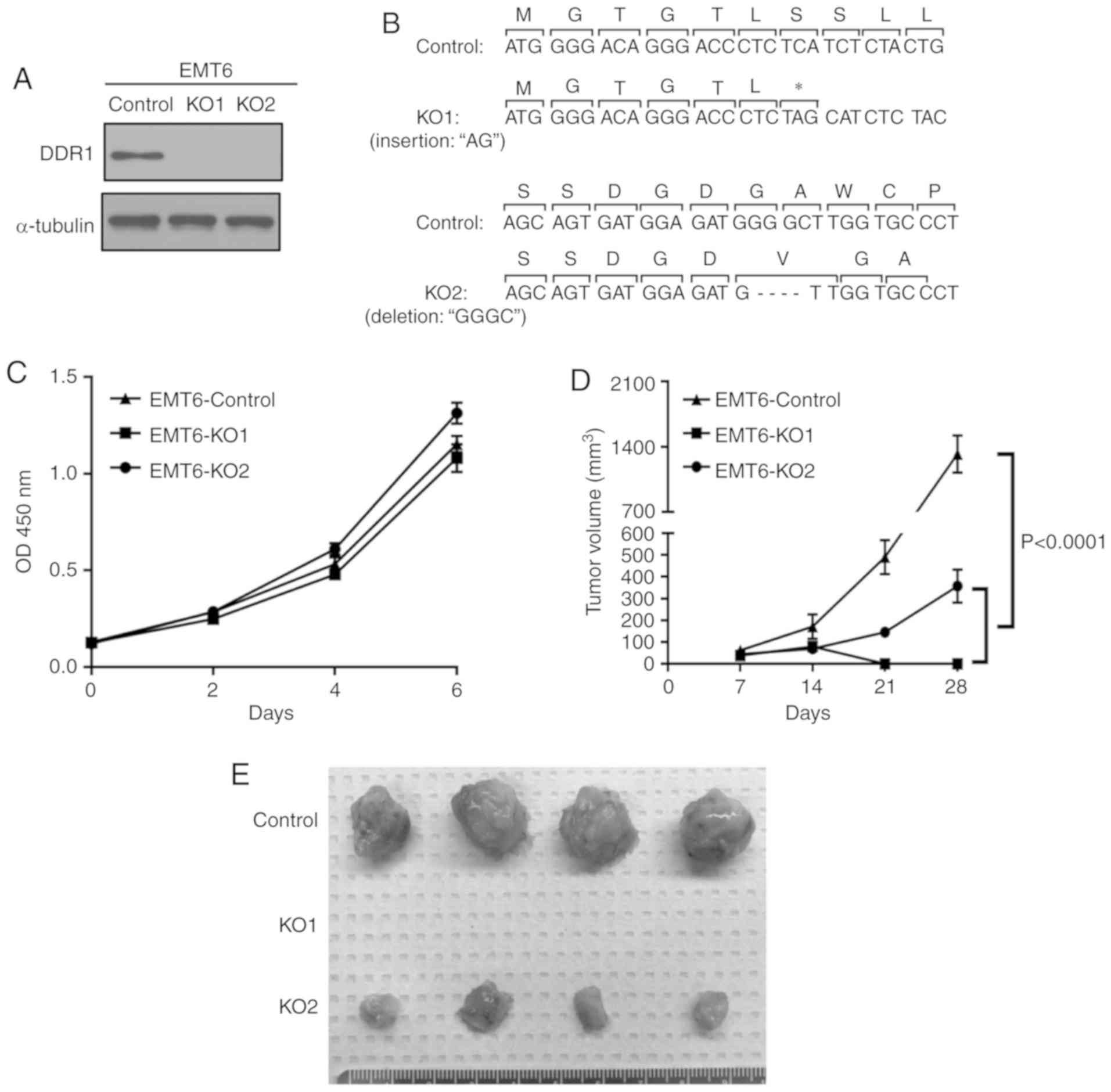
Deletion of DDR1 inhibits tumor growth
in vivo
To further elucidate the role of DDR1 in breast
cancer cell growth, DDR1-deficient EMT-6 cell lines were
established using the CRISPR/Cas9 system using two different sgRNA
sequences (Fig. 3A). DDR1 KO clone 1
has a ‘AG’ insertion, which created a stop codon shortly after
initiation (Fig. 3B). DDR1 KO clone 2
deletes four nucleotides ‘GGGC’ and results in a frame shift in the
DDR1 coding sequence (Fig. 3B). DDR1
KO did not affect tumor cell growth in vitro (Fig. 3C). Notably, the tumors derived from
the KO1 clone of EMT-6 cells regressed completely after they
reached ~100 mm3 (P<0.0001; Fig. 3D and E). The tumors derived from the
KO2 clone grew dramatically slower in the BALB/c hosts compared
with those in the control group (P<0.0001; Fig. 3D and E). Taken together, DDR1
expression in the tumor cells was demonstrated to be important for
sustained breast tumor growth in vivo.
Cytotoxic T cells mediate the
tumor-promoting activity of DDR1
In order to examine the influence of tumor cell DDR1
on antitumor immunity, the tumors derived from EMT-6/Control and
EMT-6/KO2 groups were collected, and tumor-infiltrating lymphocytes
(TILs) were assessed via flow cytometric analysis. The results
demonstrated that the percentage of CD4+ and
CD8+ TILs was significantly increased (P=0.042 and
P=0.025, respectively; Fig. 4A and B)
in the tumors derived from the EMT-6 DDR1 KO2 cells compared with
the tumors derived from the EMT-6/Control cells. DDR1 KO2-derived
tumors also exhibited increased density of early activated and
proliferating CD8+ T cells relative to control tumors
(P=0.03 and P=0.035, respectively; Fig.
4C and D). In addition, DDR1 KO2-derived tumors had increased
density of T-bet expressing CD8+ TILs compared with
control tumors (P=0.017; Fig. 4E).
T-bet, a T-box transcription factor, serves an important role in T
cell differentiation and maturation (35,36). These
data indicated that tumor DDR1 promoted breast cancer growth by
modulating antitumor immunity.
DDR1 inhibition suppresses tumor
growth in vivo by enhancing antitumor T cell activity
Structurally, DDR1 consists of an ECD, a
transmembrane domain and an intracellular kinase domain (37–40). A
recent study revealed that DDR1 had a kinase-independent function
in promoting breast cancer growth (41). Secreted soluble DDR1-ECD has been
reported in a number of previous studies (42–44), but
its biological significance in cancer has not yet been elucidated.
For these reasons, the present study first investigated whether
EMT6 cells were releasing DDR1-ECD in the media. Secreted soluble
DDR1-ECD was detected in the conditioned media from EMT6 cells
(Fig. 5A). It was, therefore,
hypothesized that treatment with a DDR1 neutralizing antibody
(41,45), which specifically targets DDR1-ECD,
could suppress tumor growth in vivo. To this end, parental
EMT-6 cells were inoculated in mice to form tumors, and then
treated with either the DDR1 neutralizing antibody (EMT6/anti-DDR1
group) or an isotype IgG1 control antibody (EMT6/anti-IgG group).
The tumors in the EMT6/anti-DDR1 group grew much slower than those
in the EMT6/anti-IgG group (P<0.001; Fig. 5B and C). In addition, there were more
CD4+ and CD8+ T cells in the tumors of the
EMT6/anti-DDR1 group compared with those in the EMT6/anti-IgG group
(P=0.03 and P=0.018, respectively; Fig.
5D and E).
DDR1 mRNA is negatively correlated
with the TIL signature gene expression
In order to further validate the impact of tumor
cell DDR1 on antitumor immunity, TIMER (Tumor Immune Estimation
Resource, http://cistrome.org/TIMER) (46), a comprehensive resource for the
clinical relevance of tumor-immune cell correlations, was used to
analyze TCGA breast cancer RNA sequencing dataset (27). It was revealed that the DDR1 mRNA
expression levels were positively correlated with tumor purity, and
negatively correlated with tumor infiltration of CD4+
and CD8+ T cells (Fig.
6A). Furthermore, DDR1 mRNA expression levels were negatively
correlated with the TIL signature genes CD4 and CD8A, and the
cytotoxic T cell marker granzyme B (Fig.
6B). Taken together, these data suggested that tumor cell DDR1
may affect tumor immunity in patients with breast cancer, which was
consistent with the conclusions obtained from the syngeneic mammary
tumor models in the present study.
 | Figure 6.Correlation between DDR1 expression
and immunity in TCGA breast cancer cohort. (A) Correlation between
DDR1 mRNA expression levels and tumor purity (cor=0.184,
P=5.16×10−9), CD4+ T cell count (cor=−0.221,
P=2.70×10−12) and CD8+ T cell count
(cor=−0.107, P=8.85×10−4). (B) Correlation between DDR1
mRNA expression levels and tumor-infiltrating lymphocyte signature
genes CD4 (cor=−0.265, P=5.99×10−19) and CD8A
(cor=−0.201, P=1.97×10−11), as well as the cytotoxic T
cell marker GZMB (cor=−0.208, P=3.22×10−12). DDR1,
discoidin domain receptor tyrosine kinase 1; TCGA, The Cancer
Genome Atlas; GZMB, granzyme B; BRCA, breast cancer. |
Discussion
DDR1 was originally identified during the search for
tyrosine kinase proteins expressed in human malignancies (47–49). DDR1
kinase is different from other receptor tyrosine kinase (RTK)
members due to a homology domain in discoidin (50). After two decades of study, it is now
clear that aberrant signaling through the DDR1 is closely
associated with different steps of tumorigenesis, although the
detailed molecular mechanism underlying the role of DDR1 remains
largely unknown. Several studies have demonstrated that knockdown
of DDR1 with small interfering RNA or short hairpin RNA in human
cancer cell lines leads to decreased cell proliferation and
metastatic potential both in vitro and in vivo with
immunodeficient mouse models (16,51).
However, mice with either whole body knockout of DDR1 or mammary
gland specific deletion of DDR1 resulted in hyper-proliferation and
abnormal branching of mammary ducts, and enhanced spontaneous
tumorigenesis and lung metastasis (52,53).
Notably, DDR1 may serve different roles in the initiation and
progression of breast cancer.
One recent study demonstrated that DDR1 mutations
were strongly associated with poor prognosis in postmenopausal
patients with breast cancer (25).
Some of these DDR1 mutations are in the ECD domain, such as Q92fs,
R93Q, R105Q and N371Y, but the biological significance of these
require further investigation.
The detailed molecular mechanism underlying how
tumor cell DDR1 promotes breast cancer progression remains unknown.
To help drive this research forward, the present study used the 4T1
and EMT6 syngeneic breast tumor models. These models are derived
from murine mammary carcinoma in BALB/c mice and take advantage of
the complete mouse immune system, serving as a powerful tool in
immuno-oncology studies. Notably, the results from the present
study demonstrated that DDR1 served a critical role in regulating
tumor-immune cell interactions, in specific CD4+ and
CD8+ T cells. However, how DDR1 regulates tumor
infiltrated T cells is currently unknown. A previous study revealed
functionally relevant interactions between DDR1 and Notch1; DDR1
was important for Notch1 activation (54). Notch signaling controls T cell
development, particularly for tumor-infiltrating CD8+ T
cells (55–57). CD8+ T cells have a critical
role in establishing a sufficient immune response against cancer.
Deletion or inhibition of DDR1 leads to inactivation of Notch
signaling and stimulates the cytotoxic activation of TILs (57). The function-blocking anti-DDR1
antibody used in the present study was originally developed and
tested in previous studies (45,58). These
previous studies found that the function-blocking anti-DDR1
antibody inhibits DDR1 signaling without interfering with collagen
binding. In addition, it was demonstrated that the crystal
structure of the monomeric DDR1-ECD bound to the Fab fragment of
the antibody (58).
To the best of our knowledge, there are currently
just two RTKs of which the ECD has been successfully allosterically
targeted by small molecules: FGFR and DDR2 (34,59,60). The
present study demonstrated that inhibition of DDR1, using an ECD
neutralizing antibody, decreased breast cancer growth in
vivo by recruiting CD4+ and CD8+ TILs.
This may represent a novel strategy to improve immunotherapy
efficacy in breast cancer, even in TNBCs. During the preparation of
the present manuscript, another study was published stating that
there were more CD8+ T cells in tumors with depleted
levels of DDR2, the other discoidin domain receptor family member
(61). Furthermore, DDR2 depletion in
isogenic murine models increased sensitivity to anti-programmed
cell death 1 treatment when compared with monotherapy (61).
According to current studies, DDR1 has both
kinase-dependent and independent functions in cancer (16,17,62–68).
The present study highlighted a thus far unreported role of DDR1 in
breast cancer progression, whereby DDR1 may elicit a
tumor-promoting activity through modulating TILs. Therefore, DDR1
may serve as a potential target to improve efficiency of cancer
immunotherapy. In future clinical studies, it would be meaningful
if DDR1 expression levels were recorded in relation to patients'
response to cancer immunotherapy in breast cancer, or other types
of cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Project of
Health Commission of Jiangxi province (grant no. 20175524).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author upon reasonable
request.
Authors' contributions
XZ conceived and supervised the project. WZ, TS and
XZ performed the experiments and analyzed the data. WZ and XZ wrote
the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Protocols involving the use of human tissue were
approved by the Ethics Committee of Jiangxi Cancer Hospital and
performed in accordance with the Declaration of Helsinki and the
guidelines of the Ethics Committee of Jiangxi cancer Hospital.
Written informed consent was obtained from all patients for the use
of their tissues. Protocols involving animals were performed in
accordance with the National Institute of Health Guide for the Care
and Use of Laboratory Animals, with the approval of the Ethics
Committee of Jiangxi Cancer Hospital.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Podo F, Buydens LM, Degani H, Hilhorst R,
Klipp E, Gribbestad IS, Van Huffel S, van Laarhoven HW, Luts J,
Monleon D, et al: Triple-negative breast cancer: Present challenges
and new perspectives. Mol Oncol. 4:209–229. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Palucka K and Banchereau J:
Dendritic-cell-based therapeutic cancer vaccines. Immunity.
39:38–48. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sahin U and Tureci O: Personalized
vaccines for cancer immunotherapy. Science. 359:1355–1360. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
June CH, O'Connor RS, Kawalekar OU,
Ghassemi S and Milone MC: CAR T cell immunotherapy for human
cancer. Science. 359:1361–1365. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hodi FS, O'Day SJ, McDermott DF, Weber RW,
Sosman JA, Haanen JB, Gonzalez R, Robert C, Schadendorf D, Hassel
JC, et al: Improved survival with ipilimumab in patients with
metastatic melanoma. N Engl J Med. 363:711–723. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wolchok JD, Kluger H, Callahan MK, Postow
MA, Rizvi NA, Lesokhin AM, Segal NH, Ariyan CE, Gordon RA, Reed K,
et al: Nivolumab plus ipilimumab in advanced melanoma. N Engl J
Med. 369:122–133. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Postow MA, Chesney J, Pavlick AC, Robert
C, Grossmann K, McDermott D, Linette GP, Meyer N, Giguere JK,
Agarwala SS, et al: Nivolumab and ipilimumab versus ipilimumab in
untreated melanoma. N Engl J Med. 372:2006–2017. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Adams S, Gray RJ, Demaria S, Goldstein L,
Perez EA, Shulman LN, Martino S, Wang M, Jones VE, Saphner TJ, et
al: Prognostic value of tumor-infiltrating lymphocytes in
triple-negative breast cancers from two phase III randomized
adjuvant breast cancer trials: ECOG 2197 and ECOG 1199. J Clin
Oncol. 32:2959–2966. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ali HR, Provenzano E, Dawson SJ, Blows FM,
Liu B, Shah M, Earl HM, Poole CJ, Hiller L, Dunn JA, et al:
Association between CD8+ T-cell infiltration and breast cancer
survival in 12,439 patients. Ann Oncol. 25:1536–1543. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Loi S, Michiels S, Salgado R, Sirtaine N,
Jose V, Fumagalli D, Kellokumpu-Lehtinen PL, Bono P, Kataja V,
Desmedt C, et al: Tumor infiltrating lymphocytes are prognostic in
triple negative breast cancer and predictive for trastuzumab
benefit in early breast cancer: Results from the FinHER trial. Ann
Oncol. 25:1544–1550. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Emens LA: Breast cancer immunotherapy:
Facts and hopes. Clin Cancer Res. 24:511–520. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Vonderheide RH, Domchek SM and Clark AS:
Immunotherapy for breast cancer: What are we missing? Clin Cancer
Res. 23:2640–2646. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nanda R, Chow LQ, Dees EC, Berger R, Gupta
S, Geva R, Pusztai L, Pathiraja K, Aktan G, Cheng JD, et al:
Pembrolizumab in patients with advanced triple-negative breast
cancer: Phase Ib KEYNOTE-012 study. J Clin Oncol. 34:2460–2467.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Weiner HL, Huang H, Zagzag D, Boyce H,
Lichtenbaum R and Ziff EB: Consistent and selective expression of
the discoidin domain receptor-1 tyrosine kinase in human brain
tumors. Neurosurgery. 47:1400–1409. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jin H, Ham IH, Oh HJ, Bae CA, Lee D, Kim
YB, Son SY, Chwae YJ, Han SU, Brekken RA and Hur H: Inhibition of
discoidin domain receptor 1 prevents stroma-induced peritoneal
metastasis in gastric carcinoma. Mol Cancer Res. 16:1590–1600.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ambrogio C, Gómez-López G, Falcone M,
Vidal A, Nadal E, Crosetto N, Blasco RB, Fernández-Marcos PJ,
Sánchez-Céspedes M, Ren X, et al: Combined inhibition of DDR1 and
Notch signaling is a therapeutic strategy for KRAS-driven lung
adenocarcinoma. Nat Med. 22:270–277. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Heinzelmann-Schwarz VA, Gardiner-Garden M,
Henshall SM, Scurry J, Scolyer RA, Davies MJ, Heinzelmann M, Kalish
LH, Bali A, Kench JG, et al: Overexpression of the cell adhesion
molecules DDR1, Claudin 3, and Ep-CAM in metaplastic ovarian
epithelium and ovarian cancer. Clin Cancer Res. 10:4427–4436. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Park HS, Kim KR, Lee HJ, Choi HN, Kim DK,
Kim BT and Moon WS: Overexpression of discoidin domain receptor 1
increases the migration and invasion of hepatocellular carcinoma
cells in association with matrix metalloproteinase. Oncol Rep.
18:1435–1441. 2007.PubMed/NCBI
|
|
20
|
Yamanaka R, Arao T, Yajima N, Tsuchiya N,
Homma J, Tanaka R, Sano M, Oide A, Sekijima M and Nishio K:
Identification of expressed genes characterizing long-term survival
in malignant glioma patients. Oncogene. 25:5994–6002. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shen Q, Cicinnati VR, Zhang X, Iacob S,
Weber F, Sotiropoulos GC, Radtke A, Lu M, Paul A, Gerken G and
Beckebaum S: Role of microRNA-199a-5p and discoidin domain receptor
1 in human hepatocellular carcinoma invasion. Mol Cancer.
9:2272010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Rudd ML, Mohamed H, Price JC, O'Hara AJ,
Le Gallo M, Urick ME; NISC Comparative Sequencing Program, ; Cruz
P, Zhang S, Hansen NF, et al: Mutational analysis of the tyrosine
kinome in serous and clear cell endometrial cancer uncovers rare
somatic mutations in TNK2 and DDR1. BMC Cancer. 14:8842014.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Loriaux MM, Levine RL, Tyner JW, Fröhling
S, Scholl C, Stoffregen EP, Wernig G, Erickson H, Eide CA, Berger
R, et al: High-throughput sequence analysis of the tyrosine kinome
in acute myeloid leukemia. Blood. 111:4788–4796. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ding L, Getz G, Wheeler DA, Mardis ER,
McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan
MB, et al: Somatic mutations affect key pathways in lung
adenocarcinoma. Nature. 455:1069–1075. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Griffith OL, Spies NC, Anurag M, Griffith
M, Luo J, Tu D, Yeo B, Kunisaki J, Miller CA, Krysiak K, et al: The
prognostic effects of somatic mutations in ER-positive breast
cancer. Nat Commun. 9:34762018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Agnihotri S, Jalali S, Wilson MR, Danesh
A, Li M, Klironomos G, Krieger JR, Mansouri A, Khan O, Mamatjan Y,
et al: The genomic landscape of schwannoma. Nat Genet.
48:1339–1348. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cancer Genome Atlas Network: Comprehensive
molecular portraits of human breast tumours. Nature. 490:61–70.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Prykhozhij SV, Rajan V, Gaston D and
Berman JN: CRISPR multitargeter: A web tool to find common and
unique CRISPR single guide RNA targets in a set of similar
sequences. PLoS One. 10:e01193722015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
O'Brien A and Bailey TL: GT-Scan:
Identifying unique genomic targets. Bioinformatics. 30:2673–2675.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xu L, Jensen H, Johnston JJ, Di Maria E,
Kloth K, Cristea I, Sapp JC, Darling TN, Huryn LA, Tranebjærg L, et
al: Recurrent, activating variants in the receptor tyrosine kinase
DDR2 cause warburg-cinotti syndrome. Am J Hum Genet. 103:976–983.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Klionsky DJ, Abdelmohsen K, Abe A, Abedin
MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD,
Adeli K, et al: Guidelines for the use and interpretation of assays
for monitoring autophagy (3rd edition). Autophagy. 12:1–222. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Grither WR and Longmore GD: Inhibition of
tumor-microenvironment interaction and tumor invasion by
small-molecule allosteric inhibitor of DDR2 extracellular domain.
Proc Natl Acad Sci USA. 115:E7786–E7794. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wherry EJ and Kurachi M: Molecular and
cellular insights into T cell exhaustion. Nat Rev Immunol.
15:486–499. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Mori H, Kubo M, Kai M, Yamada M, Kurata K,
Kawaji H, Kaneshiro K, Osako T, Nishimura R, Arima N, et al:
T-bet+ lymphocytes infiltration as an independent better
prognostic indicator for triple-negative breast cancer. Breast
Cancer Res Treat. 176:569–577. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Alves F, Vogel W, Mossie K, Millauer B,
Höfler H and Ullrich A: Distinct structural characteristics of
discoidin I subfamily receptor tyrosine kinases and complementary
expression in human cancer. Oncogene. 10:609–618. 1995.PubMed/NCBI
|
|
38
|
Vogel W: Discoidin domain receptors:
Structural relations and functional implications. FASEB J. 13
(Suppl):S77–S82. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Leitinger B: Discoidin domain receptor
functions in physiological and pathological conditions. Int Rev
Cell Mol Biol. 310:39–87. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Canning P, Tan L, Chu K, Lee SW, Gray NS
and Bullock AN: Structural mechanisms determining inhibition of the
collagen receptor DDR1 by selective and multi-targeted type II
kinase inhibitors. J Mol Biol. 426:2457–2470. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gao H, Chakraborty G, Zhang Z, Akalay I,
Gadiya M, Gao Y, Sinha S, Hu J, Jiang C, Akram M, et al:
Multi-organ site metastatic reactivation mediated by non-canonical
discoidin domain receptor 1 signaling. Cell. 166:47–62. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yeung D, Chmielewski D, Mihai C and
Agarwal G: Oligomerization of DDR1 ECD affects receptor-ligand
binding. J Struct Biol. 183:495–500. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Leitinger B: Molecular analysis of
collagen binding by the human discoidin domain receptors, DDR1 and
DDR2. Identification of collagen binding sites in DDR2. J Biol
Chem. 278:16761–16769. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Fu HL, Sohail A, Valiathan RR, Wasinski
BD, Kumarasiri M, Mahasenan KV, Bernardo MM, Tokmina-Roszyk D,
Fields GB, Mobashery S and Fridman R: Shedding of discoidin domain
receptor 1 by membrane-type matrix metalloproteinases. J Biol Chem.
288:12114–12129. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Carafoli F, Mayer MC, Shiraishi K, Pecheva
MA, Chan LY, Nan R, Leitinger B and Hohenester E: Structure of the
discoidin domain receptor 1 extracellular region bound to an
inhibitory Fab fragment reveals features important for signaling.
Structure. 20:688–697. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Li T, Fan J, Wang B, Traugh N, Chen Q, Liu
JS, Li B and Liu XS: TIMER: A web server for comprehensive analysis
of tumor-infiltrating immune cells. Cancer Res. 77:e108–e110. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Johnson JD, Edman JC and Rutter WJ: A
receptor tyrosine kinase found in breast carcinoma cells has an
extracellular discoidin I-like domain. Proc Natl Acad Sci USA.
90:5677–5681. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Di Marco E, Cutuli N, Guerra L, Cancedda R
and De Luca M: Molecular cloning of trkE, a novel trk-related
putative tyrosine kinase receptor isolated from normal human
keratinocytes and widely expressed by normal human tissues. J Biol
Chem. 268:24290–24295. 1993.PubMed/NCBI
|
|
49
|
Zerlin M, Julius MA and Goldfarb M: NEP: A
novel receptor-like tyrosine kinase expressed in proliferating
neuroepithelia. Oncogene. 8:2731–2739. 1993.PubMed/NCBI
|
|
50
|
Kiedzierska A, Smietana K, Czepczynska H
and Otlewski J: Structural similarities and functional diversity of
eukaryotic discoidin-like domains. Biochim Biophys Acta.
1774:1069–1078. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yuge R, Kitadai Y, Takigawa H, Naito T,
Oue N, Yasui W, Tanaka S and Chayama K: Silencing of discoidin
domain receptor-1 (DDR1) concurrently inhibits multiple steps of
metastasis cascade in gastric cancer. Transl Oncol. 11:575–584.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Vogel WF, Aszodi A, Alves F and Pawson T:
Discoidin domain receptor 1 tyrosine kinase has an essential role
in mammary gland development. Mol Cell Biol. 21:2906–2917. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Takai K, Drain AP, Lawson DA, Littlepage
LE, Karpuj M, Kessenbrock K, Le A, Inoue K, Weaver VM and Werb Z:
Discoidin domain receptor 1 (DDR1) ablation promotes tissue
fibrosis and hypoxia to induce aggressive basal-like breast
cancers. Genes Dev. 32:244–257. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Kim HG, Hwang SY, Aaronson SA, Mandinova A
and Lee SW: DDR1 receptor tyrosine kinase promotes prosurvival
pathway through Notch1 activation. J Biol Chem. 286:17672–17681.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Kuijk LM, Verstege MI, Rekers NV, Bruijns
SC, Hooijberg E, Roep BO, de Gruijl TD, van Kooyk Y and Unger WW:
Notch controls generation and function of human effector CD8+ T
cells. Blood. 121:2638–2646. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Cho OH, Shin HM, Miele L, Golde TE, Fauq
A, Minter LM and Osborne BA: Notch regulates cytolytic effector
function in CD8+ T cells. J Immunol. 182:3380–3389. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Yu W, Wang Y and Guo P: Notch signaling
pathway dampens tumor-infiltrating CD8+ T cells activity
in patients with colorectal carcinoma. Biomed Pharmacother.
97:535–542. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Juskaite V, Corcoran DS and Leitinger B:
Collagen induces activation of DDR1 through lateral dimer
association and phosphorylation between dimers. Elife. 6(pii):
e257162017. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Bono F, De Smet F, Herbert C, De Bock K,
Georgiadou M, Fons P, Tjwa M, Alcouffe C, Ny A, Bianciotto M, et
al: Inhibition of tumor angiogenesis and growth by a small-molecule
multi-FGF receptor blocker with allosteric properties. Cancer Cell.
23:477–488. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Herbert C, Schieborr U, Saxena K, Juraszek
J, De Smet F, Alcouffe C, Bianciotto M, Saladino G, Sibrac D,
Kudlinzki D, et al: Molecular mechanism of SSR128129E, an
extracellularly acting, small-molecule, allosteric inhibitor of FGF
receptor signaling. Cancer Cell. 23:489–501. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Tu MM, Lee FYF, Jones RT, Kimball AK,
Saravia E, Graziano RF, Coleman B, Menard K, Yan J, Michaud E, et
al: Targeting DDR2 enhances tumor response to anti-PD-1
immunotherapy. Sci Adv. 5:eaav24372019. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Tao Y, Wang R, Lai Q, Wu M, Wang Y, Jiang
X, Zeng L, Zhou S, Li Z, Yang T, et al: Targeting of DDR1 with
antibody-drug conjugates has antitumor effects in a mouse model of
colon carcinoma. Mol Oncol. May 22–2019.(Epub ahead of print).
View Article : Google Scholar
|
|
63
|
Vehlow A, Klapproth E, Jin S, Hannen R,
Hauswald M, Bartsch JW, Nimsky C, Temme A, Leitinger B and Cordes
N: Interaction of discoidin domain receptor 1 with a
14-3-3-Beclin-1-Akt1 complex modulates glioblastoma therapy
sensitivity. Cell Rep. 26:3672–3683.e7. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Aguilera KY, Huang H, Du W, Hagopian MM,
Wang Z, Hinz S, Hwang TH, Wang H, Fleming JB, Castrillon DH, et al:
Inhibition of discoidin domain receptor 1 reduces collagen-mediated
tumorigenicity in pancreatic ductal adenocarcinoma. Mol Cancer
Ther. 16:2473–2485. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Lu QP, Chen WD, Peng JR, Xu YD, Cai Q,
Feng GK, Ding K, Zhu XF and Guan Z: Antitumor activity of 7RH, a
discoidin domain receptor 1 inhibitor, alone or in combination with
dasatinib exhibits antitumor effects in nasopharyngeal carcinoma
cells. Oncol Lett. 12:3598–3608. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Shitomi Y, Thogersen IB, Ito N, Leitinger
B, Enghild JJ and Itoh Y: ADAM10 controls collagen signaling and
cell migration on collagen by shedding the ectodomain of discoidin
domain receptor 1 (DDR1). Mol Biol Cell. 26:659–673. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Gao M, Duan L, Luo J, Zhang L, Lu X, Zhang
Y, Zhang Z, Tu Z, Xu Y, Ren X and Ding K: Discovery and
optimization of
3-(2-(Pyrazolo[1,5-a]pyrimidin-6-yl)ethynyl)benzamides as novel
selective and orally bioavailable discoidin domain receptor 1
(DDR1) inhibitors. J Med Chem. 56:3281–3295. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Ali-Rahmani F, FitzGerald DJ, Martin S,
Patel P, Prunotto M, Ormanoglu P, Thomas C and Pastan I: Anticancer
effects of mesothelin-targeted immunotoxin therapy are regulated by
tyrosine kinase DDR1. Cancer Res. 76:1560–1568. 2016. View Article : Google Scholar : PubMed/NCBI
|